The Smell of Synthetic Biology: Engineering Strategies for Aroma Compound Production in Yeast
Abstract
1. Introduction
2. Yeast as a Recombinant Host for Bioflavour Production
3. Yeast Precursors Utilised
3.1. Phenylpropanoids
3.2. Terpenoids
4. Biosensing Aroma Compounds in Yeast
5. Future Outlook
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Schrader, J. Flavours and Fragrances: Chemistry, Bioprocessing and Sustainability; Berger, R.G., Ed.; Springer-Verlag: Berlin/Heidelberg, Germany, 2007; pp. 507–574. [Google Scholar] [CrossRef]
- Kempinski, C.; Jiang, Z.; Bell, S.; Chappell, J. Biotechnology of Isoprenoids; Schrader, J., Bohlmann, J., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2015; pp. 161–199. [Google Scholar] [CrossRef]
- Lanciotti, R.; Gianotti, A.; Patrignani, F.; Belletti, N.; Guerzoni, M.E.; Gardini, F. Use of natural aroma compounds to improve shelf-life and safety of minimally processed fruits. Trends Food Sci. Technol. 2004, 15, 201–208. [Google Scholar] [CrossRef]
- Belletti, N.; Kamdem, S.S.; Patrignani, F.; Lanciotti, R.; Covelli, A.; Gardini, F. Antimicrobial activity of aroma compounds against Saccharomyces cerevisiae and improvement of microbiological stability of soft drinks as assessed by logistic regression. Appl. Environ. Microbiol. 2007, 73, 5580–5586. [Google Scholar] [CrossRef] [PubMed]
- Buck, L.; Axel, R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 1991, 65, 175–187. [Google Scholar] [CrossRef]
- Swiegers, J.H.; Chambers, P.J.; Pretorius, I.S. Olfaction and taste: Human perception, physiology and genetics. Aust. J. Grape Wine Res. 2005, 11, 109–113. [Google Scholar] [CrossRef]
- Stevenson, R.J.; Boakes, R.A. A mnemonic theory of odor perception. Psychol. Rev. 2003, 110, 340–364. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.L.; Desai, S.H.; Atsumi, S. Microbial production of scent and flavor compounds. Curr. Opin. Biotechnol. 2016, 37, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Steensels, J.; Snoek, T.; Meersman, E.; Nicolino, M.P.; Voordeckers, K.; Verstrepen, K.J. Improving industrial yeast strains: Exploiting natural and artificial diversity. FEMS Microbiol. Rev. 2014, 38, 947–995. [Google Scholar] [CrossRef] [PubMed]
- Dzialo, M.C.; Park, R.; Steensels, J.; Lievens, B.; Verstrepen, K.J. Physiology, ecology and industrial applications of aroma formation in yeast. FEMS Microbiol. Rev. 2017, 41, S95–S128. [Google Scholar] [CrossRef] [PubMed]
- Hirst, M.B.; Richter, C.L. Review of aroma formation through metabolic pathways of Saccharomyces cerevisiae in beverage fermentations. Am. J. Enol. Vitic. 2016, 67, 361–370. [Google Scholar] [CrossRef]
- Peter, J.; Chiara, M.D.; Friedrich, A.; Yue, J.; Pflieger, D.; Bergström, A.; Sigwalt, A.; Barre, B.; Freel, K.; Llored, A.; et al. Genome evolution across 1011 Saccharomyces cerevisiae isolates. Nature 2018, 556, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Gellissen, G.; Kunze, G.; Gaillardin, C.; Cregg, J.M.; Berardi, E.; Veenhuis, M.; Klei, I.V.D. New yeast expression platforms based on methylotrophic Hansenula polymorpha and Pichia pastoris and on dimorphic Arxula adeninivorans and Yarrowia lipolytica—A comparison. FEMS Yeast Res. 2005, 5, 1079–1096. [Google Scholar] [CrossRef] [PubMed]
