Abstract
The existence of antibiotic-resistant bacteria in food products, particularly those carrying acquired resistance genes, has increased concerns about the transmission of these genes from beneficial microbes to human pathogens. In this study, we evaluated the antibiotic resistance-susceptibility patterns of 16 antibiotics in eight S. thermophilus strains, whose genome sequence is available, using phenotypic and genomic approaches. The minimal inhibitory concentration values collected revealed intermediate resistance to aminoglycosides, whereas susceptibility was detected for different classes of β-lactams, quinolones, glycopeptide, macrolides, and sulfonamides in all strains. A high tetracycline resistance level has been detected in strain M17PTZA496, whose genome analysis indicated the presence of the tet(S) gene and the multidrug and toxic compound extrusion (MATE) family efflux pump. Moreover, an in-depth genomic analysis revealed genomic islands and an integrative and mobilizable element (IME) in the proximity of the gene tet(S). However, despite the presence of a prophage, genomic islands, and IME, no horizontal gene transfer was detected to Lactobacillus delbrueckii subsp. lactis DSM 20355 and Lactobacillus rhamnosus GG during 24 h of skim milk fermentation, 2 weeks of refrigerated storage, and 4 h of simulated gastrointestinal transit.
1. Introduction
Antibiotics are the most important therapeutic option for treating bacterial diseases in humans and animals [1,2]. However, the overutilization of these therapeutic agents has led to the development of bacterial antibiotic resistance, which is rapidly increasing, and, thus, creating a serious global problem [3]. The presence of resistant bacteria in foods, especially, in fermented products, has increased concerns about the possible diffusion of resistance genes from beneficial bacteria to pathogens [4]. For this reason, several studies have been undertaken to assess antibiotic susceptibility-resistance profiles of food-related bacteria [3,5,6]. Generally, acquired antibiotic resistance genes are located in mobile genetic elements such as plasmids, transposons, and phages that confer on them great transferability. Recent advancements in genome sequencing technologies have made the detection of resistance genes easier and more reliable. By performing a comprehensive in silico analysis, all known mobile elements existing inside a bacterial genome can be detected [7,8]. Bacteriophages are quite widespread and abundant in many environments. They can contribute to gene transfer among the bacteria by specialized, generalized, and lateral transductions [9]. Integrative and conjugative elements (ICEs) play a vital role in bacterial horizontal gene transfer due to their self-transmissibility and fully functioning conjugation machinery among bacterial cells [10]. Moreover, IMEs also contribute to horizontal gene transfer between bacteria. They are usually genomic islands within bacterial genomes that may carry antibiotic resistance genes and encode their excision and integration in the chromosome [10]. However, they cannot be transferred by themselves due to the lack of a conjugative apparatus. These elements could be transferred to other bacteria in the presence of conjugative elements such as ICEs. They can be picked up by ICEs and subsequently transferred to other bacterial cells [11,12]. In fermented foods, microbial interactions, such as conjugation or transduction can happen during manufacturing and storage [13]. On the other hand, low temperatures and gastrointestinal conditions can provide a stressful environment for the cell, which can favor the transfer of genetic elements [14,15]. For this reason, lactic acid bacteria, as the predominant microorganisms in dairy environments [16,17], are frequently linked to antibiotic resistance [18]. Within this group, Streptococcus thermophilus is the only species of the genus with GRAS (Generally recognized as safe) status, endowed with interesting technological and probiotic properties [19,20]. This species, as a fast acidifier, can break down lactose into lactic acid, thus lowering the pH, an essential feature in dairy technology [21,22]. For its technological properties, S. thermophilus is the second most important industrial bacterium after Lactococcus lactis, since it has been estimated that around 1021 live cells are being consumed by people around the world annually [23]. Considering the tremendous usage and consumption of this interesting industrial species, we still have limited data regarding the resistance-susceptibility limit for several antibiotics. The current study aimed to determine the antibiotic resistance patterns of 16 antibiotics, among those mostly used on humans, in eight S. thermophilus strains isolated from a dairy environment, whose genomes have been sequenced, using phenotypic and genomic approaches. This study also examined the transferability of an antibiotic resistance gene found in one of the strains to other lactic acid bacteria during fermentation, two weeks of storage at refrigeration temperature, and during a simulated gastrointestinal transit.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
S. thermophilus strains used in the present study are listed in Table 1. All strains were isolated from dairy environments in Italy and are part of the Department of Agronomy Food Natural Resources Animals and Environment collection. The type strain S. thermophilus ATCC19258T was included as a reference. Strains were kept at −80 °C in 10% Skim Milk broth (Oxoid, UK) containing glycerol (20% v/v) and grown M17 medium (Oxoid, UK) containing 0.5% lactose at 37 °C for 24 h before their use. Lactobacillus delbrueckii subsp. lactis DSM 20,355 and Lactobacillus rhamnosus GG were grown in an MRS medium (Oxoid) at 37 °C for 24 h.

