Improving the Utilization of Isomaltose and Panose by Lager Yeast Saccharomyces pastorianus
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Cultivation Methods
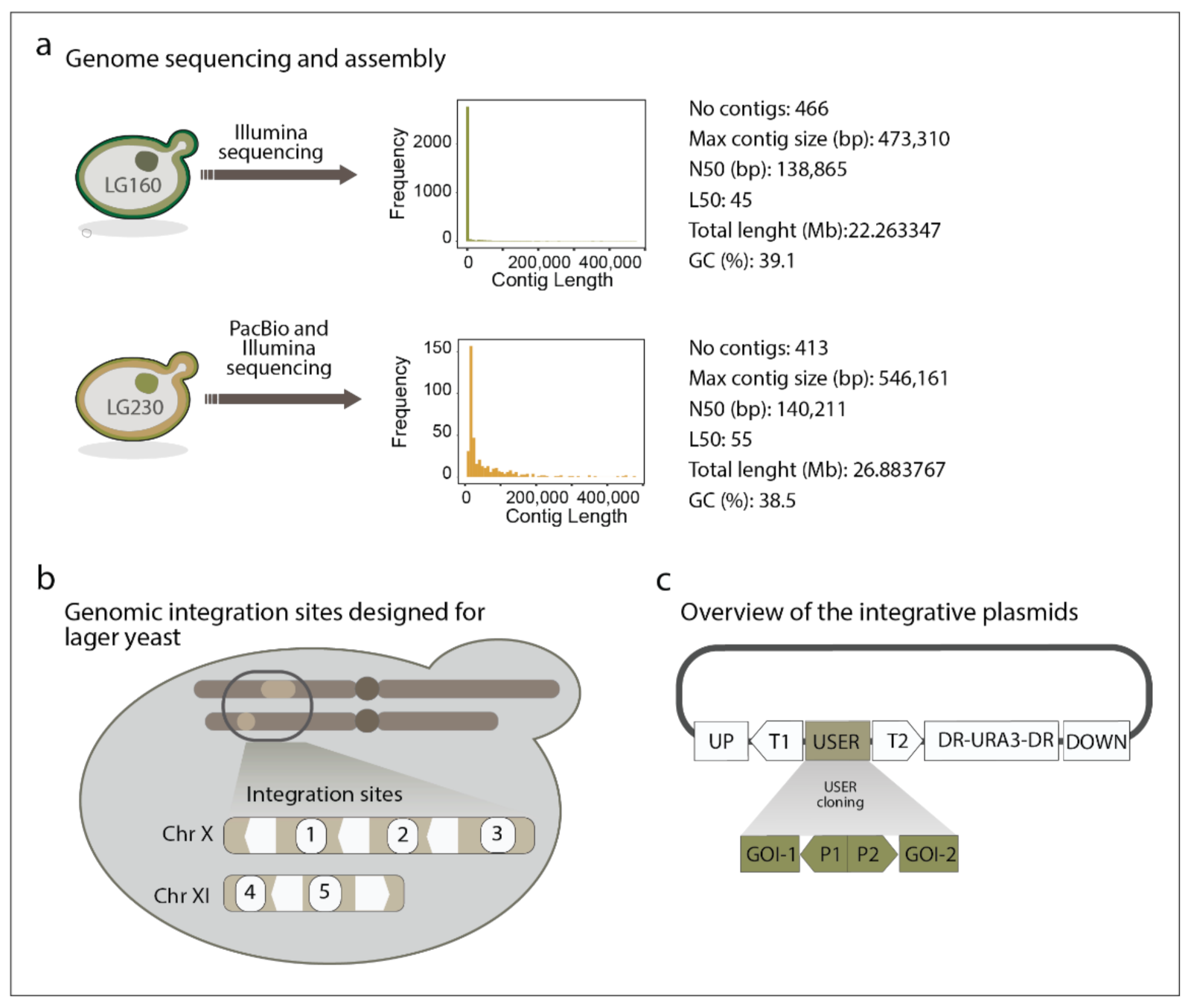
2.2. Illumina and PacBio Whole Genome Sequencing and Assembly
2.3. Design of Integration Sites
2.4. Plasmid Construction
2.5. Strain Construction
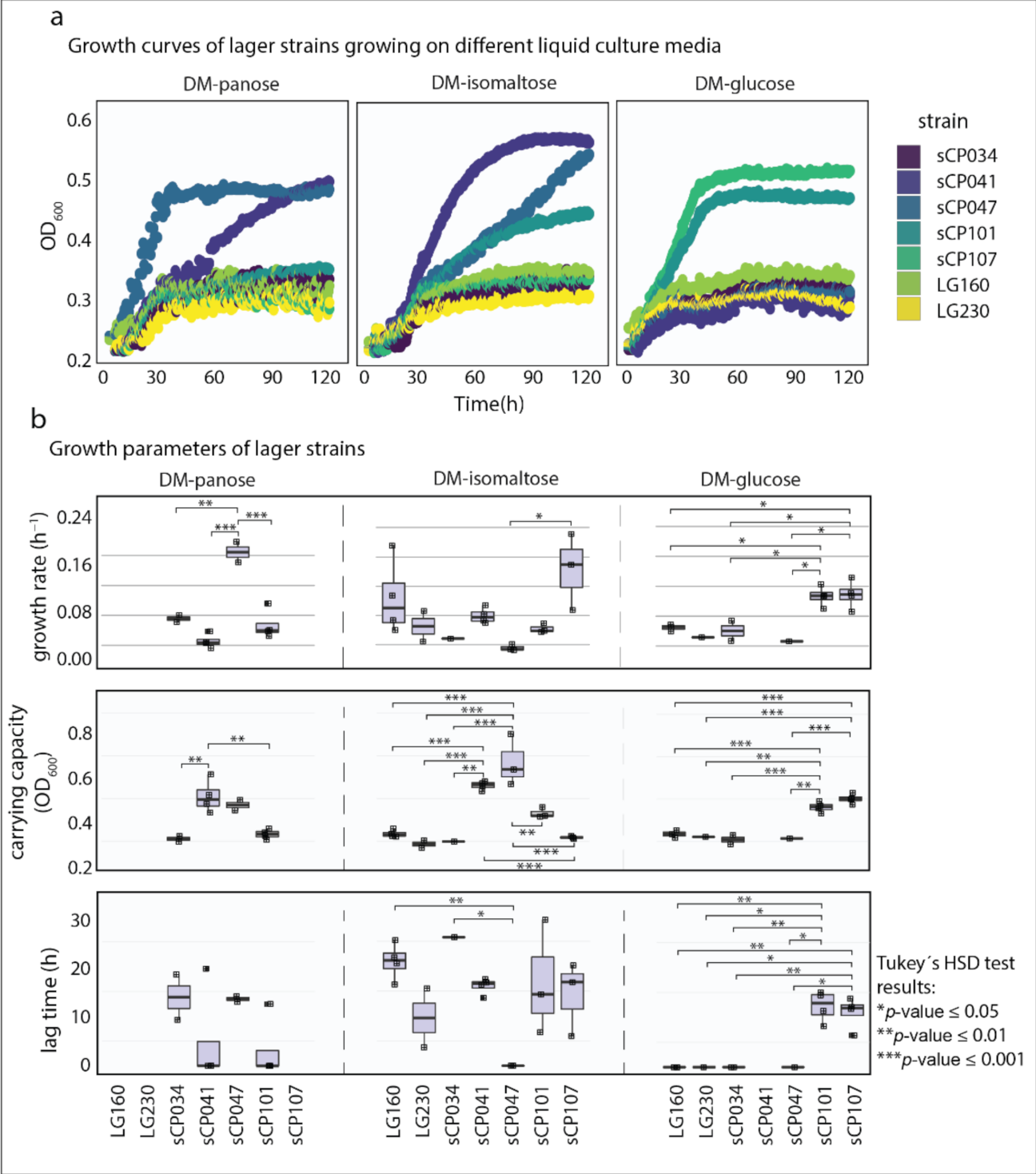
2.6. Phenotypic Characterization
2.7. Statistical Analysis
3. Results
3.1. Whole Genome Sequencing and Assembly of Genomes of Model Lager Yeast
3.2. Endogenous IMA1 and AGT1 Alleles in the Model Lager Yeast
3.3. Effect of the Integration of an Extra Copy of AGT1 and IMA1 on the Growth of Lager Yeast Saccharomyces pastorianus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carslberg Annual Report. Available online: https://www.carlsberggroup.com/media/28928/carlsberg-as-2018-annual-report.pdf (accessed on 5 June 2021).