- Forti, L.; Mauro, S.D.; Cramarossa, M.R.; Filippucci, S.; Turchetti, B.; Buzzini, P. Non-conventional yeasts whole cells as efficient biocatalysts for the production of flavors and fragrances. Molecules 2015, 20, 10377–10398. [Google Scholar] [CrossRef] [PubMed]
- Verstrepen, K.J.; Derdelinckx, G.; Dufour, J.P.; Winderickx, J.; Thevelein, J.M.; Pretorius, I.S.; Delvaux, F.R. Flavor-active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 2003, 96, 110–118. [Google Scholar] [CrossRef]
- Kroukamp, H.; den Haan, R.; la Grange, D.C.; Sibanda, N.; Foulquié-Moreno, M.R.; Thevelein, J.M.; van Zyl, W.H. Strain breeding enhanced heterologous cellobiohydrolase secretion by Saccharomyces cerevisiae in a protein specific manner. Biotechnol. J. 2017, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Hvorecny, K.; Prelich, G. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast 2010, 27, 861–865. [Google Scholar] [CrossRef] [PubMed]
- DiCarlo, J.E.; Conley, A.J.; Penttilä, M.; Jäntti, J.; Wang, H.H.; Church, G. Yeast gligo-mediated genome Engineering (YOGE). ACS Synth. Biol. 2013, 2, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, L.A.; Chuang, J.; Agmon, N.; Khunsriraksakul, C.; Phillips, N.A.; Cai, Y.; Truong, D.M.; Veerakumar, A.; Wang, Y.; Mayorga, M.; et al. Versatile genetic assembly system (VEGAS) to assemble pathways for expression in S. cerevisiae. Nucleic Acids Res. 2015, 43, 6620–6630. [Google Scholar] [CrossRef] [PubMed]
- Pennisi, E. Building the ultimate yeast genome. Science 2014, 343, 1426–1429. [Google Scholar] [CrossRef] [PubMed]
- Annaluru, N.; Muller, H.; Mitchell, L.A.; Ramalingam, S.; Stracquadanio, G.; Richardson, S.M.; Dymond, J.S.; Kuang, Z.; Scheifele, L.Z.; Cooper, E.M.; et al. Total synthesis of a functional designer eukaryotic chromosome. Science 2014, 344, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.M.; Mitchell, L.A.; Stracquadanio, G.; Yang, K.; Dymond, J.S.; DiCarlo, J.E.; Lee, D.; Huang, C.L.V.; Chandrasegaran, S.; Cai, Y.; et al. Design of a synthetic yeast genome. Science 2017, 355, 1040–1044. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Stracquadanio, G.; Wang, Y.; Yang, K.; Mitchell, L.A.; Xue, Y.; Cai, Y.; Chen, T.; Dymond, J.S.; Kang, K.; et al. SCRaMbLE generates designed combinatorial stochastic diversity in synthetic chromosomes. Genome Res. 2015, 26, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, I.S.; Boeke, J.D. Yeast 2.0—Connecting the dots in the construction of the world’s first functional synthetic eukaryotic genome. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Pickens, L.; Tang, Y.; Chooi, Y.-H. Metabolic engineering for the production of natural products. Annu. Rev. Chem. Biomol. Eng. 2014, 2, 211–236. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Wendisch, V.F. Biotechnological production of aromatic compounds of the extended shikimate pathway from renewable biomass. J. Biotechnol. 2017, 257, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Braus, G.H. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: A model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol. Rev. 1991, 55, 349–370. [Google Scholar] [PubMed]
