Recombinant Diploid Saccharomyces cerevisiae Strain Development for Rapid Glucose and Xylose Co-Fermentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Strain Construction and Adaptive Evolution
2.3. Enzyme Activity Assay
2.4. Glucose and Xylose Fermentation by the Recombinant Strains
2.5. Analytical Methods
2.6. Quantitative Reserve Transcription Polymerase Chain Reaction (RT-PCR)
2.7. Biomass Hydrolysate Fermentation Using Strain S. cerevisiae STXQ
3. Results
3.1. Expression of XIs with Various Combinations
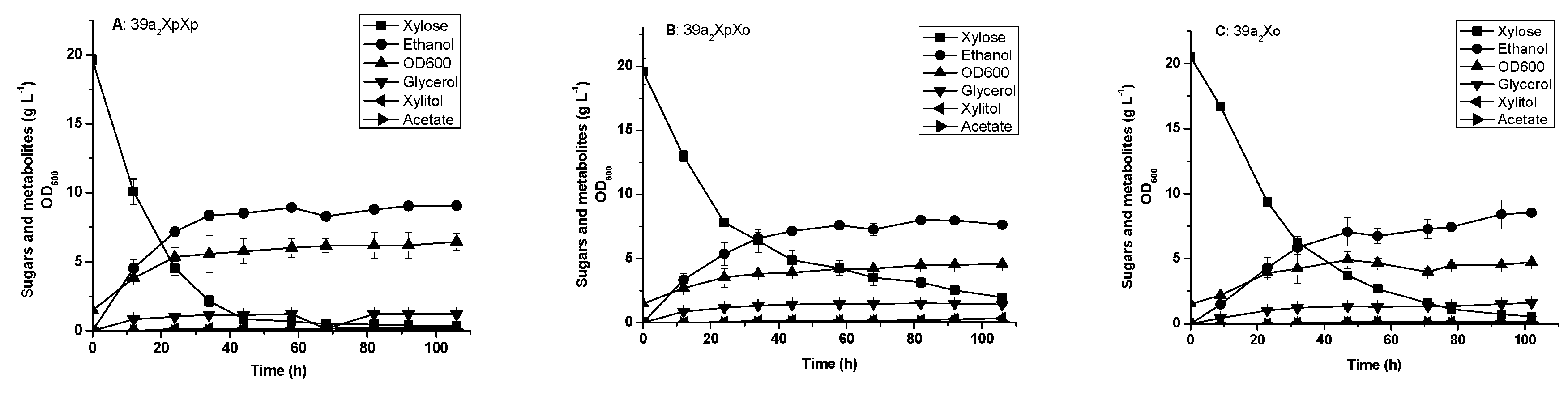
3.2. Glucose and Xylose Fermentation by the Engineered 39a2 Strains Harboring Episomal XI Genes
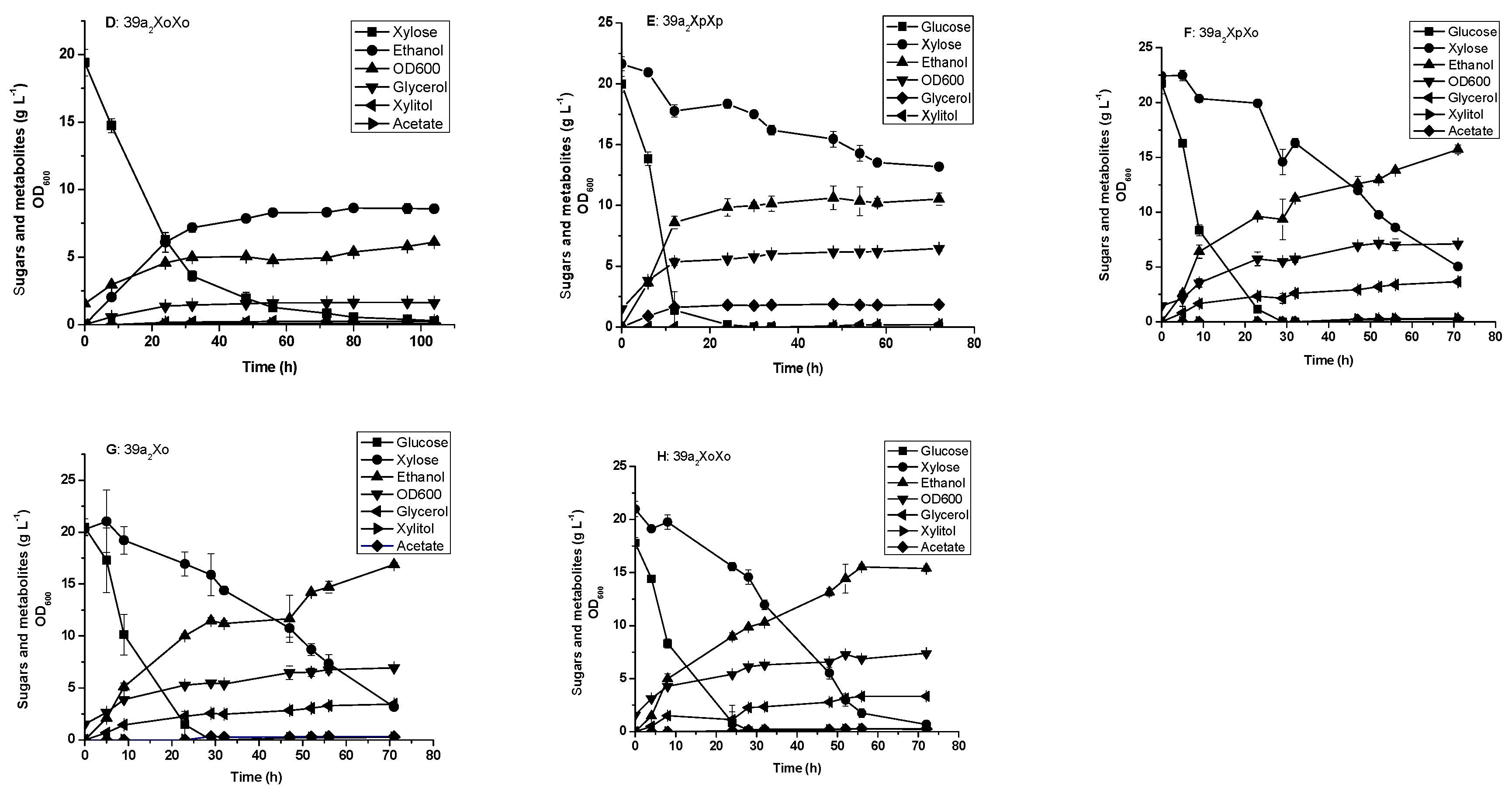
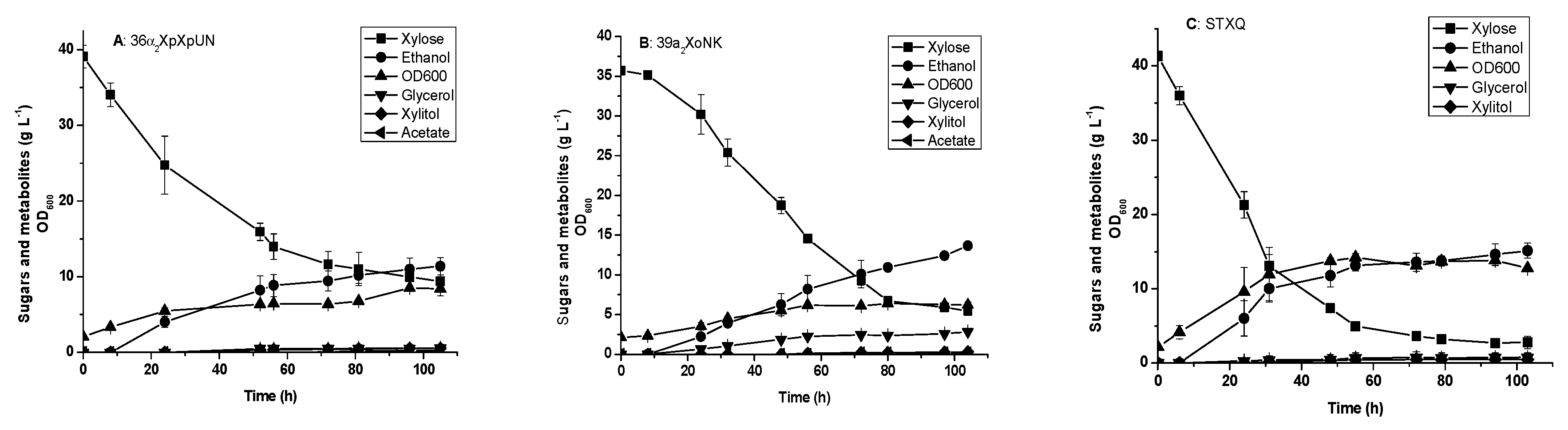
3.3. Glucose and xylose Fermentation by the Engineered Yeast Strains Harboring Integrated XI Genes
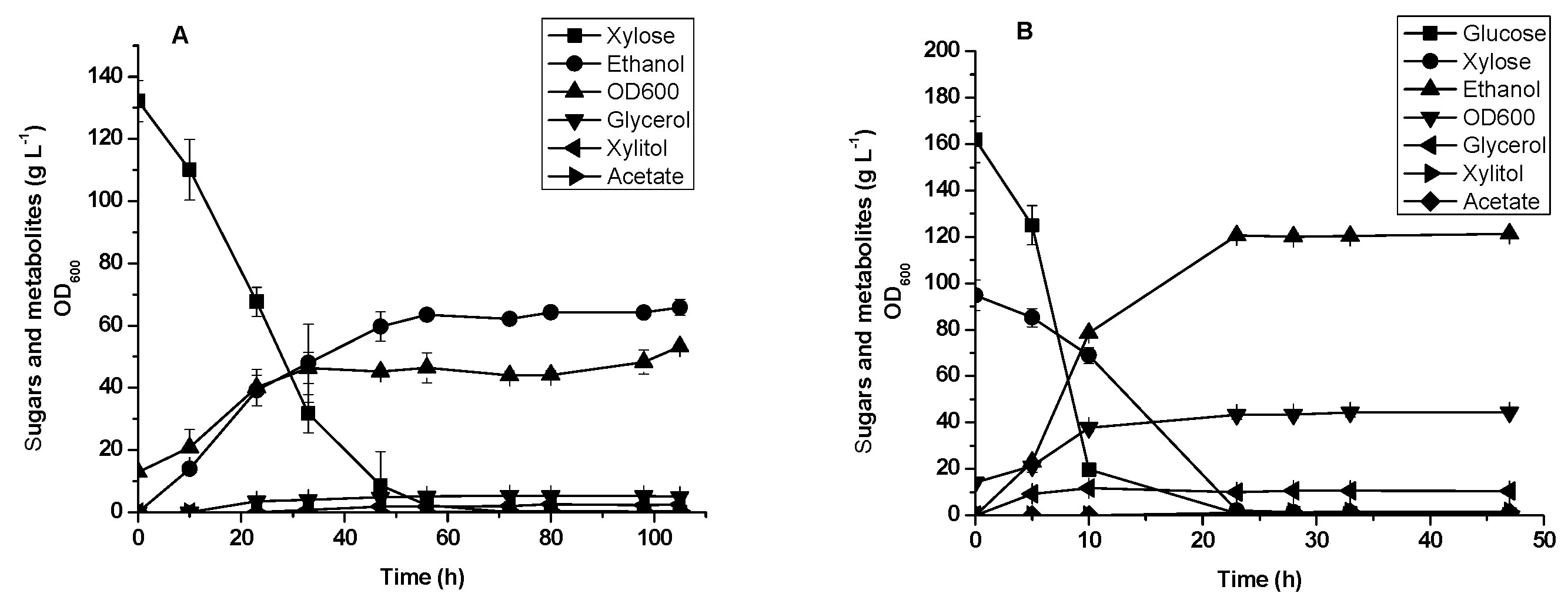
3.4. Glucose and xylose Fermentation by Diploid Recombinant Strain STXQ
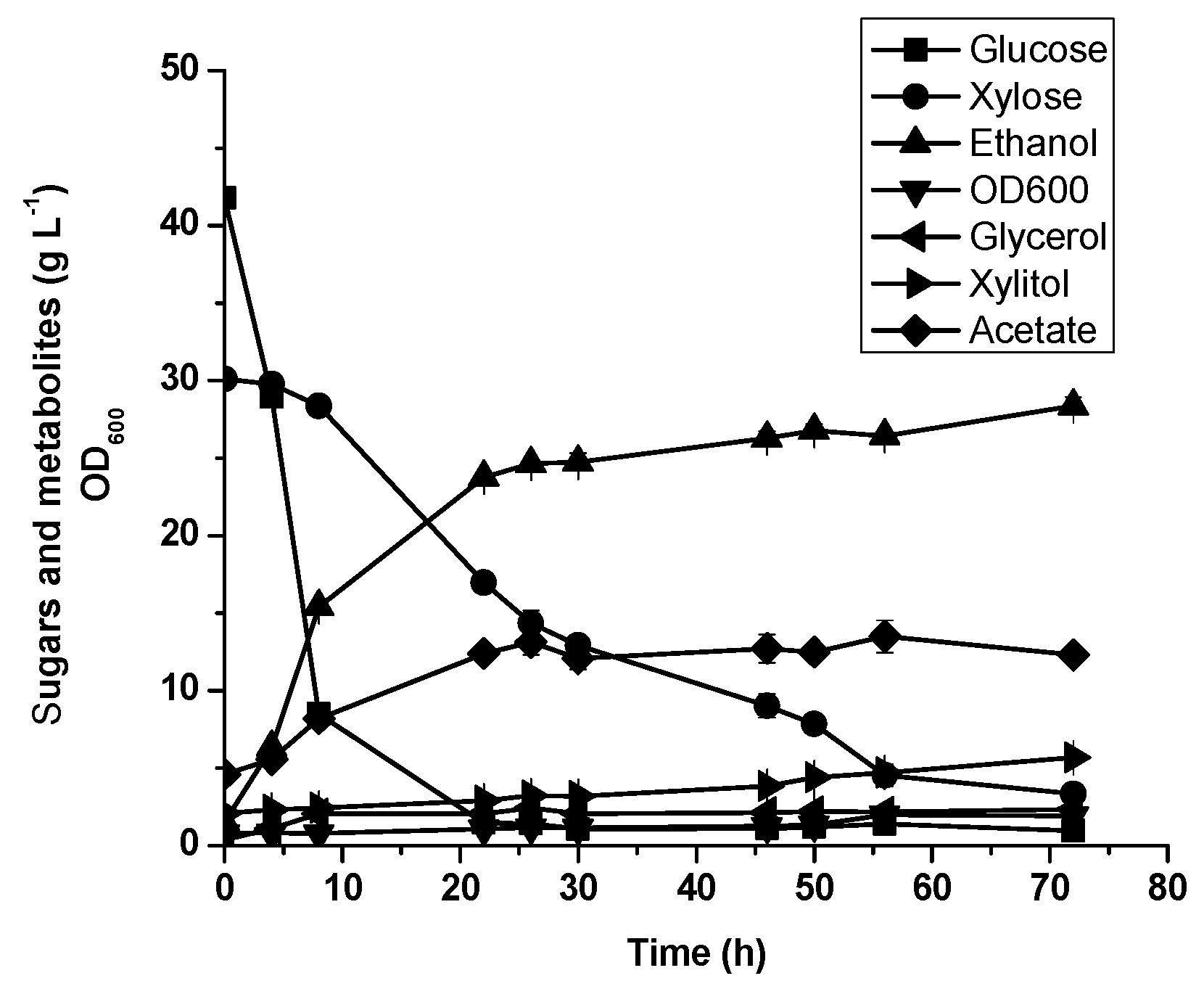
3.5. Oil Palm Empty Fruit Bunch Hydrolysate Fermentation by Diploid Recombinant Strain STXQ
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Somerville, C.; Youngs, H.; Taylor, C.; Davis, S.C.; Long, S.P. Feedstocks for lignocellulosic biofuels. Science 2010, 329, 790–792. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.; Grift, T.E.; Hansen, A.C.; Ting, K.C. An overview of lignocellulosic biomass feedstock harvest, processing and supply for biofuel production. Biofuels 2013, 4, 5–8. [Google Scholar] [CrossRef]
- Fatma, S.; Hameed, A.; Noman, M.; Ahmed, T.; Shahid, M.; Tariq, M.; Sohail, I.; Tabassum, R. Lignocellulosic biomass: A sustainable bioenergy source for the future. Protein Pept. Lett. 2018, 25, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Moyses, D.N.; Reis, V.C.; de Almeida, J.R.; de Moraes, L.M.; Torres, F.A. Xylose fermentation by Saccharomyces cerevisiae: Challenges and prospects. Int. J. Mol. Sci. 2016, 17, 207. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Gou, Z.X.; Zhang, Y.; Xia, Z.Y.; Tang, Y.Q.; Kida, K. Inhibitor tolerance of a recombinant flocculating industrial Saccharomyces cerevisiae strain during glucose and xylose co-fermentation. Braz. J. Microbiol. 2017, 48, 798–800. [Google Scholar] [CrossRef] [PubMed]