- Nilsson-Tillgren, T.; Gjermansen, C.; Holmberg, S.; Litske Petersen, J.G.; Kielland-Brandt, M.C. Analysis of chromosome V and theILV1 gene from Saccharomyces carlsbergensis. Carlsberg Res. Commun. 1986, 51, 309. [Google Scholar] [CrossRef]
- Martini, A.V.; Kurtzman, C.P. Deoxyribonucleic Acid Relatedness among Species of Saccharomyces sensu lato. Mycologia 2007, 80, 241. [Google Scholar] [CrossRef]
- Martini, A.V.; Martini, A. Three newly delimited species of Saccharomyces sensu stricto. Antonie Van Leeuwenhoek 1987, 53, 77–84. [Google Scholar] [CrossRef]
- Brickwedde, A.; Brouwers, N.; van den Broek, M.; Murillo, J.S.G.; Fraiture, J.L.; Pronk, J.T.; Daran, J.-M.G. Structural, physiological and regulatory analysis of maltose transporter genes in Saccharomyces eubayanus CBS 12357T. Front. Microbiol. 2018, 9, 1786. [Google Scholar] [CrossRef]
- Libkind, D.; Hittinger, C.T.; Valerio, E.; Goncalves, C.; Dover, J.; Johnston, M.; Goncalves, P.; Sampaio, J.P. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 14539–14544. [Google Scholar] [CrossRef]
- Baker, E.C.; Wang, B.; Bellora, N.; Peris, D.; Hulfachor, A.B.; Koshalek, J.A.; Adams, M.; Libkind, D.; Hittinger, C.T. The genome sequence of Saccharomyces eubayanus and the domestication of lager-brewing yeasts. Mol. Biol. Evol. 2015, 32, 2818–2831. [Google Scholar] [CrossRef]
- Christensen, B.E. Cross-breeding of distillers’ yeast by hybridization of spore derived clones. Carlsberg Res. Commun. 1987, 52, 253–262. [Google Scholar] [CrossRef]
- Inez, B.; Figueiredo, C.; Fontes, A.; Patrick, P.; Pimenta, D.S.; Souza, C. De crossm Crossing Techniques Using Cachaça (Brazilian Spirit) Yeasts. Appl. Environ. Microbiol. 2017, 83, e01582-17. [Google Scholar]
- Krogerus, K.; Magalhães, F.; Vidgren, V.; Gibson, B. New lager yeast strains generated by interspecific hybridization. J. Ind. Microbiol. Biotechnol. 2015, 42, 769–778. [Google Scholar] [CrossRef]
- Denby, C.M.; Li, R.A.; Vu, V.T.; Costello, Z.; Lin, W.; Chan, L.J.G.; Williams, J.; Donaldson, B.; Bamforth, C.W.; Petzold, C.J.; et al. Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Dunn, B.; Sherlock, G. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. 2008, 18, 1610–1623. [Google Scholar] [CrossRef]
- Walther, A.; Hesselbart, A.; Wendland, J. Genome sequence of Saccharomyces carlsbergensis, the world’s first pure culture lager yeast. G3 Genes Genomes Genet. 2014, 4, 783–793. [Google Scholar] [CrossRef]
- Turgeon, Z.; Sierocinski, T.; Brimacombe, C.A.; Jin, Y.; Goldhawke, B.; Swanson, J.M.; Husnik, J.I.; Dahabieh, M.S. Industrially Applicable De Novo Lager Yeast Hybrids with a Unique Genomic Architecture: Creation and Characterization. Appl. Environ. Microbiol. 2021, 87, e02434-20. [Google Scholar] [CrossRef] [PubMed]
- Hansen, E.C. Undersøgelser over alkoholgjærsvampenes fysiologi og morfologi. II. Om askosporedannelsen hos slægten Saccharomyces. Medd. F. Carlsberg Lab. 1883, 2, 29–86. [Google Scholar]
- Marilena, B.; Giacomo, Z.; Maurizio, C.; Francesca, C. Saccharomyces and Non-Saccharomyces Starter Yeasts. In Brewing Technology; IntechOpen: Rijeka, Croatia, 2017. [Google Scholar] [CrossRef]
