Abstract
In this study, we explored the diversity of yeasts and lactic acid bacteria (LAB) associated with six spontaneous sourdough fermentations from the northern part of the Apulian region (Italy). Bacterial and yeast isolates from sourdough were investigated by amplified ribosomal DNA restriction analysis (ARDRA) and restriction fragment length polymorphism (RFLP) analysis, respectively. The identification of the isolates was confirmed by sequencing bacterial 16S gene and yeast ITS1–5.8S–ITS2 rRNA gene amplicons. Microbiological analysis of all sourdough samples revealed that LAB and yeast counts ranged between 1.7 × 105 and 6.5 × 108 cfu/g, and 7.7 × 105 and 2.5 × 107 cfu/g, respectively. The molecular identification at species level revealed the occurrence of Lactobacillus plantarum as the dominant LAB and Saccharomyces cerevisiae as the dominant yeast species in all different sourdough samples. Then, the ability of all isolated strains to inhibit and/or reduce the growth of several selected fungi was valued through the overlay method. In light of their antifungal performances, ten LAB strains were inoculated, singularly and in combination, in subsequent bread-making trials. Overall, we confirmed the potential of LAB to extend the shelf life of bread through spoilage inhibition and, for the first time, we observed a synergistic effect due to the combination of several isolated LAB on the inhibition behavior against selected fungal spoilage strains. Our findings suggest the exploration of a LAB-based approach in order to extend the shelf life of bread, reducing, at the same time, the use of chemical agents for food preservation.
1. Introduction
The manufacturing of several food products is founded on starter culture inoculation. Starter cultures can be defined as microbial preparations of selected microorganisms elected to be added to the raw material in order to accelerate and steer the fermentation process [1]. Thus, on the one hand, today the food industry benefits from the numerous signs of progress made in the selection of new and increasingly active microbial starters to drive and standardize the manufacture of fermented foodstuffs, yet on the other hand we should not forget that there are some foods in which this process of microbial selection occurs naturally and gradually, leading to natural starters being independently selected over time. For example, this is the case of sourdough that is the combination of flour, water, autochthonous homofermentative and heterofermentative lactic acid bacteria (LAB), and yeasts [2].
In numerous world areas, traditional sourdoughs are passed down from generation to generation through the storage of small aliquots of bread dough that are refreshed and utilized for the production of subsequent bakery products [3]. The microbial biodiversity in sourdough is usually, but not always, counted in a ratio LAB:yeasts of 100:1 [4,5,6] and might include different species of LAB, e.g., Lactobacillus (mainly the strains Lactobacillus sanfranciscensis, Lactobacillus plantarum, Lactobacillus brevis, Lactobacillus rossiae, Lactobacillus zymae, Lactobacillus acidifarinae, Lactobacillus pontis, Lactobacillus panis), Lactococcus, Leuconostoc, Enterococcus [4,5,6], and different species of yeast, e.g., Saccharomyces cerevisiae, Saccharomyces exiguous, Candida humilis (also named Candida milleri) [2,7]. However, the sourdough ecosystem is strictly dependent on factors such as dough hydration, type of cereal source, temperature of fermentation, and geographical area of production [8,9].
The use of traditional sourdough leads to end products with ameliorated and enlarged organoleptic features, such as texture, taste, and flavor [10,11]. In fact, lactic acid bacteria (LAB) are able to enrich dough, producing organic acids, exopolysaccharides, antimicrobial compounds and enzymes [12]. Moreover, the microbial consortium could discourage the development of spoilage microbes, extending the shelf life [4,13,14,15].
In this work, the purpose was to investigate the microbial profile of six traditional sourdoughs from the northern part of the Apulian region in Italy and to screen and exploit the potential antifungal ability of isolates from the different dough matrices.
2. Materials and Methods
2.1. Sourdough Sampling, Microbial Analysis, Screening and Isolation of LAB and Yeasts
Six sourdough samples (Table 1) were collected from six different bakeries located in the Apulian territory. For all selected sourdoughs, the bakeries claimed the presence of spontaneous fermentation with a continuous back-slopping over more than 10 years.

Table 1.
Apulian sourdough samples collected in this work.
Three samples of 10 g of each sourdough were homogenized in a sterile stomacher bag containing 90 mL of sterilized saline solution (8.5 g/L NaCl) using a stomacher (BagMixer, Interscience, Saint-Nom-la-Bretèche, France) for 2 min at 260 rpm (revolutions per minute). Subsequentially, tenfold serial dilutions were carried out in order to count bacterial and yeast populations using MRS (Sigma-Aldrich, St. Louis, MO, USA) plates with cycloheximide (100 mg/L) added and Wallerstein Laboratory (WL)Nutrient (Sigma-Aldrich, St. Louis, MO, USA) plates with chloramphenicol (100 mg/L) added, respectively.
For bacteria, plates were incubated either at 37 °C for 24–48 h in aerobic conditions or at 37 °C for 7 days in anaerobic conditions in order to achieve both the growth of aerobic and anaerobic bacteria. For yeasts, the incubation of the plates was performed at 37 °C for 24–48 h.
After incubation, the colonies forming unit per g (cfu/g) were counted, and bacterial and yeast colonies (77 and 58, respectively) were randomly chosen and isolated based on phenotypic diversity. Bacterial and yeast colonies were purified by streaking four times onto the same medium used for the isolation. Each purified strain was grown, identified by alphanumeric code (Table 2) and stored at −80 °C in a broth medium supplemented with 25% w/v glycerol.

Table 2.
Alphanumeric code of bacterial isolates screened in this work.
2.2. Molecular Identification of LAB and Yeast Isolates
Microbial DNA from each purified strain of bacteria and yeast was extracted and purified using the UltraClean Microbial DNA Isolation Kit (MO BIO Laboratories, Solana Beach, CA, USA) according to the manufacturer’s protocols.
LAB strains were clustered by amplified ribosomal DNA restriction analysis (ARDRA) and yeast strains by ITS (internal transcribed spacer) region amplification and restriction fragment length polymorphism (RFLP) analysis [16].