Table 1.
Streptococcus thermophilus strains used in the present study.
2.2. Minimum Inhibitory Concentration (MIC) Determination
The MIC for sixteen antibiotics (ampicillin, chloramphenicol, ciprofloxacin, oxacillin, erythromycin, gentamycin, kanamycin, penicillin G, streptomycin, tetracycline, trimethoprim, vancomycin, neomycin, rifampicin, spectinomycin, and carbenicillin) was determined by the broth microdilution method using 96-well microtiter plates (Sigma SIAL0596, St. Louis, MO, USA) according to the Clinical and Laboratory Standards Institute (CLSI; www.clsi.org, accessed on 1 April 2021). Tests were performed in ISO-Sensitest broth (Sigma-Aldrich) containing 10% M17. All antibiotics were dissolved in the abovementioned medium and distributed as 2-fold serial dilutions in the microtiter plate wells, from 256 to 0.5 µg/mL. Each S. thermophilus strain was grown on M17 plates overnight, and some colonies were collected and dissolved in sterile phosphate-buffered saline (1.44 g/L Na2HPO4, 0.24 g/L KH2PO4, 8 g/L NaCl, 0.2 g/L KCl, pH 7.4) to obtain a turbidity corresponding to McFarland standard 1 (ca. 3 × 108 CFU/mL). This solution was further diluted 1:1000 in M17 plus ISO-Sensitest broth to a final concentration of about 3 × 105 CFU/mL. Later, 100 µL aliquots were used to inoculate the wells of a microtiter plate and incubated at 37 °C for 24 h. The test was performed with 2 individual biological replicates, and the MIC was determined as the antibiotic concentration of the first well with no visible growth [28].
2.3. Identification of Antibiotic Resistance Genes
Genomes of all S. thermophilus were retrieved from Genbank (NCBI) (Table 1), and the genomes were annotated by Rapid Annotation using Subsystems Technology (RAST) to identify antibiotics resistance genes. Subsequently, the entire protein content from the predicted genome of each strain was analyzed on the Comprehensive Antibiotic Resistance Database (CARD) server [29] using the resistance gene identifier (UGI) platform (setting on perfect, strict, and loose hits based on low/high-quality coverage) to detect the resistomes within the different genomes.
The detected resistance genes obtained by CARD were used for the confirmatory analysis using ResFinder server version 3.2 [30] to remove errors and false-positive outputs.
2.4. Identification of Genomic Islands and Mobile Elements
The IslandViewer 4 server was used to predict and detect entire genomic islands in the genomes of the strains, indicating acquired resistance genes [31]. Different methods were used, namely, IslandPick, IslandPath-DIMOB, and SIGI-HMM. Detection of the CRISPR-Cas sequence (clustered regularly interspaced short palindromic repeats) was completed using the CRISPRCasFinder server [32]. OriTfinder and PlasmidFinder 2.1 servers were used to obtain information on the origin of gene transfer in bacterial genomes [33,34], and the ICEberg2 server was used for the identification of integrative and conjugative elements inside bacterial genomes [10].
2.5. Transferability of Resistance Genes during Strain Fermentation and Storage
The transferability of the tetracycline resistance gene tet(S) from S. thermophilus M17PTZA496 to Lactobacillus rhamnosus GG and to L. delbrueckii DSM 20,355 during the milk fermentation and during storage after fermentation, were assessed separately as previously described by Garcia et al. [4] with slight modifications. Donor and recipient strains were separately grown overnight in 10% Skim Milk broth for 24 h at 37 °C. These cultures were used to perform mating trials between M17PTZA496-GG and M17PTZA496-DSM 20355. Each donor and recipient were co-cultured at a concentration of 2% in 50 mL 10% Skim Milk broth and incubated at 42 °C overnight. After incubation, tubes were stored at 4 °C for 2 weeks. Later, aliquots of fermented samples were plated on M17 and MRS (Oxoid, UK) agar containing 20 μg/mL tetracycline to detect transconjugants. The experiment was performed with 3 technical and two biological replicates.