- Morris, G.H. The Analysis of Beer, with some Remarks on the Unfermentable Reducing Residue. J. Fed. Inst. Brew. 1895, 1, 125–142. [Google Scholar] [CrossRef]
- Phillips, A.W. Utilization by yeast of the carbohydrates of wort. J. Inst. Brew. 1955, 61, 122–126. [Google Scholar] [CrossRef]
- Clapperton, J.; Macwilliam, I. Fermentation of minor wort carbohydrates by brewing yeast. J. Inst. Brew. 1971, 77, 519–522. [Google Scholar] [CrossRef]
- Buckee, G.K.; Hargitt, R. Measurement of carbohydrates in wort and beer-a review. J. Inst. Brew. 1978, 84, 13–21. [Google Scholar] [CrossRef]
- Bathgate, G.N.; Bringhurst, T.A. Letter to the Editor: Update on Knowledge Regarding Starch Structure and Degradation by Malt Enzymes (DP/DU and Limit Dextrinase). J. Inst. Brew. 2011, 117, 33–38. [Google Scholar] [CrossRef]
- Alves, S.L.; Herberts, R.A.; Hollatz, C.; Trichez, D.; Miletti, L.C.; De Araujo, P.S.; Stambuk, B.U. Molecular analysis of maltotriose active transport and fermentation by Saccharomyces cerevisiae reveals a determinant role for the AGT1 permease. Appl. Environ. Microbiol. 2008, 74, 1494–1501. [Google Scholar] [CrossRef]
- Enevoldsen, B.S.; Schmidt, F. Dextrins in brewing. J. Inst. Brew. 1974, 80, 520–533. [Google Scholar] [CrossRef]
- Sundekilde, U.K.; Meier, S. 1H–13C NMR-Based Profiling of Biotechnological Starch Utilization. Anal. Chem. 2016, 88, 9685–9690. [Google Scholar] [CrossRef] [PubMed]
- Petersen, B.O.; Nilsson, M.; Bøjstrup, M.; Hindsgaul, O.; Meier, S. 1H NMR spectroscopy for profiling complex carbohydrate mixtures in non-fractionated beer. Food Chem. 2014, 150, 65–72. [Google Scholar] [CrossRef]
- Spevacek, A.R.; Benson, K.H.; Bamforth, C.W.; Slupsky, C.M. Beer metabolomics: Molecular details of the brewing process and the differential effects of late and dry hopping on yeast purine metabolism. J. Inst. Brew. 2016, 122, 21–28. [Google Scholar] [CrossRef]
- Bathgate, G.N. Isolation and characterisation of panose and isopanose from wort and beer. Chem. Ind. 1969, 520–521. [Google Scholar]
- Bathgate, G.N. A review of malting and malt processing for whisky distillation. J. Inst. Brew. 2016, 122, 197–211. [Google Scholar] [CrossRef]
- Engan, S. Wort composition and beer flavour II. The influence of different carbohydrates on the formation of some flavour components during fermentation. J. Inst. Brew. 1972, 78, 169–173. [Google Scholar] [CrossRef]
- Langstaff, S.A.; Lewis, M.J. The mouthfeel of beer-a review. J. Inst. Brew. 1993, 99, 31–37. [Google Scholar] [CrossRef]
- He, Y.; Dong, J.; Yin, H.; Zhao, Y.; Chen, R.; Wan, X.; Chen, P.; Hou, X.; Liu, J.; Chen, L. Wort composition and its impact on the flavour-active higher alcohol and ester formation of beer–A review. J. Inst. Brew. 2014, 120, 157–163. [Google Scholar] [CrossRef]
- Hughes, P.S.; Baxter, E.D. Flavour Determinants of Beer Quality. In Beer: Quality, Safety and Nutritional Aspects; Royal Society of Chemistry: Cambridge, UK, 2001; pp. 41–42. ISBN 0854045880. [Google Scholar]
- Solodovnikova, N.Y.; Garcia Sanchez, R.; Gojkovic, Z. Yeast for Preparing Alcoholic Beverages. WO2016101960, 30 June 2016. [Google Scholar]