Each amplification reaction was carried out in a final volume of 50 μL using Taq PCR Core Kit (Qiagen, Hilden, Germany). The bacterial 16S rRNA gene was amplified using BSF8 (5′-AGAGTTTGATCCTGGCTCAG-3′) and BSR1541 (5′AAGGAGGTGATCCAGCCGCA-3′) primers [17,18]. For yeasts, the primers SC1 (5′-AACGGTGAGAGATTTCTGTGC-3′), SC2 (5′-AGCTGGCAGTATTCCCACAG-3′) were first used in order to distinguish Saccharomyces cerevisiae from the other yeasts. The PCR conditions used were: 94 °C initial denaturation for 5 min; 30 amplification cycles of 30 s at 94 °C, 1 min at 50 °C annealing temperature, 1 min at 72 °C; final extension at 72 °C for 7 min. The presence of amplicons was confirmed by electrophoresis in 1.0% w/v agarose gel stained with ethidium bromide (0.5 μg/mL) [19]. Subsequently, for all yeast strains not individuated as S. cerevisiae and for five strains presumably designated as S. cerevisiae (thanks to the aforementioned PCR analysis) the ITS regions were amplified using ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) primers [20], using the following PCR conditions: 95 °C initial denaturation for 5 min; 35 amplification cycles of 30 s at 94 °C, 30 s at 55 °C annealing temperature, 45 s at 72 °C; final extension at 72 °C for 10 min. The presence of amplicons was confirmed by electrophoresis in 1.0% w/v agarose gel stained with ethidium bromide (0.5 μg/mL).
LAB 16S rRNA gene amplicons were digested at 37 °C with the fast-restriction endonucleases AluI, HinfI, and HaeIII (Thermo Scientific, Rochford, IL), while yeast ITS amplicons were digested with HaeIII and HinfI enzymes. The restriction fragments were separated (at 50 V for 2 h) on 1.5% w/v agarose gels stained with ethidium bromide (0.5 μg/mL) in Tris-Borate-EDTA buffer (Sigma-Aldrich). All restriction profiles were digitally processed and analyzed with BioNumerics software version 7.5 (Applied Maths, St-Martens-Latem, Belgium). ARDRA and ITS–RFLP profiles from all digestions were combined, respectively for LAB and yeasts, and clustered (Ward’s method).
The identification of the isolates was confirmed by sequencing LAB 16S and yeast ITS1–5.8S–ITS2 rRNA gene amplicons (Humanizing Genomics Magrogen, Amsterdam, The Netherlands). The sequencing profiles were analyzed using BLAST software by NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.3. In Vitro Antifungal Activity
The ability of all isolated strains to inhibit and/or reduce the growth of target fungi was evaluated through the overlay method according to [21]. Briefly, 10 μL of the isolated cultures at the middle exponential phase (OD600nm = 1.0) were spotted on MRS agar plates. After 24 h of incubation at 30 °C, the plates were overlaid with 15 mL of soft malt extract agar (0.75% agar) supplemented with 100 μL of malt extract broth containing approximately 1 × 106 spores/mL of each mold (OD600 = 0.5). After 5 days of incubation at 25 °C, antifungal activity was valued based on the halo of inhibition surrounding the spots and was classified as strains of no (+/−), mild (+), or strong (++) inhibition showing inhibition zones lower than 1 mm, ranging from 1–3 mm, or more than 3 mm, respectively [21].
The target fungi, in order to test the antifungal activity, were Aspergillus flavus CECT 20802, Aspergillus niger CECT 2805, Penicillium expansum CECT 2278, Penicillium roqueforti CECT 20508, Fusarium culmorum CECT 2148, provided by the Spanish Type Culture Collection (CECT, Paterna, Spain). These fungi were chosen because they are the most frequent food contaminating species.
2.4. Sourdough Preparation, Bread Making and Antifungal Activity on Bread
For the preparation of the sourdoughs, 400 g of flour and various amounts of microbial biomass were mixed with 200 mL tap water, according to [22]. The doughs were mixed for 5–10 min and, then, sourdough fermentation was carried out at 37 °C for 16 h. The microorganisms inoculated into the dough were a strain of S. cerevisiae (1 × 108 cf/mL) and 10 strains of LAB chosen on the basis of their antifungal capability (1 × 106 cfu/mL) (see the previous paragraph). One dough was inoculated only with yeast strain, while 10 doughs were fermented by the mix of S. cerevisiae/bacterial strain (Table 3). Moreover, 9 doughs were added with combinations of a single strain of S. cerevisiae with an increasing number of LAB strains, as shown in Table 4. The obtained sourdoughs were used to produce bread by mixing 400 g flour, 200 g sourdough (50% w/w on flour basis), 6 g salt (1.5% w/w on flour basis) and 200 mL tap water, and kneading the doughs for 5 min. The fermentative process was carried out at 37 °C for 4 h and baking was carried out at 200 °C for 10 min and, then, 210 °C for 30 min. The obtained loaves of bread were opportunely contaminated by saline solution (8.5 g/L NaCl) containing approximately 1 × 105 spores/mL of each aforementioned mold (OD600 = 0.5). Bread products were packed in polyethylene bags to maintain constant moisture and incubated at room temperature [23]. The effects of microbial biodiversity on the fungal growth were evaluated by visually monitoring over time (7, 14, 21, and 28 days) the fungal growth and development. All experiments were carried out three times.

Table 3.
Dual combinations of the Saccharomyces cerevisiae strain and lactic acid bacteria (LAB) isolates for the sourdough preparation.

Table 4.
Increased biodiversity combination of the S. cerevisiae strain and LAB isolates for the sourdough preparation.
3. Results and Discussion
3.1. The Biodiversity of Bacterial and Yeast Isolates from Different Apulian Sourdoughs
Six Italian sourdoughs collected from different localities, i.e., Monte Sant’Angelo (sourdough identified in this work by the letter A), Orsara di Puglia (sourdough identified in this work by the letter B), Trani (sourdoughs identified in this work by the letters C, D, and E), and Ortanova (sourdough identified in this work by the letter F) (Table 1), placed in the northern part of Apulia (southern Italian region), were collected and analyzed. Microbiological analysis of all sourdough samples revealed LAB and yeast counts in the range between 1.7 × 105 and 6.5 × 108 cfu/g, and 7.7 × 105 and 2.5 × 107 cfu/g, respectively (Figure 1). Generally, the sourdough microbial ratio LAB:yeasts was 100:1 [9] and for some of the analyzed samples, i.e., B (Orsara di Puglia), D (Trani 02), E (Trani 03) and F (Ortanova), we found this kind of proportion. Intriguingly, for two sourdough samples, i.e., A (Monte Sant’Angelo) and C (Trani 01), the ratio LAB:yeasts was around 1:100, hence the inverse of the ratio usually found.
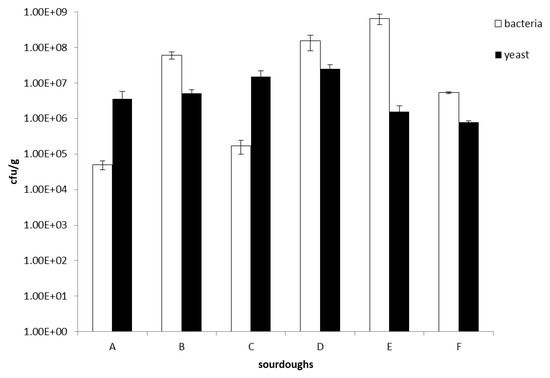
Figure 1.