- Hahn-Hägerdal, B.; Karhumaa, K.; Jeppsson, M.; Gorwa-Grauslund, M.F. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv. Biochem. Eng. Biotechnol. 2007, 108, 147–177. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, A.; Srivastava, A.; Kondo, A.; Bisaria, V.S. Bioconversion of lignocellulose-derived sugars to ethanol by engineered Saccharomyces cerevisiae. Crit. Rev. Biotechnol. 2012, 32, 22–48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.C.; Liu, J.J.; Kong, I.I.; Kwak, S.; Jin, Y.S. Combining C6 and C5 sugar metabolism for enhancing microbial bioconversion. Curr. Opin. Chem. Biol. 2015, 29, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.; Jin, Y.S. Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: A review and perspective. Microb. Cell Factories 2017, 16, 82. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Qiu, C.; Shen, Y.; Li, H.; Bao, X. Engineering of Saccharomyces cerevisiae for the efficient co-utilization of glucose and xylose. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Kuyper, M.; Hartog, M.M.; Toirkens, M.J.; Almering, M.J.; Winkler, A.A.; van Dijken, J.P.; Pronk, J.T. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 2005, 5, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Huang, S.; Liu, T.; Geng, A. Bacterial xylose isomerases from the mammal gut Bacteroidetes cluster function in Saccharomyces cerevisiae for effective xylose fermentation. Microb. Cell Factories 2015, 14, 70. [Google Scholar] [CrossRef] [PubMed]
- Johansson, B.; Hahn-Hägerdal, B. The non-oxidative pentose phosphate pathway controls the fermentation rate of xylulose but not of xylose in Saccharomyces cerevisiae TMB3001. FEMS Yeast Res. 2002, 2, 277–282. [Google Scholar] [PubMed]
- Peng, B.; Shen, Y.; Li, X.; Chen, X.; Hou, J.; Bao, X. Improvement of xylose fermentation in respiratory-deficient xylose-fermenting Saccharomyces cerevisiae. Metab. Eng. 2012, 14, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kodaki, T.; Park, Y.C.; Seo, J.H. Effects of NADH-preferring xylose reductase expression on ethanol production from xylose in xylose-metabolizing recombinant Saccharomyces cerevisiae. J. Biotechnol. 2012, 158, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Sonderegger, M.; Sauer, U. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl. Environ. Microbiol. 2003, 69, 1990–1998. [Google Scholar] [CrossRef] [PubMed]