- Mulleder, M.; Calvani, E.; Alam, M.T.; Wang, R.K.; Eckerstorfer, F.; Zelezniak, A.; Ralser, M. Functional metabolomics describes the yeast biosynthetic regulome. Cell 2016, 167, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Hansen, E.H.; Møller, B.L.; Kock, G.R.; Bünner, C.M.; Kristensen, C.; Jensen, O.R.; Okkels, F.T.; Olsen, C.E.; Motawia, M.S.; Hansen, J. De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker’s yeast (Saccharomyces cerevisiae). Appl. Environ. Microbiol. 2009, 75, 2765–2774. [Google Scholar] [CrossRef] [PubMed]
- Iwaki, A.; Ohnuki, S.; Suga, Y.; Izawa, S.; Ohya, Y. Vanillin inhibits translation and induces messenger ribonucleoprotein (mRNP) granule formation in Saccharomyces cerevisiae: Application and validation of high-content, image-based profiling. PLoS ONE 2013, 8, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Brochado, A.R.; Matos, C.; Møller, B.L.; Hansen, J.; Mortensen, U.H.; Patil, K.R. Improved vanillin production in baker’s yeast through in silico design. Microb. Cell Fact. 2010, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Brochado, A.R.; Patil, K.R. Overexpression of O-methyltransferase leads to improved vanillin production in baker’s yeast only when complemented with model-guided network engineering. Biotechnol. Bioeng. 2013, 110, 656–659. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.H.; Choon, Y.W.; Chai, L.E.; Chong, C.K.; Deris, S.; Illias, R.M.; Mohamad, M.S. Advances in Biomedical Infrastructure; Sidhu, A.S., Dhillon, S.K., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2013; Volume 477, pp. 101–116. [Google Scholar] [CrossRef]
- Vannelli, T.; Qi, W.W.; Sweigard, J.; Gatenby, A.A.; Sariaslani, F.S. Production of p-hydroxycinnamic acid from glucose in Saccharomyces cerevisiae and Escherichia coli by expression of heterologous genes from plants and fungi. Metab. Eng. 2007, 9, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Gottardi, M.; Grün, P.; Bode, H.B.; Hoffmann, T.; Schwab, W.; Oreb, M.; Boles, E. Optimisation of trans-cinnamic acid and hydrocinnamyl alcohol production with recombinant Saccharomyces cerevisiae and identification of cinnamyl methyl ketone as a by-product. FEMS Yeast Res. 2017, 9, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wood, K.V.; Morgan, J.A. Metabolic engineering of the phenylpropanoid pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 2962–2969. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lloyd, N.D.R.; Pretorius, I.S.; Borneman, A.R. Heterologous production of raspberry ketone in the wine yeast Saccharomyces cerevisiae via pathway engineering and synthetic enzyme fusion. Microb. Cell Fact. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Cho, B.R.; Hahn, J.S. Metabolic engineering of Saccharomyces cerevisiae for the production of 2-phenylethanol via Ehrlich pathway. Biotechnol. Bioeng. 2014, 111, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Vaseghi, S.; Baumeister, A.; Rizzi, M.; Reuss, M. In vivo dynamics of the pentose phosphate pathway in Saccharomyces cerevisiae. Metab. Eng. 1999, 1, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Curran, K.A.; Leavitt, J.M.; Karim, A.S.; Alper, H.S. Metabolic engineering of muconic acid production in Saccharomyces cerevisiae. Metab. Eng. 2013, 15, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Deaner, M.; Alper, H.S. Systematic