Microbiological analysis of the Apulian sourdoughs investigated in this work. All values are mean ± SD. A, sourdough sample from Monte Sant’Angelo; B, sourdough sample from Orsara di Puglia; C, sourdough sample from Trani 01; D, sourdough sample from Trani 02; E, sourdough sample from Trani 03; F, sourdough sample from Ortanova.
From MRS and WL Nutrient plates, 77 LAB and 58 yeast colonies were randomly selected among the isolates with different colony morphologies, in order to proceed with a genotypic characterization.
For all LAB isolates, the 16S rRNA gene amplicon was amplified and subjected to restriction analysis using the AluI, HaeIII, and HinfI enzymes. AluI produced five different ARDRA profiles arbitrarily identified as Ia, IIa, IIIa, IVa and Va, of which around 33% of LAB showed the profile Ia, around 47% showed the profile IIa, around 9% showed the profile IIIa, around 9% showed the profile IVa, and around 2% showed the profile Va (Figure 2a). HinfI produced three different ARDRA profiles arbitrarily identified as Ib, IIb, and IIIb, of which around 69% of LAB showed the profile Ib, around 26% with the profile IIb, and around 5% with the profile IIIb (Figure 2b). HaeIII produced four different ARDRA profiles arbitrarily identified as Ic, IIc, IIIc, and IVc, of which around 37% of LAB showed the profile Ic, around 19% showed the profile IIc, around 40% showed the profile IIIc, and around 4% showed the profile IVc (Figure 2c).
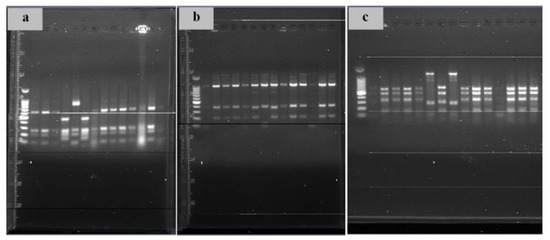
Figure 2.
Restriction analysis examples of 16S rRNA genes using the AluI (a), HaeIII (b), and HinfI (c) enzymes.
Combining the restriction profiles obtained from the three different restriction enzymes and comparing them for all isolates, all LAB isolates were grouped in eighteen different clusters (Figure 3 and Figure 4).
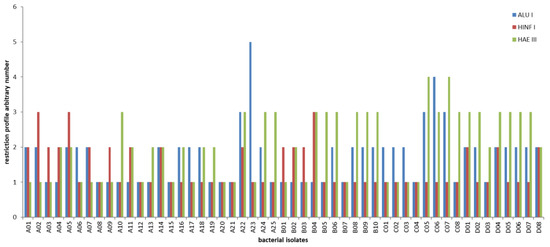
Figure 3.
Histogram representation of different restriction profiles obtained using three different restriction enzymes (AluI, HaeIII, and HinfI). An arbitrary number has been attributed to each diverse digestion profile.
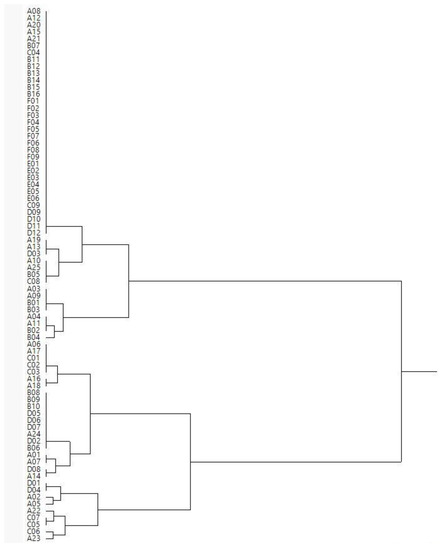
Figure 4.
Dendrogram constructed using a hierarchical cluster analysis (Ward’s method) showing the relative similarities of restriction profiles obtained using three different restriction enzymes (AluI, HaeIII, and HinfI).
Thus, five isolates, randomly selected from each cluster (where the cluster included fewer than five members, the available number of isolates was analyzed), were subjected to 16S rRNA gene sequence analysis. The molecular identification at species level revealed the occurrence of L. plantarum (around 43% of all bacterial isolates) as the dominant LAB species in all the different sourdough samples, in agreement with previous works reporting this species as the most widespread microorganism in sourdoughs [24]. The other isolates were identified as Lactobacillus sunkij (around 14% of bacterial isolates), Lactobacillus parabuchneri (around 14% of bacterial isolates), Pediococcus pentosaceus (around 14% of bacterial isolates), and Lactobacillus brevis (around 15% of bacterial isolates) (Table S1).
For yeasts, 58 isolates were molecularly identified by amplifying a target gene sequence susceptible to discriminating S. cerevisiae from other than S. cerevisiae yeast. Around 57% of the total isolates were presumably identified as Saccharomyces and grouped in a separate cluster. Five of the aforementioned isolates, presumably S. cerevisiae, and the remaining isolates were subjected to amplification of the ITS regions and restriction analysis using the HinfI and HaeIII enzymes. HinfI produced seven different restriction profiles arbitrarily identified as Ia, IIa, IIIa, IVa, Va, VIa, and VIIa, of which around 18% of yeasts showed the profile Ia, around 6% showed the profile IIa, around 12% showed the profile IIIa, around 47% showed the profile IVa, around 6% showed the profile Va, around 6% showed the profile VIa, and around 5% showed the profile VIIa (data not shown). HaeIII produced six different restriction profiles arbitrarily identified as Ib, IIb, IIIb, IVb, Vb, and VIb, comprising around 19% of yeasts in the profile Ib, around 13% in the profile IIb, around 50% in the profile IIIb, around 6% in the profile IVb, around 6% in the profile Vb, and around 6% in the profile VIb (data not shown). Combining the two restriction profiles obtained using the restriction enzymes, all isolates were compared and grouped in clusters according to the similarity of restriction profiles. Eight clusters were found and ITS amplicons from each cluster were subjected to sequence analysis. The molecular identification at species level revealed the occurrence of S. cerevisiae (around 70% of the total isolates, with an identity of around 95%) as the dominant yeast species in all the different sourdough samples, while the other isolates were identified as Candida humilis (around 17%, with an identity of around 90%) and Saccharomyces exiguous (around 13%, with an identity of around 80%) (data not shown). Altogether, these findings are in agreement with other studies on the microbial characterization of the Italian sourdough ecosystem where S. cerevisiae was identified as the prevalent yeast species [25]. However, apart from the S. cerevisiae species, other species such as C. humilis and S. exiguous, but also Candida krusei, Pichia anomala, Saturnispora saitoi, Saccharomyces delbrueckii, Candida famata, and Pichia membranifaciens, Candida lambica, Candida valida and Candida glabrata may be also identified [2,26,27].