- Scalcinati, G.; Otero, J.M.; Vleet, J.R.; Jeffries, T.W.; Olsson, L.; Nielsen, J. Evolutionary engineering of Saccharomyces cerevisiae for efficient aerobic xylose consumption. FEMS Yeast Res. 2012, 12, 582–597. [Google Scholar] [CrossRef] [PubMed]
- Demeke, M.M.; Dietz, H.; Li, Y.; Foulquie-Moreno, M.R.; Mutturi, S.; Deprez, S.; Den Abt, T.; Bonini, B.M.; Liden, G.; Dumortier, F.; et al. Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol. Biofuels 2013, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Sedlak, M.; Ho, N.W. Characterization of the effectiveness of hexose transporters for transporting xylose during glucose and xylose co-fermentation by a recombinant Saccharomyces yeast. Yeast 2004, 21, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Runquist, D.; Hahn-Hagerdal, B.; Radstrom, P. Comparison of heterologous xylose transporters in recombinant Saccharomyces cerevisiae. Biotechnol. Biofuels 2010, 17, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, D.L.; Matsushika, A.; de Sales, B.B.; Goshima, T.; Bon, E.P.; Stambuk, B.U. Xylose and xylose/glucose co-fermentation by recombinant Saccharomyces cerevisiae strains expressing individual hexose transporters. Enzyme Microb. Technol. 2014, 63, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Diao, L.; Liu, Y.; Qian, F.; Yang, J.; Jiang, Y.; Yang, S. Construction of fast xylose-fermenting yeast based on industrial ethanol-producing diploid Saccharomyces cerevisiae by rational design and adaptive evolution. BMC Biotechnol. 2013, 13, 110. [Google Scholar] [CrossRef] [PubMed]
- Demeke, M.M.; Dumortier, F.; Li, Y.; Broeckx, T.; Foulquié-Moreno, M.R.; Thevelein, J.M. Combining inhibitor tolerance and D-xylose fermentation in industrial Saccharomyces cerevisiae for efficient lignocellulose-based bioethanol production. Biotechnol. Biofuels 2013, 6, 120. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Lee, K.S.; Kong, I.I.; Lesmana, A.; Lee, W.H.; Seo, J.H.; Kweon, D.H.; Jin, Y.S. Construction of an efficient xylose-fermenting diploid Saccharomyces cerevisiae strain through mating of two engineered haploid strains capable of xylose assimilation. J. Biotechnol. 2013, 164, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shen, Y.; Wu, M.; Hou, J.; Jiao, C.; Li, Z.; Liu, X.; Bao, X. Engineering a wild-type diploid Saccharomyces cerevisiae strain for second-generation bioethanol production. Bioresour. Bioprocess. 2016, 3, 51. [Google Scholar] [CrossRef] [PubMed]