- Vidgren, V.; Huuskonen, A.; Virtanen, H.; Ruohonen, L.; Londesborough, J. Improved fermentation performance of a lager yeast after repair of its AGT1 maltose and maltotriose transporter genes. Appl. Environ. Microbiol. 2009, 75, 2333–2345. [Google Scholar] [CrossRef]
- Teste, M.A.; Marie François, J.; Parrou, J.L. Characterization of a new multigene family encoding isomaltases in the yeast Saccharomyces cerevisiae, the IMA family. J. Biol. Chem. 2010, 285, 26815–26824. [Google Scholar] [CrossRef]
- Deng, X.; Petitjean, M.; Teste, M.A.; Kooli, W.; Tranier, S.; François, J.M.; Parrou, J.L. Similarities and differences in the biochemical and enzymological properties of the four isomaltases from Saccharomyces cerevisiae. FEBS Open Bio 2014, 4, 200–212. [Google Scholar] [CrossRef]
- Day, R.E.; Higgins, V.J.; Rogers, P.J.; Dawes, I.W. Characterization of the putative maltose transporters encoded by YDL247w and YJR160c. Yeast 2002, 19, 1015–1027. [Google Scholar] [CrossRef]
- Alves-Jr, S.L.; Herberts, R.A.; Hollatz, C.; Miletti, L.C.; Stambuk, B.U. Maltose and Maltotriose Active Transport and Fermentation by Saccharomyces Cerevisiaes. J. Am. Soc. Brew. Chem. 2007, 65, 99–104. [Google Scholar] [CrossRef]
- Brouwers, N.; Brickwedde, A.; de Vries, A.R.; van den Broek, M.; Weening, S.M.; van den Eijnden, L.; Diderich, J.A.; Bai, F.-Y.; Pronk, J.T.; Daran, J.-M.G. Himalayan Saccharomyces eubayanus Genome Sequences Reveal Genetic Markers Explaining Heterotic Maltotriose Consumption by Saccharomyces pastorianus Hybrids. Appl. Environ. Microbiol. 2019, 85, e01516-19. [Google Scholar] [CrossRef]
- Cousseau, F.E.M.; Alves, S.L.; Trichez, D.; Stambuk, B.U. Characterization of maltotriose transporters from the Saccharomyces eubayanus subgenome of the hybrid Saccharomyces pastorianus lager brewing yeast strain Weihenstephan 34/70. Lett. Appl. Microbiol. 2013, 56, 21–29. [Google Scholar] [CrossRef]
- Magalhães, F.; Vidgren, V.; Ruohonen, L.; Gibson, B. Maltose and maltotriose utilisation by group I strains of the hybrid lager yeast Saccharomyces pastorianus. FEMS Yeast Res. 2016, 16, 1–11. [Google Scholar] [CrossRef]
- Naumoff, D.G.; Naumov, G.I. Discovery of a novel family of α-glucosidase IMA genes in yeast Saccharomyces cerevisiae. Dokl. Biochem. Biophys. 2010, 432, 114–116. [Google Scholar] [CrossRef]
- Bell, P.J.L.; Higgins, V.J.; Dawes, I.W.; Bissinger, P.H. Tandemly repeated 147 bp elements cause structural and functional variation in divergent MAL promoters of Saccharomyces cerevisiae. Yeast 1997, 13, 1135–1144. [Google Scholar] [CrossRef]
- Change, Y.S.; Dubin, R.A.; Perkins, E.; Forrest, D.; Michels, C.A.; Needleman, R.B. MAL63 codes for a positive regulator of maltose fermentation in Saccharomyces cerevisiae. Curr. Genet. 1988, 14, 201–209. [Google Scholar] [CrossRef]
- Hoffman, L. The Defective Sporulation of Lager Brewing Yeast. Ph.D. Thesis, University of Copenhagen, Copenhagen, Denmark, 2000. [Google Scholar]
- Gjermansen, C.; Sigsgaard, P. Construction of a hybrid brewing strain of Saccharomyces carlsbergensis by mating of meiotic segregants. Carlsberg Res. Commun. 1981, 46, 1–11. [Google Scholar] [CrossRef]
- Hahn-Hägerdal, B.; Karhumaa, K.; Larsson, C.U.; Gorwa-Grauslund, M.; Görgens, J.; van Zyl, W.H. Role of cultivation media in the development of yeast strains for large scale industrial use. Microb. Cell Fact. 2005, 4, 1–16. [Google Scholar] [CrossRef]