3.2. Antifungal Activity of Bacterial Isolates
LAB isolates from sourdough samples were screened for their antifungal activity against six molds belonging to the species A. niger, A. flavus, F. culmorum, P. roqueforti, P. expansum, and P. chrysogenum. These molds were chosen for the reason of spoilage potential in a wide range of food matrices, including bread and bakery goods, and for their potential to produce mycotoxins. In fact, filamentous fungi can be a microbiological hazard for consumer health and an economic problem for the food industry [28,29]. Many LAB are able to be highly active towards several fungal species and the possibility of exploiting these bioprotective capabilities has been considered in the food industry in order to increase shelf life [21,30,31]. The antifungal activity of LAB could be due to different factors, including a direct competition for growth substrates or, indirectly, by producing active antagonistic metabolites, such as organic acids, e.g., lactic, acetic, and phenyllactic acids [32,33], which have been shown to have synergistic effects [33,34].
In this work, on the basis of the inhibition halo (Figure 5), the LAB isolates were classified into three different groups with no, mild, or strong antifungal ability, respectively (Table 5). P. roqueforti, P. chrysogenum, and P. expansum were the most resistant fungal strains since the tested LAB isolates were unable to inhibit their growth. Conversely, the growth of A. flavus was strongly affected by several isolates, e.g., the isolate B07 showed an inhibiting alone of 11 mm, while the isolates A11, A21, A22, and B09 were able to contrast the mold by showing an inhibiting alone of 10 mm. Overall, A. niger and F. culmorum were mildly inhibited by numerous isolates. In detail, the isolate E05 showed an inhibiting alone of 11 mm and the isolates C02, D03, and D08 showed an inhibiting alone of 9 mm against A. niger, while the isolate F02 was able to contrast the growth of F. culmorum showing an inhibiting alone of 13 mm.
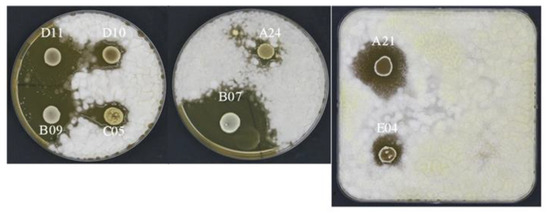
Figure 5.
Examples of halos of inhibition against Aspergillus flavus.

Table 5.
Antifungal activity of the bacterial isolates against Aspergillus flavus, Aspergillus niger, Fusarium culmorum, Penicillium chrysogenum, Penicillium expansum, and Penicillium roqueforti.
3.3. Antifungal Activity on Bread Making by Sourdough Preparation Using Microbial Isolates
On the basis of the previous results, ten LAB isolates were chosen to be used in subsequent bread-making trials. The criterion for choosing these 10 strains was to select the isolates which were able to inhibit at least three molds and which belonged to different species according to the microbial characterization. Sourdough loaves of bread were made using sourdoughs containing mixed cultures of the yeast S. cerevisiae with 10 chosen LAB isolates in i) dual and ii) increasing biodiversity combinations. Bread made only with the S. cerevisiae strain was also produced as the control trial (control without LAB strain), in order to understand the contributions of the different LAB on the antifungal activity. The loaves of bread were then artificially contaminated with the filamentous fungi A. niger, A. flavus, F. culmorum, P. roqueforti, P. expansum, and P. chrysogenum, packed in bags with standard features for all samples, and the growth of all fungi was monitored over time. After 28 days of shelf life, the presence or the absence of fungal mycelium was visually verified. Overall, the observed results indicated a longer shelf life in bread fermented with sourdough inoculated with LAB, as the fungi were strongly inhibited (Figure 6, Figure 7, Figure 8, Figure 9, Figure 10, Figure 11, Figure 12, Figure 13, Figure 14, Figure 15, Figure 16 and Figure 17). Moreover, the increase of the microbial complexity of the sourdough corresponded to an enhanced antifungal action.

Figure 6.
Examples of bread made from sourdoughs inoculated with the isolates A09 and B07, and then artificially contaminated with Aspergillus flavus and incubated at room temperature for 7, 14, 21, and 28 days.
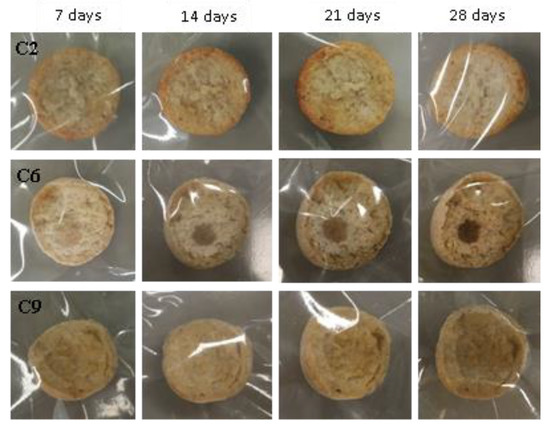
Figure 7.
Examples of bread made from sourdoughs inoculated with the combination of isolates C2, C6, and C9, and then artificially contaminated with A. flavus and incubated at room temperature for 7, 14, 21, and 28 days.
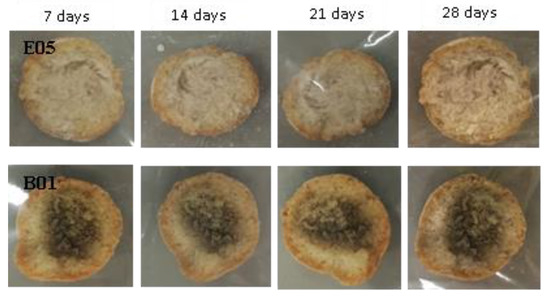
Figure 8.
Examples of bread made from sourdoughs inoculated with the dual combination of isolates E05 and B01, and then artificially contaminated with Aspergillus niger and incubated at room temperature for 7, 14, 21, and 28 days.
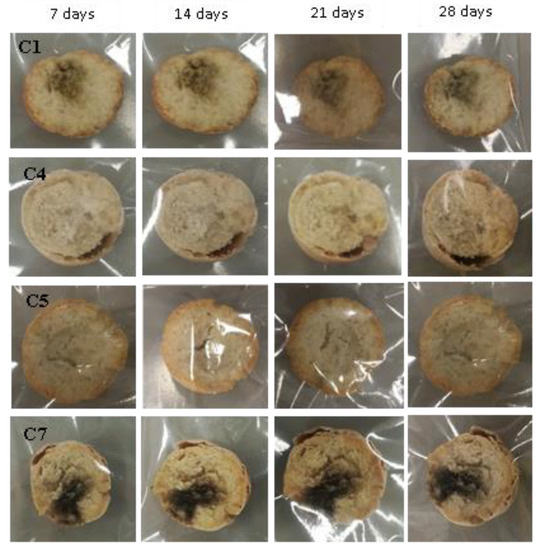
Figure 9.