2.6. Transferability of Resistance Genes during Gastrointestinal Transit
The transferability of tet(S) from the donor M17PTZA496 to recipients L. rhamnosus GG and L. delbrueckii DSM 20,355 during simulated human gastrointestinal transit was evaluated separately. The simulated gastrointestinal conditions were prepared as previously described by Tarrah et al. [28]. Briefly, the gastric juice and the intestinal juice were prepared separately. A total of 1 mL of an overnight culture of donor and recipients were transferred to 8 mL gastric juice and incubated at 37 °C for 1 h. After incubation, 10 mL of the intestinal juice was added to the mixture, and the tubes were incubated for a further 3 h at 37 °C. Finally, each tube was transferred in MRS broth, incubated at 37 °C for 24 h, and plated on an MRS agar containing 20 μg/mL of tetracycline to detect transconjugants. The experiment was performed with 3 technical and two biological replicates.
3. Results
3.1. Minimal Inhibitory Concentrations for S. thermophilus Strains
MIC values for 16 antibiotics widely used in human and veterinary therapy were determined for eight S. thermophilus strains along with the strain type as reference (Table 2). By considering, where present, the cut-off values established by the EFSA [35], all strains tested demonstrated susceptibility to ampicillin, chloramphenicol, erythromycin, gentamycin, kanamycin, vancomycin, and streptomycin. Strain M17PTZA496 showed a very high resistance level to tetracycline (128 µg/mL), compared to all others that tested susceptible (<0.25 µg/mL). Although a cut-off for the remaining antibiotics has not been established by the EFSA for S. thermophilus, the recorded MIC values of trimethoprim, neomycin, spectinomycin, and kanamycin were relatively constant throughout all strains, which can be considered a good indicator of intrinsic, rather than acquired, resistance. The MIC values for oxacillin varied from 0.5 to 8 (µg/mL), indicating considerable variability across strains. Strain TH982 scored the lowest MIC values among all strains toward aminoglycosides, namely gentamycin, chloramphenicol, kanamycin, spectinomycin, and streptomycin.

Table 2.
Minimum Inhibitory Concentration (µg/mL) for 16 antibiotics against S. thermophilus strains.
3.2. Antibiotic Resistance Genes Investigation
As expected, the analysis by the Comprehensive Antibiotic Resistance Database (CARD) server based on the predicted protein content from the entire genome of each strain revealed the presence of the gene tet(S) only in the tetracycline-resistant strain M17PTZA496. The gene was located on the chromosome from position 1,659,979 to 1,661,904 bp, with a size of 1925 bp (Figure 1A). The blastp analysis of this gene against the NCBI database revealed 100% similarity with tetracycline resistance ribosomal protection protein tet(S), isolated from multiple species (accession number: WP_000691722). Interestingly, in M17PTZA496 the tet(S) gene is flanked on both sides by some mobile element proteins, which enforces the possibility of an integrated plasmid/transposon presence nearby the tet(S) gene (Figure 1A). The ResFinder server analysis also confirmed the tet(S) presence only in M17PTZA496, while both servers established the absence of acquired resistance genes in the other strains.
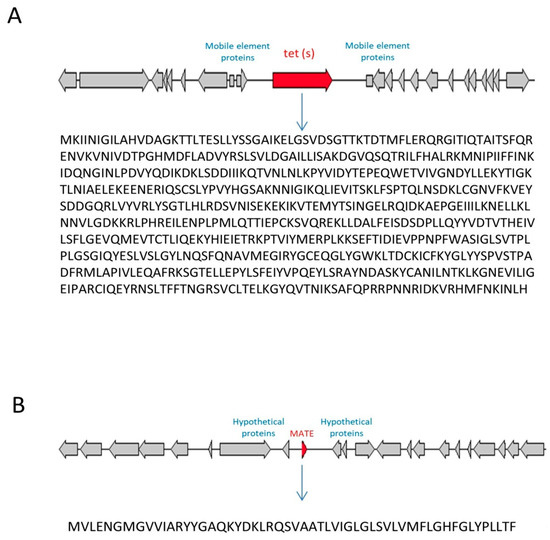
Figure 1.
Genetic location of tet(S) (A) and conserved motif of MATE (multidrug and toxic compound extrusion) (B) genes analyzed on CARD and RAST servers, the respective amino acid sequences in S. thermophilus M17PTZA496.
In addition to the presence of Tet(S) protein, the annotation analysis by RAST detected in M17PTZA496 the presence of a MATE (multidrug and toxic compound extrusion) protein, a family of MDR (multi-drug resistance efflux pump), located from position 975,639 to 975,472 with a size of 168 bp (Figure 1B).
3.3. Identification of Genomic Islands (GI) and Mobile Elements
Among all the S. thermophilus genomes studied, the tetracycline-resistant strain M17PTZA496 displayed the largest number (31) of GIs with an overall size of 317.6 Kb, corresponding approximately to 16% of its genome size, including all clusters predicted by IslandPick, IslandPath-DIMOB, and SIGI-HMM and an intact prophage region (Figure 2).