- Zhou, N.; Katz, M.; Knecht, W.; Compagno, C.; Piškur, J. Genome dynamics and evolution in yeasts: A long-term yeast-bacteria competition experiment. PLoS ONE 2018, 13, e0194911. [Google Scholar] [CrossRef]
- Gutiérrez, A.; Beltran, G.; Warringer, J.; Guillamón, J.M. Genetic Basis of Variations in Nitrogen Source Utilization in Four Wine Commercial Yeast Strains. PLoS ONE 2013, 8, e67166. [Google Scholar] [CrossRef]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Mikkelsen, M.D.; Buron, L.D.; Salomonsen, B.; Olsen, C.E.; Hansen, B.G.; Mortensen, U.H.; Halkier, B.A. Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab. Eng. 2012, 14, 104–111. [Google Scholar] [CrossRef]
- Nour-eldin, H.H.; Geu-flores, F.; Halkier, B.A. USER Cloning and USER Fusion: The Ideal Cloning Techniques for Small and Big Laboratories. In Plant Secondary Metabolism Engineering, Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2010; Volume 643, pp. 185–200. ISBN 978-1-60761-722-8. [Google Scholar]
- Gietz, R.D. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol. Biol. 2014, 1205, 1–12. [Google Scholar] [CrossRef]
- RStudio Team RStudio: Integrated Development for R. RStudio; PBC: Boston, MA, USA. Available online: https://www.rstudio.com/ (accessed on 5 July 2021).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Petzoldt, T. growthrates: Estimate Growth Rates from Experimental Data. R package version 0.8.2. Available online: https://CRAN.R-project.org/package=growthrates (accessed on 5 July 2021).
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef]
- Okuno, M.; Kajitani, R.; Ryusui, R.; Morimoto, H.; Kodama, Y.; Itoh, T. Next-generation sequencing analysis of lager brewing yeast strains reveals the evolutionary history of interspecies hybridization. DNA Res. 2016, 23, 67–80. [Google Scholar] [CrossRef]
- Nakao, Y.; Kanamori, T.; Itoh, T.; Kodama, Y.; Rainieri, S.; Nakamura, N.; Shimonaga, T.; Hattori, M.; Ashikari, T. Genome Sequence of the Lager Brewing Yeast, an Interspecies Hybrid. DNA Res. 2009, 16, 115–129. [Google Scholar] [CrossRef]
- Yue, J.-X.; Li, J.; Aigrain, L.; Hallin, J.; Persson, K.; Oliver, K.; Bergström, A.; Coupland, P.; Warringer, J.; Lagomarsino, M.C.; et al. Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat. Genet. 2017, 49, 913. [Google Scholar] [CrossRef]
- Yue, J.-X.; Liti, G. Long-read sequencing data analysis for yeasts. Nat. Protoc. 2018, 13, 1213. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Jensen, N.B.; Strucko, T.; Kildegaard, K.R.; David, F.; Maury, J.; Mortensen, U.H.; Forster, J.; Nielsen, J.; Borodina, I. EasyClone: Method for iterative chromosomal integration of multiple genes in Saccharomyces cerevisiae. FEMS Yeast Res. 2014, 14, 238–248. [Google Scholar] [CrossRef]
- Maury, J.; Germann, S.M.; Baallal Jacobsen, S.A.; Jensen, N.B.; Kildegaard, K.R.; Herrgàrd, M.J.; Schneider, K.; Koza, A.; Forster, J.; Nielsen, J.; et al. EasyCloneMulti: A set of vectors for simultaneous and multiple genomic integrations in Saccharomyces cerevisiae. PLoS ONE 2016, 11, e0150394. [Google Scholar] [CrossRef]