Examples of bread made from sourdoughs inoculated with the combination of isolates C1, C4, C5, and C7, and then artificially contaminated with Aspergillus niger and incubated at room temperature for 7, 14, 21, and 28 days.
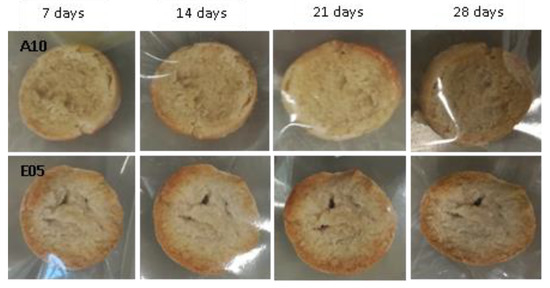
Figure 10.
Examples of bread made from sourdoughs inoculated with the dual combination of isolates A10 and E05, and then artificially contaminated with Fusarium culmorum and incubated at room temperature for 7, 14, 21, and 28 days.
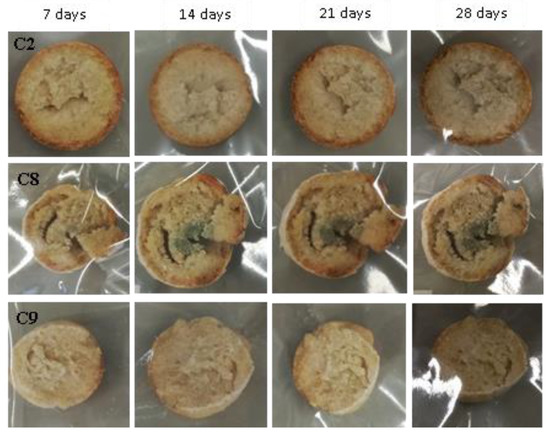
Figure 11.
Examples of bread made from sourdoughs inoculated with the combination of isolates C2, C8, and C9, and then artificially contaminated with Fusarium culmorum and incubated at room temperature for 7, 14, 21, and 28 days.
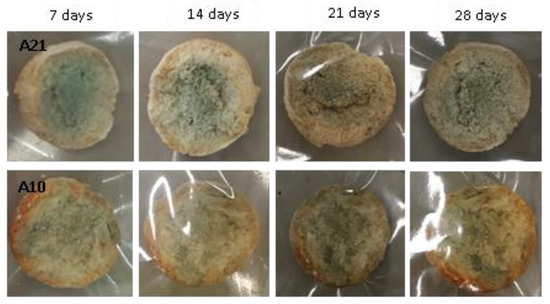
Figure 12.
Examples of bread made from sourdoughs inoculated with the dual combination of isolates A21 and A10, and then artificially contaminated with Penicillium chrysogenum and incubated at room temperature for 7, 14, 21, and 28 days.
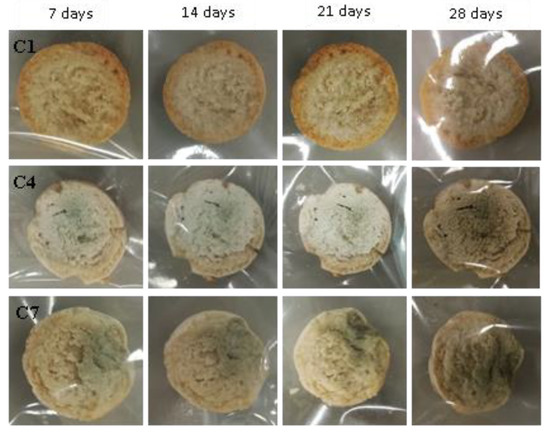
Figure 13.
Examples of bread made from sourdoughs inoculated with the combination of isolates C1, C4, and C7, then artificially contaminated with Penicillium chrysogenum and incubated at room temperature for 7, 14, 21, and 28 days.
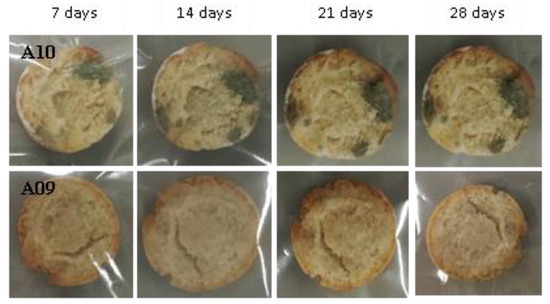
Figure 14.
Examples of bread made from sourdoughs inoculated with the dual combination of isolates A10 and A09, then artificially contaminated with Penicillium expansum and incubated at room temperature for 7, 14, 21, and 28 days.
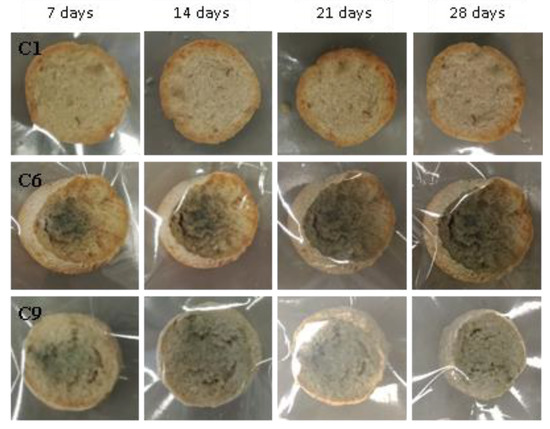
Figure 15.
Examples of bread made from sourdoughs inoculated with the combination of isolates C1, C6, and C9, then artificially contaminated with Penicillium expansum and incubated at room temperature for 7, 14, 21, and 28 days.
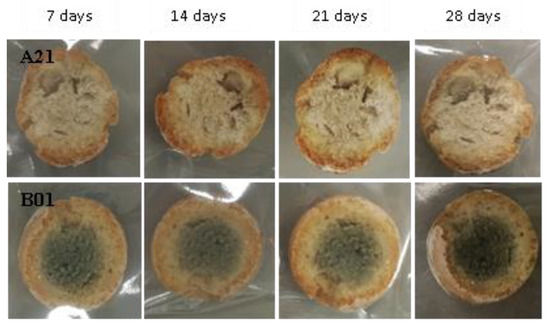
Figure 16.
Examples of bread made from sourdoughs inoculated with the dual combination of isolates A21 and B01, then artificially contaminated with Penicillium roqueforti and incubated at room temperature for 7, 14, 21, and 28 days.