- Tanino, T.; Hotta, A.; Ito, T.; Ishii, J.; Yamada, R.; Hasunuma, T.; Ogino, C.; Ohmura, N.; Ohshima, T.; Kondo, A. Construction of a xylose-metabolizing yeast by genome integration of xylose isomerase gene and investigation of the effect of xylitol on fermentation. Appl. Microbiol. Biotechnol. 2010, 88, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Bamba, T.; Hasunuma, T.; Kondo, A. Disruption of PHO13 improves ethanol production via the xylose isomerase pathway. AMB Express 2016, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Cheng, J.S.; Wang, B.L.; Fink, G.R.; Stephanopoulos, G. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab. Eng. 2012, 14, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.K.; Um, Y.; Woo, H.M.; Kim, K.H.; Lee, S.M. Ethanol production from lignocellulosic hydrolysates using engineered Saccharomyces cerevisiae harboring xylose isomerase-based pathway. Bioresour. Technol. 2016, 209, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Güldener, U.; Heck, S.; Fiedler, T.; Beinhauer, J.; Hegemann, J.H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996, 24, 2519–2524. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Schiestl, R.H. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 38–41. [Google Scholar] [CrossRef] [PubMed]
- Kersters-Hilderson, H.; Callens, M.; Opstal, O.V.; Vangrysperre, W.; Bruyne, C.K.D. Kinetic characterization of d-xylose isomerases by enzymatic assays using d-sorbitol dehydrogenase. Enzyme Microb. Technol. 1987, 9, 145–148. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ong, H.X.; Geng, A. Cellulase production and oil palm empty fruit bunch saccharification by a new isolate of Trichoderma koningii D-64. Proc. Biochem. 2012, 47, 1564–1571. [Google Scholar] [CrossRef]
- Qi, X.; Zha, J.; Liu, G.G.; Zhang, W.; Li, B.Z.; Yuan, Y.J. Heterologous xylose isomerase pathway and evolutionary engineering improve xylose utilization in Saccharomyces cerevisiae. Front. Microbiol. 2015, 6, 1165. [Google Scholar] [CrossRef] [PubMed]