- Jessop-Fabre, M.M.; Jakočiūnas, T.; Stovicek, V.; Dai, Z.; Jensen, M.K.; Keasling, J.D.; Borodina, I. EasyClone-MarkerFree: A vector toolkit for marker-less integration of genes into Saccharomyces cerevisiae via CRISPR-Cas9. Biotechnol. J. 2016, 11, 1110–1117. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Raman, M.; Martin, K. One solution for cloning and mutagenesis: In-Fusion® HD Cloning Plus. Nat. Methods 2014, 11, 972. [Google Scholar] [CrossRef]
- Weber, E.; Engler, C.; Gruetzner, R.; Werner, S.; Marillonnet, S. A Modular Cloning System for Standardized Assembly of Multigene Constructs. PLoS ONE 2011, 6, e16765. [Google Scholar] [CrossRef]
- Brown, C.A.; Murray, A.W.; Verstrepen, K.J. Rapid Expansion and Functional Divergence of Subtelomeric Gene Families in Yeasts. Curr. Biol. 2010, 20, 895–903. [Google Scholar] [CrossRef]
- Brouwers, N.; Gorter de Vries, A.R.; van den Broek, M.; Weening, S.M.; Elink Schuurman, T.D.; Kuijpers, N.G.A.; Pronk, J.T.; Daran, J.-M.G. In vivo recombination of Saccharomyces eubayanus maltose-transporter genes yields a chimeric transporter that enables maltotriose fermentation. PLoS Genet. 2019, 15, e1007853. [Google Scholar] [CrossRef]
- Marques, W.L.; Mans, R.; Marella, E.R.; Cordeiro, R.L.; van den Broek, M.; Daran, J.M.G.; Pronk, J.T.; Gombert, A.K.; van Maris, A.J.A. Elimination of sucrose transport and hydrolysis in Saccharomyces cerevisiae: A platform strain for engineering sucrose metabolism. FEMS Yeast Res. 2017, 17, fox006. [Google Scholar] [CrossRef]
| Strain | Parental Strain | Integration Site | Genotype | Source |
|---|---|---|---|---|
| LG160 | N.A. | N.A. | S. pastorianus (MATa/MATa ura3 ura3) | Carlsberg collection |
| LG230 | N.A. | N.A. | S. pastorianus (MATalpha/MATalpha ura3-ca-Δ2 ura3-KpnI met2-ca-Δ) | Carlsberg collection |
| sCP034 | LG230 | XI-I | PCCW12 -> IMA1 | This study |
| sCP041 | LG160 | XI-II | PPGI1 -> AGT1, PPGK1 -> IMA1 | This study |
| sCP047 | LG230 | XI-I | PPGI1 -> AGT1, PPGK1 -> IMA1 | This study |
| sCP101 | LG160 | X3 | PPGI1 -> AGT1 | This study |
| sCP107 | LG230 | X3 | PPGI1 -> AGT1 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porcayo Loza, J.; Chailyan, A.; Forster, J.; Katz, M.; Mortensen, U.H.; Garcia Sanchez, R. Improving the Utilization of Isomaltose and Panose by Lager Yeast Saccharomyces pastorianus. Fermentation 2021, 7, 107. https://doi.org/10.3390/fermentation7030107
Porcayo Loza J, Chailyan A, Forster J, Katz M, Mortensen UH, Garcia Sanchez R. Improving the Utilization of Isomaltose and Panose by Lager Yeast Saccharomyces pastorianus. Fermentation. 2021; 7(3):107. https://doi.org/10.3390/fermentation7030107
Chicago/Turabian StylePorcayo Loza, Javier, Anna Chailyan, Jochen Forster, Michael Katz, Uffe Hasbro Mortensen, and Rosa Garcia Sanchez. 2021. "Improving the Utilization of Isomaltose and Panose by Lager Yeast Saccharomyces pastorianus" Fermentation 7, no. 3: 107. https://doi.org/10.3390/fermentation7030107
APA StylePorcayo Loza, J., Chailyan, A., Forster, J., Katz, M., Mortensen, U. H., & Garcia Sanchez, R. (2021). Improving the Utilization of Isomaltose and Panose by Lager Yeast Saccharomyces pastorianus. Fermentation, 7(3), 107. https://doi.org/10.3390/fermentation7030107






