Yeast Life Span and its Impact on Food Fermentations
Abstract
1. Introduction
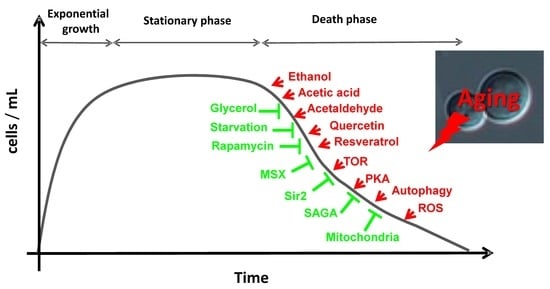
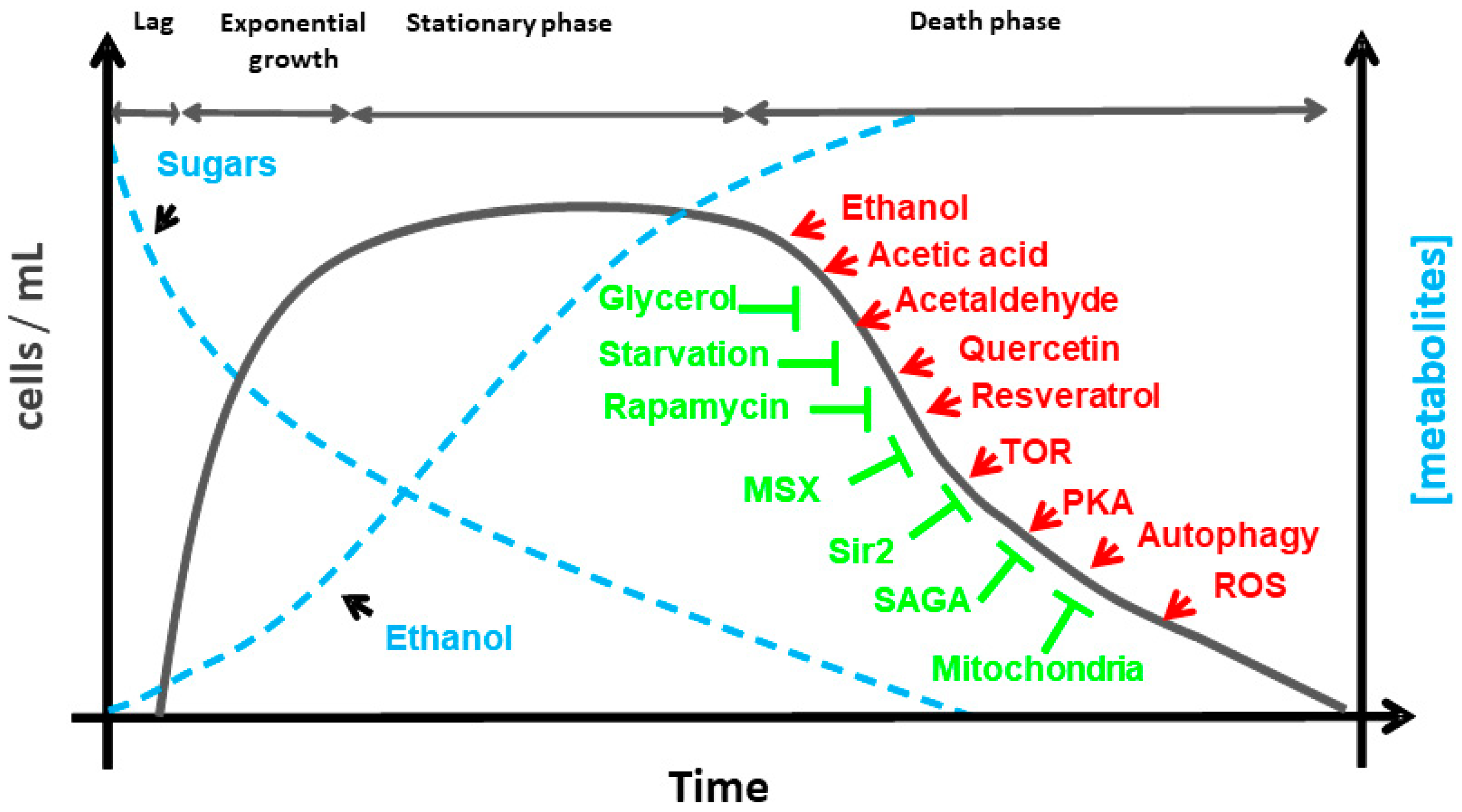
2. Aging in Saccharomyces cerevisiae
3. The Genes, Pathways and Molecules that Regulate Life Span
4. Chronological Life Span in Wine Yeasts
5. Replicative Life Span in Brewing Yeasts
6. Molecular Regulation of the Stationary Phase during Sake Fermentation
7. Aging in other Yeasts
Author Contributions
Funding
Conflicts of Interest
References
- Longo, V.D.; Shadel, G.S.; Kaeberlein, M.; Kennedy, B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012, 16, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Shima, J.; Takagi, H. Stress-tolerance of baker’s-yeast (Saccharomyces cerevisiae) cells: Stress-protective molecules and genes involved in stress tolerance. Biotechnol. Appl. Biochem. 2009, 53 Pt 3, 155–164. [Google Scholar] [CrossRef]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending healthy life span--from yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef]
- Kaeberlein, M. Lessons on longevity from budding yeast. Nature 2010, 464, 513–519. [Google Scholar] [CrossRef]
- Fabrizio, P.; Longo, V.D. The chronological life span of Saccharomyces cerevisiae. Aging Cell 2003, 2, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, K.; Sinclair, D.; Gordon, J.I.; Guarente, L. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1999, 96, 9100–9105. [Google Scholar] [CrossRef]
- Mirisola, M.G.; Braun, R.J.; Petranovic, D. Approaches to study yeast cell aging and death. FEMS Yeast Res. 2014, 14, 109–118. [Google Scholar] [CrossRef]
- Chen, C.; Contreras, R. Identifying genes that extend life span using a high-throughput screening system. Methods Mol. Biol. 2007, 371, 237–248. [Google Scholar]
- Powers, R.W., 3rd; Kaeberlein, M.; Caldwell, S.D.; Kennedy, B.K.; Fields, S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006, 20, 174–184. [Google Scholar] [CrossRef]
- Harman, D. The Free Radical Theory of Aging: Effect of Age on Serum Copper Levels. J. Gerontol. 1965, 20, 151–153. [Google Scholar] [CrossRef]
- Aguilaniu, H.; Gustafsson, L.; Rigoulet, M.; Nystrom, T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 2003, 299, 1751–1753. [Google Scholar] [CrossRef]
- Zhou, C.; Slaughter, B.D.; Unruh, J.R.; Guo, F.; Yu, Z.; Mickey, K.; Narkar, A.; Ross, R.T.; McClain, M.; Li, R. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell 2014, 159, 530–542. [Google Scholar] [CrossRef]
- Sinclair, D.A.; Guarente, L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell 1997, 91, 1033–1042. [Google Scholar] [CrossRef]