- Vilela Lde, F.; de Araujo, V.P.; Paredes Rde, S.; Bon, E.P.; Torres, F.A.; Neves, B.C.; Eleutherio, E.C. Enhanced xylose fermentation and ethanol production by engineered Saccharomyces cerevisiae strain. AMB Express 2015, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, A.; Tamalampudi, S.; Srivastava, A.; Fukuda, H.; Bisaria, V.S.; Kondo, A. Alcoholic fermentation of xylose and mixed sugars using recombinant Saccharomyces cerevisiae engineered for xylose utilization. Appl. Microbiol. Biotechnol. 2009, 82, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Perez-Traves, L.; Lopes, C.A.; Barrio, E.; Querol, A. Evaluation of different genetic procedures for the generation of artificial hybrids in Saccharomyces genus for winemaking. Int. J. Food Microbiol. 2012, 156, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Ho, N.W.; Chen, Z.; Brainard, A.P. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl. Environ. Microbiol. 1998, 64, 1852–1859. [Google Scholar] [PubMed]
- Kuyper, M.; Harhangi, H.R.; Stave, A.K.; Winkler, A.A.; Jetten, M.S.; de Laat, W.T.; den Ridder, J.J.; Op den Camp, H.J.; van Dijken, J.P.; et al. High-level functional expression of a fungal xylose isomerase: The key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res. 2003, 4, 69–78. [Google Scholar] [CrossRef]
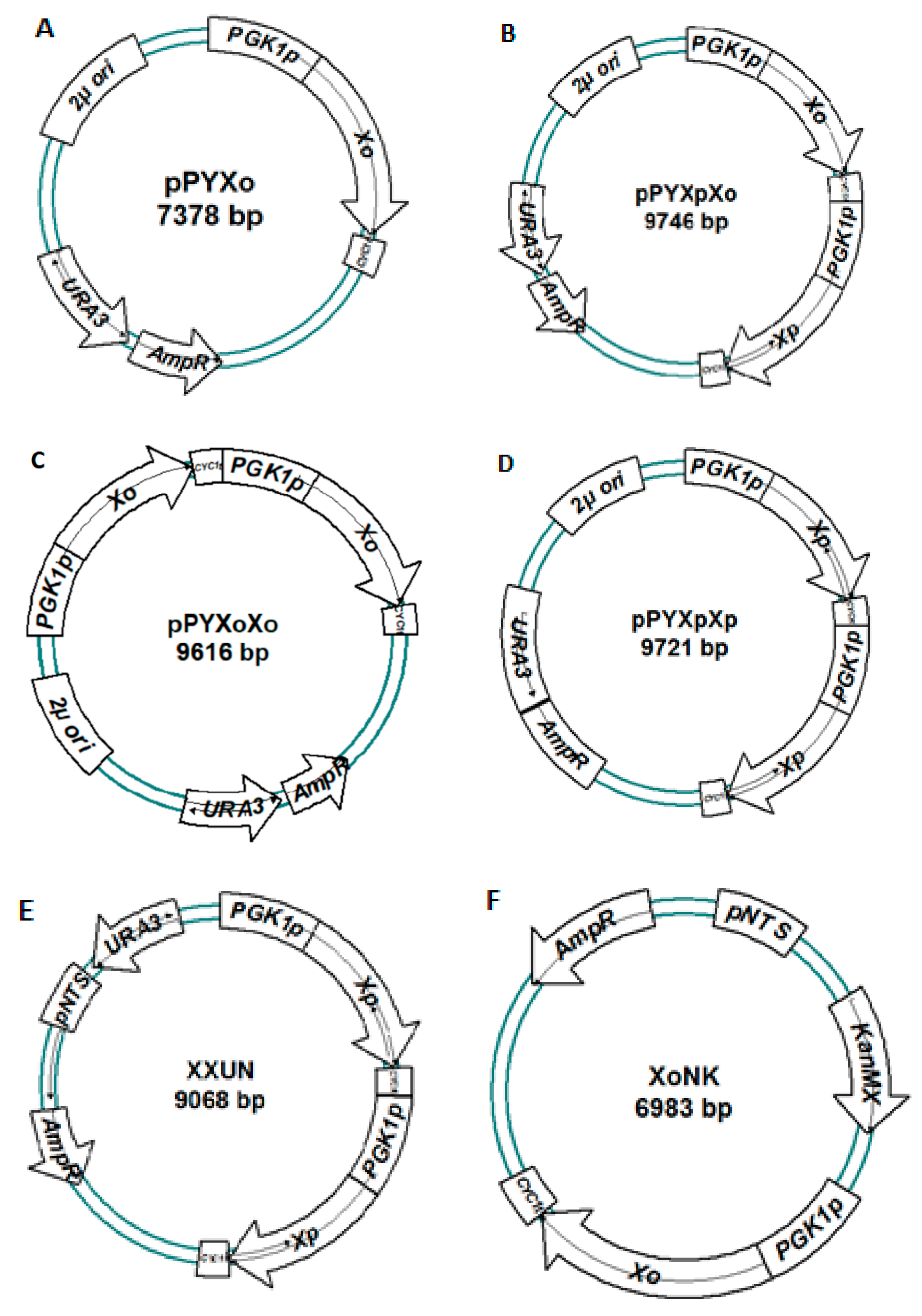
| Strains/Plasmid | Genotype/Phenotype | References |
|---|---|---|
| Saccharomyces cerevisiae strains | ||
| Saccharomyces cerevisiae ATCC 24860 | Obtained from American Type Culture Collection (ATCC) | |
| JUK36α | S. cerevisiae ATCC 24860 segregant; MAT; ura3::loxP; TKL1::RKIt-RKI1-ADH1p-RPE1t-RPE1-TPI1p-loxP-XKS1t-XKS1-PGK1p-PDC1p-TAL1-TAL1t-FBA1p; gre3::loxP | [12] |
| JUK39a | S. cerevisiae ATCC 24860 segregant; MATa; ura3::loxP; TKL1::RKIt-RKI1-ADH1p-RPE1t-RPE1-TPI1p-loxP-XKS1t-XKS1-PGK1p-PDC1p-TAL1-TAL1t-FBA1p; gre3::loxP; cyc3::loxP | [12] |
| JUK51a_2 | JUK36α derivative; {pJFX11}/(BvuXylA, XK, PPP, gre3∆) | [12] |
| 39aXpXp | JUK39a derivative; {pPYXpXp}/(two-copy PirXylA, XK, PPP, gre3∆, cyc3∆) | This work |
| 39aXpXp2415 | 39aXpXp derivative; {pPYXpXp}/(two-copy PirXylA, XK, PPP, gre3∆, cyc3∆, AE) | This work |
| 36α2 | Isolate from chemostat anaerobic and adaptive evolution at a dilution rate of 0.15 h−1 on xylose of JUK51a_2 and loss of plasmid pJFX11 | This work |