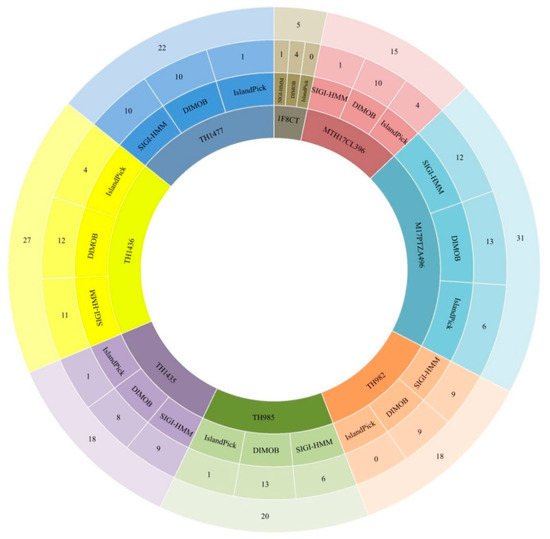
Figure 2.
Distribution of genomic islands predicted by IslandPick, IslandPath-DIMOB, and SIGI-HMM on IslandViewer 4 server in the studied S. thermophilus strains.
Moreover, this strain has the largest genome size among all other strains, which is a good indicator of large horizontal gene transfer (HGT) [36]. On the other hand, strain 1F8CT revealed only five GIs, the lowest number among the strains tested (Figure 2). Among all the GIs detected in M17PTZA496, one is located close to the tet(S) gene, from 1,644,596 to 1,651,123 with an approximate size of 6.5 Kb. This GI carries genes associated with plasmids, beta-lactamase class C-like, and penicillin-binding proteins (PBPs) superfamily. Interestingly, analysis by CRISPRCasFinder revealed that the tetracycline-resistant strain M17PTZA496 is the only one that is missing the CRISPR-CaslllA, which can explain the number of mobile elements detected in this strain [37]. All strains, including M17PTZA496, also possess the CRISPR-CasllC inside their genomes.
Besides, an investigation by the ICEberg2 server for the detection of integrative and conjugative elements within the genome of strain M17PTZA496 revealed an integrative and mobilizable element (IME) located from 1,768,635 to 1,776,232 with an approximate size of 7.5 Kb, including seven putative open reading frames (Figure 3). However, the investigation of the OriTfinder and PlasmidFinder servers did not show any gene transfer origin or actual plasmid integrated with the strain M17PTZA496 genome.
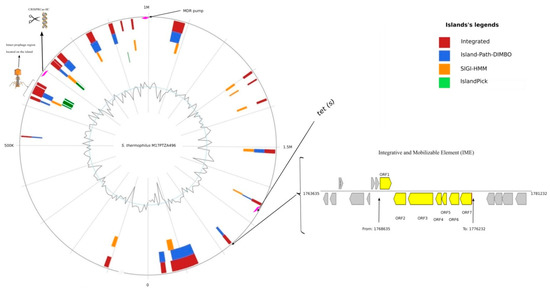
Figure 3.
Circular visualization and translocation of predicted genomic islands (using IslandViewer 4 server), GC content (using IslandViewer 4 server), resistance genes (using CARD and RAST servers), prophage (using PHASTER server), CRISPR-CasllC (using CRISPRCasFinder server), and integrative and conjugative elements (Using ICEberg2 server)related to tetracycline-resistant S. thermophilus M17PTZA496.
3.4. Horizontal Transfer of the Tet(S) Gene
The transferability of tet(S) from S. thermophilus M17PTZA496 to L. rhamnosus GG and to L. delbrueckii DSM 20,355 was studied during growth in skimmed milk, during storage at low temperature and during incubation under simulated gastrointestinal conditions. No transconjugant Lactobacillus colonies were detected on MRS agar plates containing 20 μg/mL tetracycline, neither after storage for 2 weeks at 4 °C nor after 4 h of incubation (1 h gastric juice + 3 h intestinal juice) and the transfer in MSR broth for 24 h at 37 °C. This indicates that, under the conditions tested, no transmission of the tet(S) gene took place between S. thermophilus M17PTZA496 and L. rhamnosus GG or L. delbrueckii DSM 20355.