- Fabrizio, P.; Gattazzo, C.; Battistella, L.; Wei, M.; Cheng, C.; McGrew, K.; Longo, V.D. Sir2 blocks extreme life-span extension. Cell 2005, 123, 655–667. [Google Scholar] [CrossRef]
- Burtner, C.R.; Murakami, C.J.; Kennedy, B.K.; Kaeberlein, M. A molecular mechanism of chronological aging in yeast. Cell Cycle 2009, 8, 1256–1270. [Google Scholar] [CrossRef]
- Wei, M.; Fabrizio, P.; Madia, F.; Hu, J.; Ge, H.; Li, L.M.; Longo, V.D. Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet. 2009, 5, e1000467. [Google Scholar] [CrossRef]
- Mohammad, K.; Dakik, P.; Medkour, Y.; McAuley, M.; Mitrofanova, D.; Titorenko, V.I. Some Metabolites Act as Second Messengers in Yeast Chronological Aging. Int. J. Mol. Sci. 2018, 19, 860. [Google Scholar] [CrossRef]
- Kapahi, P.; Kaeberlein, M.; Hansen, M. Dietary restriction and lifespan: Lessons from invertebrate models. Ageing Res. Rev. 2017, 39, 3–14. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Kennedy, B.K. Large-scale identification in yeast of conserved ageing genes. Mech. Ageing Dev. 2005, 126, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef]
- Fabrizio, P.; Pozza, F.; Pletcher, S.D.; Gendron, C.M.; Longo, V.D. Regulation of longevity and stress resistance by Sch9 in yeast. Science 2001, 292, 288–290. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, M.; Powers, R.W., 3rd; Steffen, K.K.; Westman, E.A.; Hu, D.; Dang, N.; Kerr, E.O.; Kirkland, K.T.; Fields, S.; Kennedy, B.K. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 2005, 310, 1193–1196. [Google Scholar] [CrossRef]
- Steffen, K.K.; MacKay, V.L.; Kerr, E.O.; Tsuchiya, M.; Hu, D.; Fox, L.A.; Dang, N.; Johnston, E.D.; Oakes, J.A.; Tchao, B.N.; et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 2008, 133, 292–302. [Google Scholar] [CrossRef]
- Wei, M.; Fabrizio, P.; Hu, J.; Ge, H.; Cheng, C.; Li, L.; Longo, V.D. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008, 4, e13. [Google Scholar] [CrossRef]
- Fabrizio, P.; Liou, L.L.; Moy, V.N.; Diaspro, A.; Valentine, J.S.; Gralla, E.B.; Longo, V.D. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics 2003, 163, 35–46. [Google Scholar] [PubMed]
- Pan, Y.; Schroeder, E.A.; Ocampo, A.; Barrientos, A.; Shadel, G.S. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 2011, 13, 668–678. [Google Scholar] [CrossRef]
- Lin, S.J.; Kaeberlein, M.; Andalis, A.A.; Sturtz, L.A.; Defossez, P.A.; Culotta, V.C.; Fink, G.R.; Guarente, L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 2002, 418, 344–348. [Google Scholar] [CrossRef]