testing of enzyme perturbation sensitivities via graded dCas9 modulation in Saccharomyces cerevisiae. Metab. Eng. 2017, 40, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Ravasio, D.; Wendland, J.; Walther, A. Major contribution of the Ehrlich pathway for 2-phenylethanol/rose flavor production in Ashbya gossypii. FEMS Yeast Res. 2014, 14, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.Y.; Lee, S.W.; Oh, M.K. Biosynthesis of 2-phenylethanol from glucose with genetically engineered Kluyveromyces marxianus. Enzym. Microb. Technol. 2014, 61–62, 44–47. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wang, Y.; Zong, H.; Ji, H.; Zhuge, B.; Dong, Z. Bioconversion of l-phenylalanine to 2-phenylethanol by the novel stress-tolerant yeast Candida glycerinogenes WL2002-5. Bioengineered 2016, 7, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Mierzejewska, J.; Tymoszewska, A.; Chreptowicz, K.; Krol, K. Mating of 2 laboratory Saccharomyces cerevisiae strains resulted in enhanced production of 2-phenylethanol by biotransformation of l-Phenylalanine. J. Mol. Microbiol. Biotechnol. 2017, 27, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Eshkol, N.; Sendovski, M.; Bahalul, M.; Katz-Ezov, T.; Kashi, Y.; Fishman, A. Production of 2-phenylethanol from l-phenylalanine by a stress tolerant Saccharomyces cerevisiae strain. J. Appl. Microbiol. 2009, 106, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Zwenger, S.; Basu, C. Plant terpenoids : Applications and future potentials. Biotechnol. Mol. Biol. Rev. 2008, 3, 1–7. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; Low, C.; Bottema, C.D.K.; Parks, L.W. Multiple functions for sterols in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1985, 837, 336–343. [Google Scholar] [CrossRef]
- Liao, P.; Hemmerlin, A.; Bach, T.J.; Chye, M.L. The potential of the mevalonate pathway for enhanced isoprenoid production. Biotechnol. Adv. 2016, 34, 697–713. [Google Scholar] [CrossRef] [PubMed]
- Paramasivan, K.; Mutturi, S. Progress in terpene synthesis strategies through engineering of Saccharomyces cerevisiae. Crit. Rev. Biotechnol. 2017, 37, 974–989. [Google Scholar] [CrossRef] [PubMed]
- Ignea, C.; Pontini, M.; Maffei, M.E.; Makris, A.M.; Kampranis, S.C. Engineering monoterpene production in yeast using a synthetic dominant negative geranyl diphosphate synthase. ACS Synth. Biol. 2014, 3, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Vickers, C.E.; Williams, T.C.; Peng, B.; Cherry, J. Recent advances in synthetic biology for engineering isoprenoid production in yeast. Curr. Opin. Chem. Biol. 2017, 40, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Wriessnegger, T.; Pichler, H. Yeast metabolic engineering—Targeting sterol metabolism and terpenoid formation. Prog. Lipid Res. 2013, 52, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Asadollahi, M.A.; Maury, J.; Schalk, M.; Clark, A.; Nielsen, J. Enhancement of farnesyl diphosphate pool as direct precursor of sesquiterpenes through metabolic engineering of the mevalonate pathway in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2010, 106, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Plan, M.R.; Chrysanthopoulos, P.; Hodson, M.P.; Nielsen, L.K.; Vickers, C.E. A squalene synthase protein degradation method for improved sesquiterpene production in Saccharomyces cerevisiae. Metab. Eng. 2017, 39, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Polakowski, T.; Stahl, U. Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Appl. Microbiol. Biotechnol. 1998, 8, 66–71. [Google Scholar] [CrossRef]