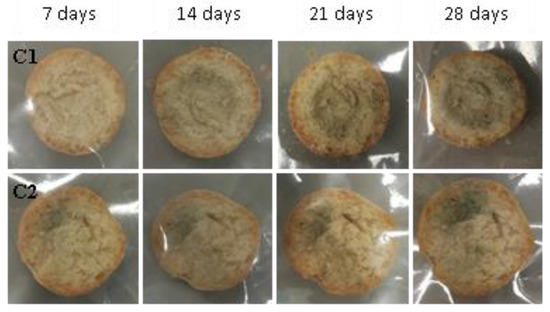
Figure 17.
Examples of bread made from sourdoughs inoculated with the combination of isolates C1 and C2, then artificially contaminated with Penicillium roqueforti and incubated at room temperature for 7, 14, 21, and 28 days.
Specifically, the best results were achieved when sourdough was made with microbial combinations containing more than five LAB isolates. In particular, while the presence of a single LAB isolate determined, in most of the cases, the inhibition of A. flavus mycelium, breads obtained with sourdough inoculated with the dual combination of S. cerevisiae and the isolates A09, A21, B01, D08, E05, and F07 extended the shelf life more than 28 days. In contrast, the fungal strain was apparently unaffected by the dual combination of S. cerevisiae and the isolates D11, A10, C02, and B07 (Figure 6).
As the biodiversity of the LAB inoculum into the sourdough increased (e.g., combinations of C1, C2, C3, C4, and C5), no mycelium was found on the surfaces of the slices of bread up to 28 days (Figure 7).
When loaves of bread were contaminated with the A. niger species, bread made with sourdough obtained with the dual combination S. cerevisiae and single isolates did not have any inhibitory effects on mycelium growth (Figure 8).
Interestingly, also in this case, the increased biodiversity due to a combination of S. cerevisiae and LAB isolates (C1, C2, C3, C4, and C5) showed an increasing inhibiting activity against fungal growth (Figure 9).
The same inhibitory results were observed when slices of bread were contaminated with the F. culmorum species (Figure 10 and Figure 11).
Regarding the species P. chrysogenum, P. expansum, and P. roqueforti, that were, in vitro, unaffected by LAB isolates (halo plate experiment, please see above), the surface of bread contaminated with the aforementioned filamentous fungi was totally covered by mycelium when the bread was made using the dual combination S. cerevisiae and single LAB isolates. Interestingly, as the microbial biodiversity increased (i.e., combination C1, C2, and C3), the ability of fungal growth and development decreased considerably (up to a maximum 14 days of shelf life) (Figure 12, Figure 13, Figure 14, Figure 15, Figure 16 and Figure 17).
Filamentous fungi contamination remains a major threat to the grain market. Food losses due to fungal deterioration are difficult to estimate [35]. However, the contamination of bakery goods normally occurs after cooking and storage, especially when the end product is subjected to cutting and packaging because cutting surfaces are an ideal fungal growth substrate and packaging prevents loss of moisture [36]. Some of the most widely used strategies to inhibit and/or reduce the mold growth and, thus, extend the shelf life, are physical and chemical actions [37]. However, several studies in the field of plant protection aim to reduce and/or replace pesticides with environmentally acceptable biological preparations, highlighting the high potential, inexpensiveness, and renewability features of the use of microbial metabolites such as bactericidal, fungicidal, and nematodocidal agents [38,39]. Here, we provide new insights into the biological control of fungal spoilage. Among the future perspectives, we might envisage further studies on the sensory impact of the tested bacteria [40,41], the possible additional biotechnological exploitation in bread (e.g., improving nutritional and functional properties) [42,43], and the necessary supplementary investigations to elucidate the biological mechanisms behind the antifungal activities [44,45].
4. Conclusions
In this work, we aim to investigate the microbial biodiversity of sourdough collected from the Apulian region (southern Italy), where the tradition of using sourdough instead of industrial starter to make bread is strongly recommended in bakeries, most of which are family-run. Moreover, we have been able to select LAB strains useful for improving shelf life and inhibiting fungal contaminants [21].
In terms of biocontrol potential, our study indicates that, beyond the application of a single antagonistic lactic acid bacteria, the association between different LAB strains represents a great resource in order to inhibit fungal contamination [46,47,48]. Thus, moving toward a green food industry, advantageous interactions within the microbial starter culture could help in preventing the growth of spoilage organisms, resulting in a longer shelf life [13,14,49,50,51].
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/2311-5637/5/4/97/s1.
Author Contributions
Conceptualization, M.P.A., P.R., G.S., and V.C.; methodology, M.P.A., P.R., and V.C.; validation, M.P.A., P.R., G.S., and V.C.; formal analysis, M.P.A. and P.R.; investigation, M.P.A.; resources, G.S. and V.C.; data curation, M.P.A., P.R., and V.C.; writing—original draft preparation, M.P.A.; writing—review and editing, P.R., G.S., and V.C.; supervision, G.S. and V.C.; project administration, V.C.; funding acquisition, V.C.
Funding
Vittorio Capozzi was supported by Fondo per lo sviluppo e la coesione 2007–2013—APQ Ricerca Regione Puglia “Programma regionale a sostegno della specializzazione intelligente e della sostenibilità sociale ed ambientale—FutureInResearch.” Pasquale Russo is the beneficiary of a grant by MIUR in the framework of AIM: Attraction and International Mobility (PON R&I 2014–2020) (practice code D74I18000190001).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Behera, S.S.; Ray, R.C.; Ray, R.C. Sourdough Bread. Available online: https://www.taylorfrancis.com/ (accessed on 26 September 2019).