- Molin, M.; Yang, J.; Hanzen, S.; Toledano, M.B.; Labarre, J.; Nystrom, T. Life span extension and H(2)O(2) resistance elicited by caloric restriction require the peroxiredoxin Tsa1 in Saccharomyces cerevisiae. Mol. Cell 2011, 43, 823–833. [Google Scholar] [CrossRef]
- Kruegel, U.; Robison, B.; Dange, T.; Kahlert, G.; Delaney, J.R.; Kotireddy, S.; Tsuchiya, M.; Tsuchiyama, S.; Murakami, C.J.; Schleit, J.; et al. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet. 2011, 7, e1002253. [Google Scholar] [CrossRef]
- Alvers, A.L.; Fishwick, L.K.; Wood, M.S.; Hu, D.; Chung, H.S.; Dunn, W.A., Jr.; Aris, J.P. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell 2009, 8, 353–369. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, M.; McVey, M.; Guarente, L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999, 13, 2570–2580. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.; Armstrong, C.M.; Kaeberlein, M.; Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000, 403, 795–800. [Google Scholar] [CrossRef]
- Erjavec, N.; Larsson, L.; Grantham, J.; Nystrom, T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007, 21, 2410–2421. [Google Scholar] [CrossRef]
- Crespo, J.L.; Powers, T.; Fowler, B.; Hall, M.N. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 2002, 99, 6784–6789. [Google Scholar] [CrossRef] [PubMed]
- Lushchak, O.; Strilbytska, O.; Piskovatska, V.; Storey, K.B.; Koliada, A.; Vaiserman, A. The role of the TOR pathway in mediating the link between nutrition and longevity. Mech. Ageing Dev. 2017, 164, 127–138. [Google Scholar] [CrossRef]
- Madeo, F.; Carmona-Gutierrez, D.; Hofer, S.J.; Kroemer, G. Caloric Restriction Mimetics against Age-Associated Disease: Targets, Mechanisms, and Therapeutic Potential. Cell Metab. 2019, 29, 592–610. [Google Scholar] [CrossRef] [PubMed]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.L.; et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 2003, 425, 191–196. [Google Scholar] [CrossRef]
- Goldberg, A.A.; Richard, V.R.; Kyryakov, P.; Bourque, S.D.; Beach, A.; Burstein, M.T.; Glebov, A.; Koupaki, O.; Boukh-Viner, T.; Gregg, C.; et al. Chemical genetic screen identifies lithocholic acid as an anti-aging compound that extends yeast chronological life span in a TOR-independent manner, by modulating housekeeping longevity assurance processes. Aging (Albany NY) 2010, 2, 393–414. [Google Scholar] [CrossRef] [PubMed]
- Carrascosa Santiago, A.V.; Muñoz, R.; González Garcia, R.N. Molecular Wine Microbiology, 1st ed.; Academic Press: Amsterdam, The Netherlands; Boston, MA, USA; p. vii. 363p.