4. Discussion
Resistance to antibiotics in bacteria is an issue of primary importance as it has been estimated that it will be a primary source of death by 2050. For this reason, it is of great importance to evaluate this property in microbes that can come in contact with the human body, particularly those that can be introduced with foods. In the present work, we studied the resistance/susceptibility patterns of 16 antibiotics, among those most widely used for human and veterinary therapy, in eight S. thermophilus strains, plus the species type strain, by both phenotypic and genomic approaches. S. thermophilus is the second most important technological bacterial species in terms of sales volume, used for a huge variety of dairy productions worldwide. For this reason, it appears very important to gain information on the possible presence of transmissible antibiotic resistances inside this species.
The evaluation of the MIC values obtained in this study evidence intermediate resistance to aminoglycosides (kanamycin, streptomycin, spectinomycin, neomycin, and gentamycin) for all strains. Resistance to this class of drugs is known to be generally intrinsic in S. thermophilus strains and, therefore, not transmissible [38,39].
Conversely, all tested strains showed susceptibility to β-lactams, quinolones, glycopeptides, macrolides, and sulfonamides. Values for chloramphenicol resistance were always below the breakpoint and were low for Rifampicin, although a breakpoint for S. thermophilus is lacking. Strain TH1435 evidenced a MIC value for oxacillin considerably higher than that of the other strains tested; however, no resistance genes for ß-lactam drug resistance were found in its genome, so this resistance should be linked to a non-specific cellular modification. Again, the absence of a breakpoint value for this drug makes a reliable attribution of resistance difficult. Strain M17PTZA496 was the only one that demonstrated a very high resistance level to tetracycline. This strain possesses the tet(S) gene and genes for proteins belonging to the MATE family. Several genes that code for ribosomal protection proteins have been found for tetracycline resistance in S. thermophilus strains, including genes tet(S), tet(M), tet(L), and tet(A) [40,41,42]. The tet(S) gene has been found in some Gram-positive bacteria such as Listeria monocytogenes, Enterococcus faecalis, and Lactococcus lactis [43]. This gene encodes a tetracycline resistance protein Tet(S), which abolishes the inhibitory effect of tetracycline on protein synthesis by a non-covalent modification of the ribosomes [44]. Moreover, the bacterial MATE is a family of proteins that function as antiporters and can confer resistance to different drugs, including antibiotics and other DNA-damaging agents, by constantly pumping the toxic agents out of the cytoplasm [45,46]. A comparison of the tetracycline MIC values between M17PTZA496 and other tetracycline-resistant S. thermophilus strains in the literature showed a higher resistance level in M17PTZA496, that can be associated with the simultaneous presence of the protein Tet(S) and the MATE family [42,47,48].
Data on antibiotic resistance among streptococci indicate a high tetracycline resistance rate among pathogenic Streptococcus, such as S. agalactiae and S. pyogenes, that can be linked to the level of antibiotic usage in humans and horizontal gene transfer from other bacteria [4,49]. However, the presence of acquired tetracycline resistance genes among S. thermophilus strains is rare [50] and few studies have reported tetracycline-resistant strains among S. thermophilus in food (mainly dairy) environments [47,51]. Two studies reported the presence of the resistance gene tet(S) in a few S. thermophilus strains; however, there was no report of horizontal gene transfer from S. thermophilus to other bacteria [47,51]. Interestingly, the Tet(S) amino acid sequence of M17PTZA496 reported in this study indicates 100% similarity with the other two S. thermophilus reported as Tet(S) carriers [4].
In previous studies, we evaluated some properties and safety aspects of S. thermophilus M17PTZA496 [51]. This strain revealed interesting probiotic properties in vitro, cytotoxic activity against HT-29 cancer cells line, and a considerable folate production level [52]. As a safety aspect, the potential release of the prophage present in the genome of M17PTZA496 was evaluated using different phage-inducing agents, such as drugs, H2O2, and NaCl; however, the study revealed that the phage was non-inducible under any of the conditions tested [51].
For a potential probiotic strain, the possibility of antibiotic resistance gene(s) transmission is a serious issue. In some strains, resistance traits are on genomic islands inside the genome that encode their excision and integration into the chromosome [10]. However, they cannot be transferred by themselves to other bacteria, due to the lack of a conjugative apparatus, but only mobilized in the presence of conjugative elements such as ICEs that can pick them up and transfer them to other bacterial cells [11,12].
5. Conclusions
The importance of transmissible antibiotic resistance genes in food-related bacteria is related to their possible transmission to pathogens during food manufacturing and storage or in the course of the human gastrointestinal transit. In this study, despite the presence of an acquired tetracycline resistance gene tet(S) in S. thermophilus M17PTZA496, no transfer of tet(S) was detected under the conditions tested, which can be ascribed to the chromosomal location of the gene rather than the mobile elements. Moreover, the presence of a genomic island and IME in the proximity of tet(S) should not raise any concerns of possible horizontal transfer due to the lack of a conjugal apparatus and the origin of transfer in M17PTZA496, which are essential for mating between bacterial cells.