- Gobbetti, M.; Gänzle, M. Handbook on Sourdough Biotechnology; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; ISBN 978-1-4614-5425-0. [Google Scholar]
- De Vuyst, L.; Vrancken, G.; Ravyts, F.; Rimaux, T.; Weckx, S. Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota. Food Microbiol. 2009, 26, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Komlenić, D.K.; Ugarčić-Hardi, Ž.; Jukić, M.; Planinić, M.; Bucić-Kojić, A.; Strelec, I. Wheat dough rheology and bread quality effected by Lactobacillus brevis preferment, dry sourdough and lactic acid addition. Int. J. Food Sci. Technol. 2010, 45, 1417–1425. [Google Scholar] [CrossRef]
- Hammes, W.P.; Brandt, M.J.; Francis, K.L.; Rosenheim, J.; Seitter, M.F.H.; Vogelmann, S.A. Microbial ecology of cereal fermentations. Trends Food Sci. Technol. 2005, 16, 4–11. [Google Scholar] [CrossRef]
- De Vuyst, L.; Van Kerrebroeck, S.; Harth, H.; Huys, G.; Daniel, H.-M.; Weckx, S. Microbial ecology of sourdough fermentations: Diverse or uniform? Food Microbiol. 2014, 37, 11–29. [Google Scholar] [CrossRef]
- Rizzello, C.G.; Calasso, M.; Campanella, D.; De Angelis, M.; Gobbetti, M. Use of sourdough fermentation and mixture of wheat, chickpea, lentil and bean flours for enhancing the nutritional, texture and sensory characteristics of white bread. Int. J. Food Microbiol. 2014, 180, 78–87. [Google Scholar] [CrossRef]
- Gobbetti, M. The sourdough microflora: Interactions of lactic acid bacteria and yeasts. Trends Food Sci. Technol. 1998, 9, 267–274. [Google Scholar] [CrossRef]
- Mariotti, M.; Garofalo, C.; Aquilanti, L.; Osimani, A.; Fongaro, L.; Tavoletti, S.; Hager, A.-S.; Clementi, F. Barley flour exploitation in sourdough bread-making: A technological, nutritional and sensory evaluation. LWT Food Sci. Technol. 2014, 59, 973–980. [Google Scholar] [CrossRef]
- Corsetti, A.; De Angelis, M.; Dellaglio, F.; Paparella, A.; Fox, P.F.; Settanni, L.; Gobbetti, M. Characterization of sourdough lactic acid bacteria based on genotypic and cell-wall protein analyses. J. Appl. Microbiol. 2003, 94, 641–654. [Google Scholar] [CrossRef]
- Torrieri, E.; Pepe, O.; Ventorino, V.; Masi, P.; Cavella, S. Effect of sourdough at different concentrations on quality and shelf life of bread. LWT Food Sci. Technol. 2014, 56, 508–516. [Google Scholar] [CrossRef]
- Capozzi, V.; Russo, P.; Ladero, V.; Fernandez, M.; Fiocco, D.; Alvarez, M.A.; Grieco, F.; Spano, G. Biogenic Amines Degradation by Lactobacillus plantarum: Toward a Potential Application in Wine. Front. Microbiol. 2012, 3, 122. [Google Scholar] [CrossRef] [PubMed]
- Smid, E.J.; Lacroix, C. Microbe-microbe interactions in mixed culture food fermentations. Curr. Opin. Biotechnol. 2013, 24, 148–154. [Google Scholar] [CrossRef]
- Russo, P.; Fares, C.; Longo, A.; Spano, G.; Capozzi, V. Lactobacillus plantarum with Broad Antifungal Activity as a Protective Starter Culture for Bread Production. Foods 2017, 6, 110. [Google Scholar] [CrossRef] [PubMed]
- Palla, M.; Cristani, C.; Giovannetti, M.; Agnolucci, M. Identification and characterization of lactic acid bacteria and yeasts of PDO Tuscan bread sourdough by culture dependent and independent methods. Int. J. Food Microbiol. 2017, 250, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Ghariani, M.; Hamdi, M.; Beneduce, L.; Capozzi, V.; Massa, S. Identification of acetic acid bacteria isolated from Tunisian palm sap. Afr. J. Microbiol. Res. 2017, 11, 596–602. [Google Scholar]
- Wilmotte, A.; Van der Auwera, G.; De Wachter, R. Structure of the 16 S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (‘Mastigocladus laminosus HTF’) strain PCC7518, and phylogenetic analysis. FEBS Lett. 1993, 317, 96–100. [Google Scholar] [CrossRef]
- Josepa, S.; Guillamon, J.M.; Cano, J. PCR differentiation of Saccharomyces cerevisiae from Saccharomyces bayanus/Saccharomyces pastorianus using specific primers. FEMS Microbiol. Lett. 2000, 193, 255–259. [Google Scholar] [CrossRef]
- Garofalo, C.; Tristezza, M.; Grieco, F.; Spano, G.; Capozzi, V. From grape berries to wine: Population dynamics of cultivable yeasts associated to “Nero di Troia” autochthonous grape cultivar. World J. Microbiol. Biotechnol. 2016, 32, 59. [Google Scholar] [CrossRef]
- Russo, P.; Arena, M.P.; Fiocco, D.; Capozzi, V.; Drider, D.; Spano, G. Lactobacillus plantarum with broad antifungal activity: A promising approach to increase safety and shelf-life of cereal-based products. Int. J. Food Microbiol. 2017, 247, 48–54. [Google Scholar] [CrossRef]
- Plessas, S.; Fisher, A.; Koureta, K.; Psarianos, C.; Nigam, P.; Koutinas, A.A. Application of Kluyveromyces marxianus, Lactobacillus delbrueckii ssp. bulgaricus and L. helveticus for sourdough bread making. Food Chem. 2008, 106, 985–990. [Google Scholar] [CrossRef]
- Rizzello, C.G.; Cassone, A.; Coda, R.; Gobbetti, M. Antifungal activity of sourdough fermented wheat germ used as an ingredient for bread making. Food Chem. 2011, 127, 952–959. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Tu, J.; Sadiq, F.A.; Zhang, W.; Wang, W. Prevalence, Genetic Diversity, and Technological Functions of the Lactobacillus sanfranciscensis in Sourdough: A Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1209–1226. [Google Scholar] [CrossRef]
- Corsetti, A.; Lavermicocca, P.; Morea, M.; Baruzzi, F.; Tosti, N.; Gobbetti, M. Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of Southern Italy. Int. J. Food Microbiol. 2001, 64, 95–104. [Google Scholar] [CrossRef]
- Gullo, M.; Romano, A.D.; Pulvirenti, A.; Giudici, P. Candida humilis—Dominant species in sourdoughs for the production of durum wheat bran flour bread. Int. J. Food Microbiol. 2003, 80, 55–59. [Google Scholar] [CrossRef]
- Succi, M.; Reale, A.; Andrighetto, C.; Lombardi, A.; Sorrentino, E.; Coppola, R. Presence of yeasts in southern Italian sourdoughs from Triticum aestivum flour. FEMS Microbiol. Lett. 2003, 225, 143–148. [Google Scholar] [CrossRef]