- Matallana, E.; Aranda, A. Biotechnological impact of stress response on wine yeast. Lett. Appl. Microbiol. 2017, 64, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Ribéreau-Gayon, P.; Dubourdieu, D.; Donèche, B. Handbook of Enology, 2nd ed.; John Wiley: Chichester, UK; Hoboken, NJ, USA, 2006. [Google Scholar]
- Boulton, R.B. Principles and Practices of Winemaking; Chapman & Hall: New York, NY, USA, 1996. [Google Scholar]
- Orozco, H.; Matallana, E.; Aranda, A. Oxidative stress tolerance, adenylate cyclase, and autophagy are key players in the chronological life span of Saccharomyces cerevisiae during winemaking. Appl. Environ. Microbiol. 2012, 78, 2748–2757. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Lu, M. Natural variation in replicative and chronological life spans of Saccharomyces cerevisiae. Exp. Gerontol. 2006, 41, 448–456. [Google Scholar] [CrossRef]
- Orozco, H.; Matallana, E.; Aranda, A. Two-carbon metabolites, polyphenols and vitamins influence yeast chronological life span in winemaking conditions. Microb. Cell Fact. 2012, 11, 104. [Google Scholar] [CrossRef]
- Orozco, H.; Matallana, E.; Aranda, A. Wine yeast sirtuins and Gcn5p control aging and metabolism in a natural growth medium. Mech. Ageing Dev. 2012, 133, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, B.; Picazo, C.; Orozco, H.; Matallana, E.; Aranda, A. Herbicide glufosinate inhibits yeast growth and extends longevity during wine fermentation. Sci. Rep. 2017, 7, 12414. [Google Scholar] [CrossRef] [PubMed]
- Picazo, C.; Orozco, H.; Matallana, E.; Aranda, A. Interplay among Gcn5, Sch9 and mitochondria during chronological aging of wine yeast is dependent on growth conditions. PLoS ONE 2015, 10, e0117267. [Google Scholar] [CrossRef]
- Tesniere, C.; Delobel, P.; Pradal, M.; Blondin, B. Impact of nutrient imbalance on wine alcoholic fermentations: Nitrogen excess enhances yeast cell death in lipid-limited must. PLoS ONE 2013, 8, e61645. [Google Scholar] [CrossRef] [PubMed]
- Duc, C.; Pradal, M.; Sanchez, I.; Noble, J.; Tesniere, C.; Blondin, B. A set of nutrient limitations trigger yeast cell death in a nitrogen-dependent manner during wine alcoholic fermentation. PLoS ONE 2017, 12, e0184838. [Google Scholar] [CrossRef]
- Picazo, C.; McDonagh, B.; Peinado, J.; Barcena, J.A.; Matallana, E.; Aranda, A. Saccharomyces cerevisiae Cytosolic Thioredoxins Control Glycolysis, Lipid Metabolism, and Protein Biosynthesis under Wine-Making Conditions. Appl. Environ. Microbiol. 2019. [Google Scholar] [CrossRef]
- Picazo, C.; Gamero-Sandemetrio, E.; Orozco, H.; Albertin, W.; Marullo, P.; Matallana, E.; Aranda, A. Mitochondria inheritance is a key factor for tolerance to dehydration in wine yeast production. Lett. Appl. Microbiol. 2015, 60, 217–222. [Google Scholar] [CrossRef]
- Orozco, H.; Matallana, E.; Aranda, A. Genetic manipulation of longevity-related genes as a tool to regulate yeast life span and metabolite production during winemaking. Microb. Cell Fact. 2013, 12, 1. [Google Scholar] [CrossRef]
- Orozco, H.; Sepulveda, A.; Picazo, C.; Matallana, E.; Aranda, A. RNA binding protein Pub1p regulates glycerol production and stress tolerance by controlling Gpd1p activity during winemaking. Appl. Microbiol. Biotechnol. 2016, 100, 5017–5027. [Google Scholar] [CrossRef]