| 39a2 | Isolate from 39aXpXp2415 and loss of plasmid pPYXpXp | This work |
| 39a2XpXp | 39a2 derivative; {pPYXpXp}/(two-copy PirXylA, XK, PPP, gre3∆, cyc3∆) | This work |
| 39a2XpXo | 39a2 derivative; {pPYXpXo}/(OrpXylA, PirXylA, XK, PPP, gre3∆, cyc3∆) | This work |
| 39a2Xo | 39a2 derivative; {pPYXo}/(OrpXylA, XK, PPP, gre3∆, cyc3∆) | This work |
| 39a2XoXo | 39a2 derivative; {pPYXoXo}/}/(two-copy OrpXylA, XK, PPP, gre3∆, cyc3∆) | This work |
| 36α2XpXpUN | 36α2 derivative;NTS2-2::two-copy PirXylA, ura3, XK, PPP, gre3∆ | This work |
| 36α2XoNK | 36α2 derivative;NTS2-2::OrpXylA-KanMX4, XK, PPP, gre3∆, ura3∆ | This work |
| 39a2XpXpUN | 39a2 derivative; NTS2-2::two-copy PirXylA, ura3, XK, PPP, gre3∆, cyc3∆ | This work |
| 39a2XoNK | 39a2 derivative; NTS2-2::OrpXylA-KanMX4, XK, PPP, gre3∆, cyc3∆, ura3∆ | This work |
| STXQ | Isolate from mating of 36α2XpXpUN with 39a2XoNK | This work |
| Plasmids | ||
| pUG6 | E. coli plasmid with segment loxP–KanMX4–loxP | [30] |
| pJFX11 | YEp, TEF1p-BvuXylA-CYC1t | [12] |
| pPY1 | pPYES2; GAL1p replaced by PGK1p | This work |
| pPYXo | pPY1; PGK1p-OrpXylA-CYC1t | This work |
| pPYXpXp | pPY1; 2 copies of PGK1p-PirXylA-CYC1t in tandem | This work |
| pPYXpXo | pPY1; PGK1p-PirXylA-CYC1t-PGK1p-OrpXylA-CYC1t | This work |
| pPYXoXo | pPY1; 2 copies of PGK1p-OrpXylA-CYC1t in tandem | This work |
| XXUN | pPYXpXp-based yeast integration plasmid; 2 μ and ura3 were replaced with ura3 and NTS2-2 partial fragment | This work |
| XoNK | pUC19-based yeast integration plasmid; loxP-KanMX4-loxP-pNTS-PGK1p-OrpXylA-CYC1t | This work |
| Strains | Specific Activity (U mg−1 Protein) |
|---|---|
| 39a2XpXp | 0.10 ± 0.003 |
| 39a2XpXo | 0.11 ± 0.019 |
| 39a2Xo | 0.12 ± 0.041 |
| 39a2XoXo | 0.30 ± 0.079 |
| 36α2XpXpUN | 0.11 ± 0.007 |
| 36α2 XoNK | 0.04 ± 0.005 |
| 39a2XpXpUN | 0.26 ± 0.004 |
| 39a2XoNK | 0.72 ± 0.006 |
| Strain | Fold-Change a | |
|---|---|---|
| PirXylA | OrpXylA | |
| 39a2XpXp | 50.21 (47.81–52.74) | nil |
| 39a2XpXo | 29.86 (28.43–31.36) | 11.71 (10.62–12.92) |
| 39a2Xo | nil | 59.71 (51.98–68.59) |
| 39a2XoXo | nil | 59.71 (55.72–64) |
| 36α2XpXpUN | 51.98 (49.50–54.60) | nil |
| 39a2XoNK | nil | 84.45 (59.71–119.43) |