- Snyder, A.B.; Worobo, R.W. Fungal Spoilage in Food Processing. J. Food Prot. 2018, 81, 1035–1040. [Google Scholar] [CrossRef]
- Oliveira, P.M.; Mauch, A.; Jacob, F.; Waters, D.M.; Arendt, E.K. Fundamental study on the influence of Fusarium infection on quality and ultrastructure of barley malt. Int. J. Food Microbiol. 2012, 156, 32–43. [Google Scholar] [CrossRef]
- Pawlowska, A.M.; Zannini, E.; Coffey, A.; Arendt, E.K. “Green preservatives”: Combating fungi in the food and feed industry by applying antifungal lactic acid bacteria. Adv. Food Nutr. Res. 2012, 66, 217–238. [Google Scholar]
- Ryan, L.A.M.; Zannini, E.; Dal Bello, F.; Pawlowska, A.; Koehler, P.; Arendt, E.K. Lactobacillus amylovorus DSM 19280 as a novel food-grade antifungal agent for bakery products. Int. J. Food Microbiol. 2011, 146, 276–283. [Google Scholar] [CrossRef]
- Lavermicocca, P.; Valerio, F.; Visconti, A. Antifungal Activity of Phenyllactic Acid against Molds Isolated from Bakery Products. Appl. Environ. Microbiol. 2003, 69, 634–640. [Google Scholar] [CrossRef]
- Gerez, C.L.; Torino, M.I.; Obregozo, M.D.; Font de Valdez, G. A ready-to-use antifungal starter culture improves the shelf life of packaged bread. J. Food Prot. 2010, 73, 758–762. [Google Scholar] [CrossRef] [PubMed]
- Gerez, C.L.; Torres, M.J.; Font de Valdez, G.; Rollán, G. Control of spoilage fungi by lactic acid bacteria. Biol. Control 2013, 64, 231–237. [Google Scholar] [CrossRef]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 2009; ISBN 978-0-387-92206-5. [Google Scholar]
- Bosmans, G.M.; Lagrain, B.; Fierens, E.; Delcour, J.A. The impact of baking time and bread storage temperature on bread crumb properties. Food Chem. 2013, 141, 3301–3308. [Google Scholar] [CrossRef] [PubMed]
- Axel, C.; Zannini, E.; Arendt, E.K. Mold spoilage of bread and its biopreservation: A review of current strategies for bread shelf life extension. Crit. Rev. Food Sci. Nutr. 2017, 57, 3528–3542. [Google Scholar] [CrossRef] [PubMed]
- Kamzolova, S.V.; Vinokurova, N.G.; Shemshura, O.N.; Bekmakhanova, N.E.; Lunina, J.N.; Samoilenko, V.A.; Morgunov, I.G. The production of succinic acid by yeast Yarrowia lipolytica through a two-step process. Appl. Microbiol. Biotechnol. 2014, 98, 7959–7969. [Google Scholar] [CrossRef] [PubMed]
- Morgunov, I.G.; Kamzolova, S.V.; Dedyukhina, E.G.; Chistyakova, T.I.; Lunina, J.N.; Mironov, A.A.; Stepanova, N.N.; Shemshura, O.N.; Vainshtein, M.B. Application of organic acids for plant protection against phytopathogens. Appl. Microbiol. Biotechnol. 2017, 101, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, V.; Makhoul, S.; Aprea, E.; Romano, A.; Cappellin, L.; Sanchez Jimena, A.; Spano, G.; Gasperi, F.; Scampicchio, M.; Biasioli, F. PTR-MS Characterization of VOCs Associated with Commercial Aromatic Bakery Yeasts of Wine and Beer Origin. Molecules 2016, 21, 483. [Google Scholar] [CrossRef]
- Capozzi, V.; Yener, S.; Khomenko, I.; Farneti, B.; Cappellin, L.; Gasperi, F.; Scampicchio, M.; Biasioli, F. PTR-ToF-MS Coupled with an Automated Sampling System and Tailored Data Analysis for Food Studies: Bioprocess Monitoring, Screening and Nose-space Analysis. JoVE J. Vis. Exp. 2017, e54075. [Google Scholar] [CrossRef]
- Capozzi, V.; Menga, V.; Digesù, A.M.; De Vita, P.; van Sinderen, D.; Cattivelli, L.; Fares, C.; Spano, G. Biotechnological Production of Vitamin B2-Enriched Bread and Pasta. J. Agric. Food Chem. 2011, 59, 8013–8020. [Google Scholar] [CrossRef]
- Pérez-Ramos, A.; Mohedano, M.L.; López, P.; Spano, G.; Fiocco, D.; Russo, P.; Capozzi, V. In Situ β-Glucan Fortification of Cereal-Based Matrices by Pediococcus parvulus 2.6: Technological Aspects and Prebiotic Potential. Int. J. Mol. Sci. 2017, 18, 1588. [Google Scholar] [CrossRef]
- Arena, M.P.; Capozzi, V.; Spano, G.; Fiocco, D. The potential of lactic acid bacteria to colonize biotic and abiotic surfaces and the investigation of their interactions and mechanisms. Appl. Microbiol. Biotechnol. 2017, 101, 2641–2657. [Google Scholar] [CrossRef] [PubMed]
- Arena, M.P.; Capozzi, V.; Russo, P.; Drider, D.; Spano, G.; Fiocco, D. Immunobiosis and probiosis: Antimicrobial activity of lactic acid bacteria with a focus on their antiviral and antifungal properties. Appl. Microbiol. Biotechnol. 2018, 102, 9949–9958. [Google Scholar] [CrossRef] [PubMed]
- Cheirsilp, B.; Shoji, H.; Shimizu, H.; Shioya, S. Interactions between Lactobacillus kefiranofaciens and Saccharomyces cerevisiae in mixed culture for kefiran production. J. Biosci. Bioeng. 2003, 96, 279–284. [Google Scholar] [CrossRef]
- Franco, W.; Pérez-Díaz, I.M. Microbial interactions associated with secondary cucumber fermentation. J. Appl. Microbiol. 2013, 114, 161–172. [Google Scholar] [CrossRef]
- Sieuwerts, S.; de Bok, F.A.M.; Hugenholtz, J.; van Hylckama Vlieg, J.E.T. Unraveling microbial interactions in food fermentations: From classical to genomics approaches. Appl. Environ. Microbiol. 2008, 74, 4997–5007. [Google Scholar] [CrossRef]
- Alexandre, H.; Costello, P.J.; Remize, F.; Guzzo, J.; Guilloux-Benatier, M. Saccharomyces cerevisiae-Oenococcus oeni interactions in wine: Current knowledge and perspectives. Int. J. Food Microbiol. 2004, 93, 141–154. [Google Scholar] [CrossRef]
- Sieuwerts, S.; Molenaar, D.; van Hijum, S.A.F.T.; Beerthuyzen, M.; Stevens, M.J.A.; Janssen, P.W.M.; Ingham, C.J.; de Bok, F.A.M.; de Vos, W.M.; van Hylckama Vlieg, J.E.T. Mixed-culture transcriptome analysis reveals the molecular basis of mixed-culture growth in Streptococcus thermophilus and Lactobacillus bulgaricus. Appl. Environ. Microbiol. 2010, 76, 7775–7784. [Google Scholar] [CrossRef]
- Viljoen, B.C. The interaction between yeasts and bacteria in dairy environments. Int. J. Food Microbiol. 2001, 69, 37–44. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).