- Jackson, R.S. Wine Science: Principles, Practice, Perception, 2nd ed.; Academic Press: San Diego, MA, USA, 2000; 648p. [Google Scholar]
- Cebollero, E.; Gonzalez, R. Autophagy: From basic research to its application in food biotechnology. Biotechnol. Adv. 2007, 25, 396–409. [Google Scholar] [CrossRef]
- Cebollero, E.; Gonzalez, R. Induction of autophagy by second-fermentation yeasts during elaboration of sparkling wines. Appl. Environ. Microbiol. 2006, 72, 4121–4127. [Google Scholar] [CrossRef]
- Preiss, R.; Tyrawa, C.; van der Merwe, G. Autophagy gene overexpression in Saccharomyces cerevisiae perturbs subcellular organellar function and accumulates ROS to accelerate cell death with relevance to sparkling wine production. Appl. Microbiol. Biotechnol. 2018, 102, 8447–8464. [Google Scholar] [CrossRef]
- Tabera, L.; Munoz, R.; Gonzalez, R. Deletion of BCY1 from the Saccharomyces cerevisiae genome is semidominant and induces autolytic phenotypes suitable for improvement of sparkling wines. Appl. Environ. Microbiol. 2006, 72, 2351–2358. [Google Scholar] [CrossRef]
- Gibson, B.R.; Lawrence, S.J.; Leclaire, J.P.; Powell, C.D.; Smart, K.A. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol. Rev. 2007, 31, 535–569. [Google Scholar] [PubMed]
- Xu, W.; Wang, J.; Li, Q. Microarray studies on lager brewer’s yeasts reveal cell status in the process of autolysis. FEMS Yeast Res 2014, 14, 714–728. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, W.; Wang, J.; Li, Q. Comparative proteome and transcriptome analysis of lager brewer’s yeast in the autolysis process. FEMS Yeast Res. 2014, 14, 1273–1285. [Google Scholar] [CrossRef] [PubMed]
- Powell, C.D.; Van Zandycke, S.M.; Quain, D.E.; Smart, K.A. Replicative ageing and senescence in Saccharomyces cerevisiae and the impact on brewing fermentations. Microbiology 2000, 146 Pt 5, 1023–1034. [Google Scholar] [CrossRef]
- Powell, C.D.; Quain, D.E.; Smart, K.A. The impact of brewing yeast cell age on fermentation performance, attenuation and flocculation. FEMS Yeast Res. 2003, 3, 149–157. [Google Scholar] [CrossRef]
- Maskell, D.L.; Kennedy, A.I.; Hodgson, J.A.; Smart, K.A. Chronological and replicative lifespan of polyploid Saccharomyces cerevisiae (syn. S. pastorianus). FEMS Yeast Res. 2003, 3, 201–209. [Google Scholar] [CrossRef]
- Kuřec, M.; Baszczyňski, M.; Lehnert, R.; Mota, A.; Teixeira, J.A.; Brányik, T. Flow Cytometry for Age Assessment of a Yeast Population and its Application in Beer Fermentations. J. Inst. Brew. 2009, 115, 253–258. [Google Scholar] [CrossRef]
- Powell, C.D.; Diacetis, A.N. Long Term Serial Repitching and the Genetic and Phenotypic Stability of Brewer’s Yeast. J. Inst. Brew. 2007, 113, 67–74. [Google Scholar] [CrossRef]
- Buhligen, F.; Lindner, P.; Fetzer, I.; Stahl, F.; Scheper, T.; Harms, H.; Muller, S. Analysis of aging in lager brewing yeast during serial repitching. J. Biotechnol. 2014, 187, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.R.; Prescott, K.A.; Smart, K.A. Petite mutation in aged and oxidatively stressed ale and lager brewing yeast. Lett. Appl. Microbiol. 2008, 46, 636–642. [Google Scholar] [CrossRef]