| Strains | Initial glucose (g L−1) | Initial Xylose (g L−1) | Xylose Conversion (%) | Ethanol Yield (g g−1) | Ethanol Productivity (g h−1 L−1) | Specific Growth Rate (h−1) |
|---|---|---|---|---|---|---|
| 39a2xpXp a | - | 20 | 98% ± 0.004 | 0.472 ± 0.013 | 0.098 ± 0.004 | 0.014 ± 0.001 |
| 39a2XpXo a | - | 20 | 89.8% ± 0.011 | 0.434 ± 0.017 | 0.098 ± 0.004 | 0.011 ± 5.89 × 10−6 |
| 39a2Xo a | - | 20 | 97.3% ± 0.0138 | 0.428 ± 0.026 | 0.084 ± 0.001 | 0.011 ± 0.0003 |
| 39a2XoXo a | - | 20 | 98.5% ± 0.0057 | 0.449 ± 0.038 | 0.108 ± 0.001 | 0.013 ± 2.29 × 10−5 |
| 36α2XpXpUN a | - | 20 | 86.4% ± 0.052 | 0.318 ± 0.01 | 0.069 ± 0.007 | 0.014 ± 0.0003 |
| 36α2XoNK a | - | 20 | 6.42% ± 0.057 | ND | ND | 0.003 ± 0.0006 |
| 39a2XpXpUNa | - | 20 | 67.0% ± 0.089 | 0.398 ± 0.048 | 0.067 ± 0.001 | 0.013 ± 0.0002 |
| 39a2XoNK a | - | 20 | 99.0% ± 0.002 | 0.368 ± 0.029 | 0.107 ± 0.007 | 0.0098 ± 0.0003 |
| 39a2XpXp a | 20 | 20 | 22.0% ± 0.003 | 0.444 ± 0.029 | 0.221 ± 0.020 | 0.020 ± 0.0007 |
| 39a2XpXo a | 20 | 20 | 77.5% ± 0.017 | 0.392 ± 0.001 | 0.081 ± 0.006 | 0.022 ± 0.0001 |
| 39a2Xo a | 20 | 20 | 84.3% ± 0.0007 | 0.428 ± 0.01 | 0.238 ± 0.003 | 0.021 ± 0.0004 |
| 39a2XoXo a | 20 | 20 | 96.7% ± 0.011 | 0.449 ± 0.022 | 0.213 ± 0.004 | 0.021 ± 0.001 |
| 36α2XpXpUN a | 20 | 20 | 73.8% ± 0.011 | 0.348 ± 0.049 | 0.225 ± 0.007 | 0.029 ± 0.0006 |
| 36α2XoNK a | 20 | 20 | 2.62% ± 0.000 | 0.390 ± 0.043 | 0.222 ± 0.003 | 0.016 ± 0.0013 |
| 39a2XpXpUN a | 20 | 20 | 25.6% ± 0.097 | 0.405 ± 0.049 | 0.213 ± 0.002 | 0.013 ± 0.0002 |
| 39a2XoNK a | 20 | 20 | 97.3% ± 0.006 | 0.387 ± 0.002 | 0.243 ± 0.003 | 0.020 ± 0.0005 |
| 36α2XpXpUN b | - | 40 | 75.9% ± 0.041 | 0.384 ± 0.033 | 0.108 ± 0.011 | 0.013 ± 0.0004 |
| 39a2XoNK b | - | 40 | 81.6% ± 0.004 | 0.421 ± 0.004 | 0.131 ± 0.000 | 0.010 ± 0.0003 |
| STXQ b | - | 40 | 93.3% ± 0.019 | 0.393 ± 0.024 | 0.146 ± 0.009 | 0.017 ± 0.0002 |
| STXQ b | - | 132 | 100% | 0.498 ± 0.006 | 1.13 ± 0.01 | 0.014 ± 0.0004 |
| STXQ c | 162 | 95 | 99.27% ± 0.002 | 0.475 ± 0.01 | 5.24 ± 0.02 | 0.024 ± 0.0001 |
| Strain | Description | Inoculum Biomass (g DCW L−1) | Initial Glucose (g L−1) | Initial Xylose (g L−1) | Final Ethanol (g L−1) | Ethanol Yield (g g−1) | Volumetric Ethanol Productivity (g L−1·h−1) | Reference |
|---|---|---|---|---|---|---|---|---|