Author Contributions
Conceptualization A.T. and A.G.; investigation, A.T. and S.P., data curation A.T. and S.P.; writing—original draft preparation, A.T.; writing—review and editing, A.T., A.G. and V.C.; supervision, A.G.; funding acquisition, A.G. and V.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work has been funded in part by the Ministero dell’Università e della Ricerca Scientifica (MIUR).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Watve, M.G.; Tickoo, R.; Jog, M.M.; Bhole, B.D. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 2001, 176, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R. History of antimicrobial drug discovery: Major classes and health impact. Biochem. Pharmacol. 2017, 133, 4–19. [Google Scholar] [CrossRef]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-Wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2019, 85, e01738-18. [Google Scholar]
- Flórez, A.B.; Mayo, B. Antibiotic Resistance-Susceptibility profiles of Streptococcus thermophilus isolated from raw milk and genome analysis of the genetic basis of acquired resistances. Front. Microbiol. 2017, 8, 2608. [Google Scholar] [CrossRef]
- Devirgiliis, C.; Caravelli, A.; Coppola, D.; Barile, S.; Perozzi, G. Antibiotic resistance and microbial composition along the manufacturing process of Mozzarella di Bufala Campana. Int. J. Food Microbiol. 2008, 128, 378–384. [Google Scholar] [CrossRef]
- Soares-Santos, V.; Barreto, A.S.; Semedo-Lemsaddek, T. Characterization of Enterococci from Food and Food-Related Settings. J. Food Prot. 2015, 78, 1320–1326. [Google Scholar] [CrossRef]
- Tarrah, A.; Pakroo, S.; Junior, W.J.F.L.; Guerra, A.F.; Corich, V.; Giacomini, A. Complete Genome Sequence and Carbohydrates-Active EnZymes (CAZymes) Analysis of Lactobacillus paracasei DTA72, a Potential Probiotic Strain with Strong Capability to Use Inulin. Curr. Microbiol. 2020, 77, 2867–2875. [Google Scholar] [CrossRef]
- Tarrah, A.; Pakroo, S.; Corich, V.; Giacomini, A. Whole-Genome sequence and comparative genome analysis of Lactobacillus paracasei DTA93, a promising probiotic lactic acid bacterium. Arch. Microbiol. 2020, 202, 1997–2003. [Google Scholar] [CrossRef]
- Gómez-Gómez, C.; Blanco-Picazo, P.; Brown-Jaque, M.; Quirós, P.; Rodríguez-Rubio, L.; Cerdà-Cuellar, M.; Muniesa, M. Infectious phage particles packaging antibiotic resistance genes found in meat products and chicken feces. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Liu, M.; Li, X.; Xie, Y.; Bi, D.; Sun, J.; Li, J.; Tai, C.; Deng, Z.; Ou, H.-Y. ICEberg 2.0: An updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019, 47, D660–D665. [Google Scholar] [CrossRef]
- Guédon, G.; Libante, V.; Coluzzi, C.; Payot-Lacroix, S.; Leblond-Bourget, N. The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems. Genes 2017, 8, 337. [Google Scholar] [CrossRef] [PubMed]
- Bellanger, X.; Payot, S.; Leblond-Bourget, N.; Guédon, G. Conjugative and mobilizable genomic islands in bacteria: Evolution and diversity. FEMS Microbiol. Rev. 2014, 38, 720–760. [Google Scholar] [CrossRef]
- Irlinger, F.; Mounier, J. Microbial interactions in cheese: Implications for cheese quality and safety. Curr. Opin. Biotechnol. 2009, 20, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.; Novak, J.; Knocke, W.; Pruden, A. Elevation of antibiotic resistance genes at cold temperatures: Implications for winter storage of sludge and biosolids. Lett. Appl. Microbiol. 2014, 59, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Schjørring, S.; Krogfelt, K.A. Assessment of Bacterial Antibiotic Resistance Transfer in the Gut. Int. J. Microbiol. 2011, 2011, 1–10. [Google Scholar] [CrossRef]
- Tarrah, A.; Noal, V.; Giaretta, S.; Treu, L.; Duarte, V.D.S.; Corich, V.; Giacomini, A. Effect of different initial pH on the growth of Streptococcus macedonicus and Streptococcus thermophilus strains. Int. Dairy J. 2018, 86, 65–68. [Google Scholar] [CrossRef]