- Watanabe, D.; Wu, H.; Noguchi, C.; Zhou, Y.; Akao, T.; Shimoi, H. Enhancement of the initial rate of ethanol fermentation due to dysfunction of yeast stress response components Msn2p and/or Msn4p. Appl. Environ. Microbiol. 2011, 77, 934–941. [Google Scholar] [CrossRef] [PubMed]
- Urbanczyk, H.; Noguchi, C.; Wu, H.; Watanabe, D.; Akao, T.; Takagi, H.; Shimoi, H. Sake yeast strains have difficulty in entering a quiescent state after cell growth cessation. J. Biosci. Bioeng. 2011, 112, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, D.; Araki, Y.; Zhou, Y.; Maeya, N.; Akao, T.; Shimoi, H. A loss-of-function mutation in the PAS kinase Rim15p is related to defective quiescence entry and high fermentation rates of Saccharomyces cerevisiae sake yeast strains. Appl. Environ. Microbiol. 2012, 78, 4008–4016. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, D.; Kaneko, A.; Sugimoto, Y.; Ohnuki, S.; Takagi, H.; Ohya, Y. Promoter engineering of the Saccharomyces cerevisiae RIM15 gene for improvement of alcoholic fermentation rates under stress conditions. J. Biosci. Bioeng. 2017, 123, 183–189. [Google Scholar] [CrossRef]
- Oomuro, M.; Kato, T.; Zhou, Y.; Watanabe, D.; Motoyama, Y.; Yamagishi, H.; Akao, T.; Aizawa, M. Defective quiescence entry promotes the fermentation performance of bottom-fermenting brewer’s yeast. J. Biosci. Bioeng. 2016, 122, 577–582. [Google Scholar] [CrossRef]
- Kessi-Perez, E.I.; Araos, S.; Garcia, V.; Salinas, F.; Abarca, V.; Larrondo, L.F.; Martinez, C.; Cubillos, F.A. RIM15 antagonistic pleiotropy is responsible for differences in fermentation and stress response kinetics in budding yeast. FEMS Yeast Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Roux, A.E.; Chartrand, P.; Ferbeyre, G.; Rokeach, L.A. Fission yeast and other yeasts as emergent models to unravel cellular aging in eukaryotes. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2010, 65, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, G.A.; Tahara, E.B.; Gombert, A.K.; Barros, M.H.; Kowaltowski, A.J. Increased aerobic metabolism is essential for the beneficial effects of caloric restriction on yeast life span. J. Bioenerg. Biomembr. 2008, 40, 381–388. [Google Scholar] [CrossRef]
- Saini, P.; Beniwal, A.; Vij, S. Comparative Analysis of Oxidative Stress During Aging of Kluyveromyces marxianus in Synthetic and Whey Media. Appl. Biochem. Biotechnol. 2017, 183, 348–361. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aranda, A.; Orozco, H.; Picazo, C.; Matallana, E. Yeast Life Span and its Impact on Food Fermentations. Fermentation 2019, 5, 37. https://doi.org/10.3390/fermentation5020037
Aranda A, Orozco H, Picazo C, Matallana E. Yeast Life Span and its Impact on Food Fermentations. Fermentation. 2019; 5(2):37. https://doi.org/10.3390/fermentation5020037
Chicago/Turabian StyleAranda, Agustín, Helena Orozco, Cecilia Picazo, and Emilia Matallana. 2019. "Yeast Life Span and its Impact on Food Fermentations" Fermentation 5, no. 2: 37. https://doi.org/10.3390/fermentation5020037
APA StyleAranda, A., Orozco, H., Picazo, C., & Matallana, E. (2019). Yeast Life Span and its Impact on Food Fermentations. Fermentation, 5(2), 37. https://doi.org/10.3390/fermentation5020037