| CIBTS0735 | PirXylA; XKS1; PPP; ciGXF1; adaptive evolution | Rich medium, initial inoculum size at 0.63 g DCW L−1 | 40 | 17.47 | 0.44 | 1.09 | [18] | |
| Rich medium, initial inoculum size at 0.63 g DCW L−1 | 80 | 40 | 53 | 0.45 | 2.22 | |||
| RWB218 | PirXylA; XKS1; PPP; gre3∆; adaptive evolution) | defined synthetic medium, initial inoculum size at 1.1 g DCW L−1 | 100 | 25 | 47.5 | 0.38 | 1.98 | [40] |
| 1400 (pLNH32) | XR; XDH; XK; adaptive evolution | Rich medium; OD 40–45 | 50 | 24 | 0.48 | 0.52 | [39] | |
| Rich medium; OD 40–45 | 80 | 40 | 60 | 0.45 | 1.3 | |||
| H31-A3-ALCS | PirXylA; XKS1; PPP; gre3∆; adaptive evolution | Defined medium;initial inoculum size at 0.05 g DCW L−1 | 40 | 16.4 | 0.41 | 0.55 | [28] | |
| GS1.11-26 | cpXylA; XKS1; PPP; HXT7; AraT; AraA; AraB; AraD; TAL2; TKL2 mutagenesis; genome shuffling; adaptive evolution | Rich medium; initial inoculum size at 1.3 g DCW L−1 | 36 | 37 | 33.6 | 0.46 | 2.58 | [18] |
| LF1 | Ru-XylA, XKS1, PPP, gre3::MGT05196N360F, adaptive evolution | Rich medium, initial inoculum size at 1.00 g DCW L−1 | 80 | 42 | 58.0 | 0.47 | 3.60 | [25] |
| STXQ | OrpXylA, PirXylA, XKS1, PPP, gre3∆; adaptive evolution | Rich medium, initial inoculum size at 3.13 g DCW L−1 | 132 | 65.8 | 0.50 | 1.13 | This study | |
| Rich medium, initial inoculum size at 3.43 g DCW L−1 | 162 | 95 | 120.6 | 0.48 | 5.24 | This study |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Huang, S.; Geng, A. Recombinant Diploid Saccharomyces cerevisiae Strain Development for Rapid Glucose and Xylose Co-Fermentation. Fermentation 2018, 4, 59. https://doi.org/10.3390/fermentation4030059
Liu T, Huang S, Geng A. Recombinant Diploid Saccharomyces cerevisiae Strain Development for Rapid Glucose and Xylose Co-Fermentation. Fermentation. 2018; 4(3):59. https://doi.org/10.3390/fermentation4030059
Chicago/Turabian StyleLiu, Tingting, Shuangcheng Huang, and Anli Geng. 2018. "Recombinant Diploid Saccharomyces cerevisiae Strain Development for Rapid Glucose and Xylose Co-Fermentation" Fermentation 4, no. 3: 59. https://doi.org/10.3390/fermentation4030059
APA StyleLiu, T., Huang, S., & Geng, A. (2018). Recombinant Diploid Saccharomyces cerevisiae Strain Development for Rapid Glucose and Xylose Co-Fermentation. Fermentation, 4(3), 59. https://doi.org/10.3390/fermentation4030059