- Rico, J.; Pardo, E.; Orejas, M. Enhanced production of a plant monoterpene by overexpression of the 3-hydroxy-3-methylglutaryl coenzyme a reductase catalytic domain in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2010, 76, 6449–6454. [Google Scholar] [CrossRef] [PubMed]
- Pardo, E.; Rico, J.; Gil, J.V.; Orejas, M. De novo production of six key grape aroma monoterpenes by a geraniol synthase-engineered S. cerevisiae wine strain. Microb. Cell Fact. 2015, 14. [Google Scholar] [CrossRef] [PubMed]
- Vik, A.; Rine, J. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 6395–6405. [Google Scholar] [CrossRef] [PubMed]
- Meadows, A.L.; Hawkins, K.M.; Tsegaye, Y.; Antipov, E.; Kim, Y.; Raetz, L.; Dahl, R.H.; Tai, A.; Mahatdejkul-meadows, T.; Xu, L.; et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature 2016, 537, 694–697. [Google Scholar] [CrossRef] [PubMed]
- Farhi, M.; Marhevka, E.; Masci, T.; Marcos, E.; Eyal, Y.; Ovadis, M.; Abeliovich, H.; Vainstein, A. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metab. Eng. 2011, 13, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Wriessnegger, T.; Augustin, P.; Engleder, M.; Leitner, E.; Müller, M.; Kaluzna, I.; Schürmann, M.; Mink, D.; Zellnig, G.; Schwab, H.; et al. Production of the sesquiterpenoid (+)-nootkatone by metabolic engineering of Pichia pastoris. Metab. Eng. 2014, 24, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Verwaal, R.; Wang, J.; Meijnen, J.P.; Visser, H.; Sandmann, G.; Van Den Berg, J.A.; Van Ooyen, A.J.J. High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous. Appl. Environ. Microbiol. 2007, 73, 4342–4350. [Google Scholar] [CrossRef] [PubMed]
- López, J.; Essus, K.; Kim, I.-K.; Pereira, R.; Herzog, J.; Siewers, V.; Nielsen, J.; Agosin, E. Production of β-ionone by combined expression of carotenogenic and plant CCD1 genes in Saccharomyces cerevisiae. Microb. Cell Fact. 2015, 14, 84. [Google Scholar] [CrossRef] [PubMed]
- Beekwilder, J.; van Rossum, H.M.; Koopman, F.; Sonntag, F.; Buchhaupt, M.; Schrader, J.; Hall, R.D.; Bosch, D.; Pronk, J.T.; van Maris, A.J.A.; et al. Polycistronic expression of a β-carotene biosynthetic pathway in Saccharomyces cerevisiae coupled to β-ionone production. J. Biotechnol. 2014, 192, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Kirby, J.; Dietzel, K.L.; Wichmann, G.; Chan, R.; Antipov, E.; Moss, N.; Baidoo, E.E.K.; Jackson, P.; Gaucher, S.P.; Gottlieb, S.; et al. Engineering a functional 1-deoxy-d-xylulose 5-phosphate (DXP) pathway in Saccharomyces cerevisiae. Metab. Eng. 2016, 38, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.S.; Thodey, K.; Trenchard, I.; Smolke, C.D. Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res. 2012, 12, 144–170. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Keasling, J.D. Recent applications of synthetic biology tools for yeast metabolic engineering. FEMS Yeast Res. 2015, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Deloache, W.C.; Russ, Z.N.; Narcross, L.; Gonzales, A.M.; Martin, V.J.J.; Dueber, J.E. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nat. Chem. Biol. 2015, 11, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Leavitt, J.M.; Wagner, J.M.; Tu, C.C.; Tong, A.; Liu, Y.; Alper, H.S. Biosensor-enabled directed evolution to improve muconic acid production in Saccharomyces cerevisiae. Biotechnol. J. 2017, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Adeniran, A.; Sherer, M.; Tyo, K.E.J. Yeast-based biosensors: Design and applications. FEMS Yeast Res. 2015, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ravasio, D.; Walther, A.; Trost, K.; Vrhovsek, U.; Wendland, J. An indirect assay for volatile compound production in yeast strains. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.H.; Keasling, J.D. Programming adaptive control to evolve increased metabolite production. Nat. Commun. 2013, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Si, T.; Wang, M.; Zhao, H. Development of a synthetic malonyl-CoA sensor in Saccharomyces cerevisiae for intracellular metabolite monitoring and genetic screening. ACS Synth. Biol. 2015, 4, 1308–1315. [Google Scholar] [CrossRef] [PubMed]
- David, F.; Nielsen, J.; Siewers, V. Flux Control at the Malonyl-CoA node through hierarchical dynamic pathway regulation in Saccharomyces cerevisiae. ACS Synth. Biol. 2016, 5, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Skjoedt, M.L.; Snoek, T.; Kildegaard, K.R.; Arsovska, D.; Eichenberger, M.; Goedecke, T.J.; Rajkumar, A.S.; Zhang, J.; Kristensen, M.; Lehka, B.J.; et al. Engineering prokaryotic transcriptional activators as metabolite biosensors in yeast. Nat. Chem. Biol. 2016, 12, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.C.H.; Pawar, S.V.; Hallam, S.J.; Yadav, V.G. An improved whole-cell biosensor for the discovery of lignin-transforming enzymes in functional metagenomic screens. ACS Synth. Biol. 2018, 7, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, G.; Ronca, R.; Bartolucci, S. A novel E. coli biosensor for detecting aromatic aldehydes based on a responsive inducible archaeal promoter fused to the green fluorescent protein. Appl. Microbiol. Biotechnol. 2009, 82, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Siedler, S.; Khatri, N.K.; Zsohár, A.; Kjærbølling, I.; Vogt, M.; Hammar, P.; Nielsen, C.F.; Marienhagen, J.; Sommer, M.O.A.; Joensson, H.N. Development of a bacterial biosensor for rapid screening of yeast p-coumaric Acid production. ACS Synth. Biol. 2017, 6, 1860–1869. [Google Scholar] [CrossRef] [PubMed]
- West, R.W.; Chen, S.M.; Putz, H.; Butler, G.; Banerjee, M. GAL1-GAL10 divergent promoter region of Saccharomyces cerevisiae contains negative control elements in addition to functionally separate and possibly overlapping upstream activating sequences. Genes Dev. 1987, 1, 1118–1131. [Google Scholar] [CrossRef] [PubMed]
- Chao, R.; Mishra, S.; Si, T.; Zhao, H. Engineering biological systems using automated biofoundries. Metab. Eng. 2017, 42, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.; Keasling, J.D. Engineering cellular metabolism. Cell 2016, 164, 1185–1197. [Google Scholar] [CrossRef] [PubMed]