- Tarrah, A.; Noal, V.; Treu, L.; Giaretta, S.; Duarte, V.D.S.; Corich, V.; Giacomini, A. Short communication: Comparison of growth kinetics at different temperatures of Streptococcus macedonicus and Streptococcus thermophilus strains of dairy origin. J. Dairy Sci. 2018, 101, 7812–7816. [Google Scholar] [CrossRef]
- Huddleston, J.R. Horizontal gene transfer in the human gastrointestinal tract: Potential spread of antibiotic resistance genes. Infect. Drug Resist. 2014, 7, 167. [Google Scholar] [CrossRef]
- Mater, D.D.G.; Bretigny, L.; Firmesse, O.; Flores, M.-J.; Mogenet, A.; Bresson, J.-L.; Corthier, G. Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus survive gastrointestinal transit of healthy volunteers consuming yogurt. FEMS Microbiol. Lett. 2005, 250, 185–187. [Google Scholar] [CrossRef]
- Tarrah, A.; Treu, L.; Giaretta, S.; Duarte, V.; Corich, V.; Giacomini, A. Differences in Carbohydrates Utilization and Antibiotic Resistance Between Streptococcus macedonicus and Streptococcus thermophilus Strains Isolated from Dairy Products in Italy. Curr. Microbiol. 2018, 75, 1334–1344. [Google Scholar] [CrossRef] [PubMed]
- Blaiotta, G.; Sorrentino, A.; Ottombrino, A.; Aponte, M. Short communication: Technological and genotypic comparison between Streptococcus macedonicus and Streptococcus thermophilus strains coming from the same dairy environment. J. Dairy Sci. 2011, 94, 5871–5877. [Google Scholar] [CrossRef]
- Zhalnina, K.; Dias, R.; Quadros, P.D.D.; Davis-Richardson, A.; Camargo, F.A.O.; Clark, I.M.; McGrath, S.P.; Hirsch, P.R.; Triplett, E.W. Soil pH Determines Microbial Diversity and Composition in the Park Grass Experiment. Microb. Ecol. 2015, 69, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.; Tomar, S.; Maheswari, T.U.; Singh, R. Streptococcus thermophilus strains: Multifunctional lactic acid bacteria. Int. Dairy J. 2010, 20, 133–141. [Google Scholar] [CrossRef]
- Cho, H.; Park, K.-E.; Kim, K.-S. Genome analysis of Streptococcus salivarius subsp. thermophilus type strain ATCC 19258 and its comparison to equivalent strain NCTC 12958. Arch. Microbiol. 2021, 203, 1843–1849. [Google Scholar] [CrossRef]
- Treu, L.; Vendramin, V.; Bovo, B.; Campanaro, S.; Corich, V.; Giacomini, A. Genome Sequences of Four Italian Streptococcus thermophilus Strains of Dairy Origin. Genome Announc. 2014, 2, e00126-14. [Google Scholar] [CrossRef]
- Treu, L.; Vendramin, V.; Bovo, B.; Campanaro, S.; Corich, V.; Giacomini, A. Genome Sequences of Streptococcus thermophilus Strains MTH17CL396 and M17PTZA496 from Fontina, an Italian PDO Cheese. Genome Announc. 2014, 2, e00067-14. [Google Scholar] [CrossRef]
- Treu, L.; Vendramin, V.; Bovo, B.; Campanaro, S.; Corich, V.; Giacomini, A. Whole-Genome Sequences of Streptococcus thermophilus Strains TH1435 and TH1436, Isolated from Raw Goat Milk. Genome Announc. 2014, 2, e01129-13. [Google Scholar] [CrossRef]
- Tarrah, A.; Duarte, V.D.S.; Pakroo, S.; Corich, V.; Giacomini, A. Genomic and phenotypic assessments of safety and probiotic properties of Streptococcus macedonicus strains of dairy origin. Food Res. Int. 2020, 130, 108931. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. Simon Fraser University Research Computing Group IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In SilicoDetection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Li, X.; Xie, Y.; Liu, M.; Tai, C.; Sun, J.; Deng, Z.; Ou, H.-Y. oriTfinder: A web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 2018, 46, W229–W234. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel On Additives And Products Or Substances Used in Animal Feed FEEDAP. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2704. [Google Scholar] [CrossRef]
- Vendramin, V.; Treu, L.; Campanaro, S.; Lombardi, A.; Corich, V.; Giacomini, A. Genome comparison and physiological characterization of eight Streptococcus thermophilus strains isolated from Italian dairy products. Food Microbiol. 2017, 63, 47–57. [Google Scholar] [CrossRef]
- Palmer, K.L.; Gilmore, M.S. Multidrug-Resistant Enterococci Lack CRISPR-cas. MBio 2010, 1, e00227-10. [Google Scholar] [CrossRef]
- Ammor, M.S.; Flórez, A.B.; Mayo, B. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 2007, 24, 559–570. [Google Scholar] [CrossRef]
- Panel, E.F. Technical Guidance for assessing the safety of feed additives for the environment. EFSA J. 2008, 6, 842. [Google Scholar]
- Arioli, S.; Guglielmetti, S.; Amalfitano, S.; Viti, C.; Marchi, E.; Decorosi, F.; Giovannetti, L.; Mora, D. Characterization of tetA-like gene encoding for a major facilitator superfamily efflux pump in Streptococcus thermophilus. FEMS Microbiol. Lett. 2014, 355, 61–70. [Google Scholar] [CrossRef]
- Ge, B.; Jiang, P.; Han, F.; Saleh, N.K.; Dhiman, N.; Fedorko, D.P.; Nelson, N.A.; Meng, J. Identification and Antimicrobial Susceptibility of Lactic Acid Bacteria from Retail Fermented Foods. J. Food Prot. 2007, 70, 2606–2612. [Google Scholar] [CrossRef]
- Domig, K.J.; Zycka-Krzesinska, J.; Bardowski, J.; Morelli, L. Molecular assessment of erythromycin and tetracycline resistance genes in lactic acid bacteria and bifidobacteria and their relation to the phenotypic resistance. Int. J. Probiotics Prebiotics 2008, 3, 271–280. [Google Scholar]
- Kim, S.-R.; Nonaka, L.; Suzuki, S. Occurrence of tetracycline resistance genes tet(M) and tet(S) in bacteria from marine aquaculture sites. FEMS Microbiol. Lett. 2004, 237, 147–156. [Google Scholar] [CrossRef]
- Hedayatianfard, K.; Akhlaghi, M.; Sharifiyazdi, H. Detection of tetracycline resistance genes in bacteria isolated from fish farms using polymerase chain reaction. In Veterinary Research Forum; Faculty of Veterinary Medicine, Urmia University: Urmia, Iran, 2014; p. 269. [Google Scholar]
- Mishra, M.N.; Daniels, L. Characterization of the MSMEG_2631 Gene (mmp) Encoding a Multidrug and Toxic Compound Extrusion (MATE) Family Protein in Mycobacterium smegmatis and Exploration of Its Polyspecific Nature Using Biolog Phenotype MicroArray. J. Bacteriol. 2013, 195, 1610–1621. [Google Scholar] [CrossRef]
- Dridi, L.; Tankovic, J.; Petit, J.-C. CdeA of Clostridium difficile, a new multidrug efflux transporter of the MATE family. Microb. Drug Resist. 2004, 10, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Jasir, A.; Tanna, A.; Noorani, A.; Mirsalehian, A.; Efstratiou, A.; Schalen, C. High rate of tetracycline resistance inStreptococcus pyogenes in Iran: An epidemiological study. J. Clin. Microbiol. 2000, 38, 2103–2107. [Google Scholar] [CrossRef] [PubMed]
- Emaneini, M.; Mirsalehian, A.; Beigvierdi, R.; Fooladi, A.A.I.; Asadi, F.; Jabalameli, F.; Taherikalani, M. High Incidence of Macrolide and Tetracycline Resistance among Streptococcus Agalactiae Strains Isolated from Clinical Samples in Tehran, Iran. MAEDICA J. Clin. Med. 2014, 9, 157–161. [Google Scholar]
- Mathur, S.; Singh, R. Antibiotic resistance in food lactic acid bacteria—A review. Int. J. Food Microbiol. 2005, 105, 281–295. [Google Scholar] [CrossRef]
- Rizzotti, L.; Gioia, F.L.; Dellaglio, F.; Torriani, S. Characterization of Tetracycline-Resistant Streptococcus thermophilus Isolates from Italian Soft Cheeses. Appl. Environ. Microbiol. 2009, 75, 4224–4229. [Google Scholar] [CrossRef]
- Duarte, V.D.S.; Giaretta, S.; Campanaro, S.; Treu, L.; Armani, A.; Tarrah, A.; Paula, S.O.D.; Giacomini, A.; Corich, V. A Cryptic Non-Inducible Prophage Confers Phage-Immunity on the Streptococcus thermophilus M17PTZA496. Viruses 2018, 11, 7. [Google Scholar] [CrossRef]
- Tarrah, A.; De Castilhos, J.; Rossi, R.C.; Duarte, V.D.S.; Ziegler, D.R.; Corich, V.; Giacomini, A. In Vitro Probiotic Potential and Anti-Cancer Activity of Newly Isolated Folate-Producing Streptococcus thermophilus Strains. Front. Microbiol. 2018, 9, 2214. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).