- Hollywood, K.A.; Schmidt, K.; Takano, E.; Breitling, R. Metabolomics tools for the synthetic biology of natural products. Curr. Opin. Biotechnol. 2018, 54, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Dacres, H.; Wang, J.; Leitch, V.; Horne, I.; Anderson, A.R.; Trowell, S.C. Greatly enhanced detection of a volatile ligand at femtomolar levels using bioluminescence resonance energy transfer (BRET). Biosens. Bioelectron. 2011, 29, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Berna, A. Metal oxide sensors for electronic noses and their application to food analysis. Sensors 2010, 10, 3882–3910. [Google Scholar] [CrossRef] [PubMed]
- Denby, C.M.; Li, R.A.; Vu, V.T.; Costello, Z.; Lin, W.; Chan, L.J.G.; Williams, J.; Donaldson, B.; Bamforth, C.W.; Petzold, C.J.; et al. Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Erb, T.J.; Jones, P.R.; Bar-Even, A. Synthetic metabolism: metabolic engineering meets enzyme design. Curr. Opin. Chem. Biol. 2017, 37, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Mantzouridou, F.; Tsimidou, M.Z. Observations on squalene accumulation in Saccharomyces cerevisiae due to the manipulation of HMG2 and ERG6. FEMS Yeast Res. 2010, 10, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.J.C.; Meyer, S.; Claudel, P.; Bergdoll, M.; Karst, F. Metabolic engineering of monoterpene synthesis in yeast. Biotechnol. Bioeng. 2011, 108, 1883–1892. [Google Scholar] [CrossRef] [PubMed]
- Drew, D.P.; Andersen, T.B.; Sweetman, C.; Møller, B.L.; Ford, C.; Simonsen, H.T. Two key polymorphisms in a newly discovered allele of the Vitis vinifera TPS24 gene are responsible for the production of the rotundone precursor α-guaiene. J. Exp. Bot. 2016, 67, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Schatz, M.C.; Witkowski, J.; McCombie, W.R. Current challenges in de novo plant genome sequencing and assembly. Genome Biol. 2012, 13. [Google Scholar] [CrossRef]
- Claros, M.G.; Bautista, R.; Guerrero-Fernández, D.; Benzerki, H.; Seoane, P.; Fernández-Pozo, N. Why Assembling plant genome sequences is so challenging. Biology 2012, 1, 439–459. [Google Scholar] [CrossRef] [PubMed]
- Van Wyk, N.; Trollope, K.M.; Steenkamp, E.T.; Wingfield, B.D.; Volschenk, H. Identification of the gene for β-fructofuranosidase from Ceratocystis moniliformis CMW 10134 and characterization of the enzyme expressed in Saccharomyces cerevisiae. BMC Biotechnol. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.M.; Alper, H.S. Synthetic biology and molecular genetics in non-conventional yeasts: Current tools and future advances. Fungal Genet. Biol. 2016, 89, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, U.B.; Jadhav, J.P.; Bapat, V.A.; Pretorius, I.S. Synthetic biology stretching the realms of possibility in wine yeast research. Int. J. Food Microbiol. 2017, 252, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, I.S. Solving yeast jigsaw puzzles over a glass of wine. EMBO Rep. 2017, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, I. Conducting wine symphonics with the aid of yeast genomics. Beverages 2016, 2. [Google Scholar] [CrossRef]
- Moore, S.J.; Tosi, T.; Hleba, Y.B.; Bell, D.; Polizzi, K.; Freemont, P. A cell-free synthetic biochemistry platform for raspberry ketone production. bioRxiv 2017. [Google Scholar] [CrossRef]
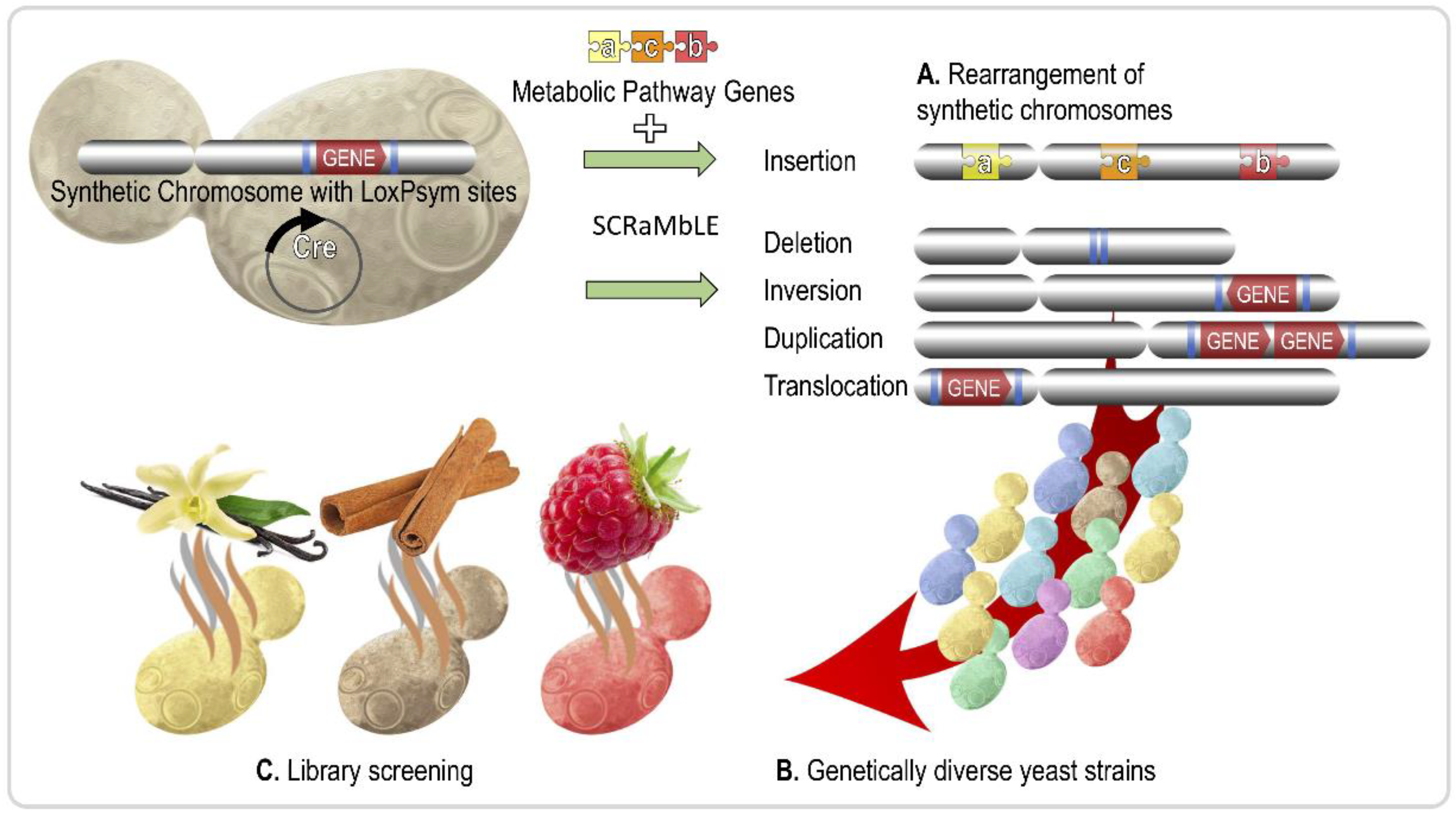
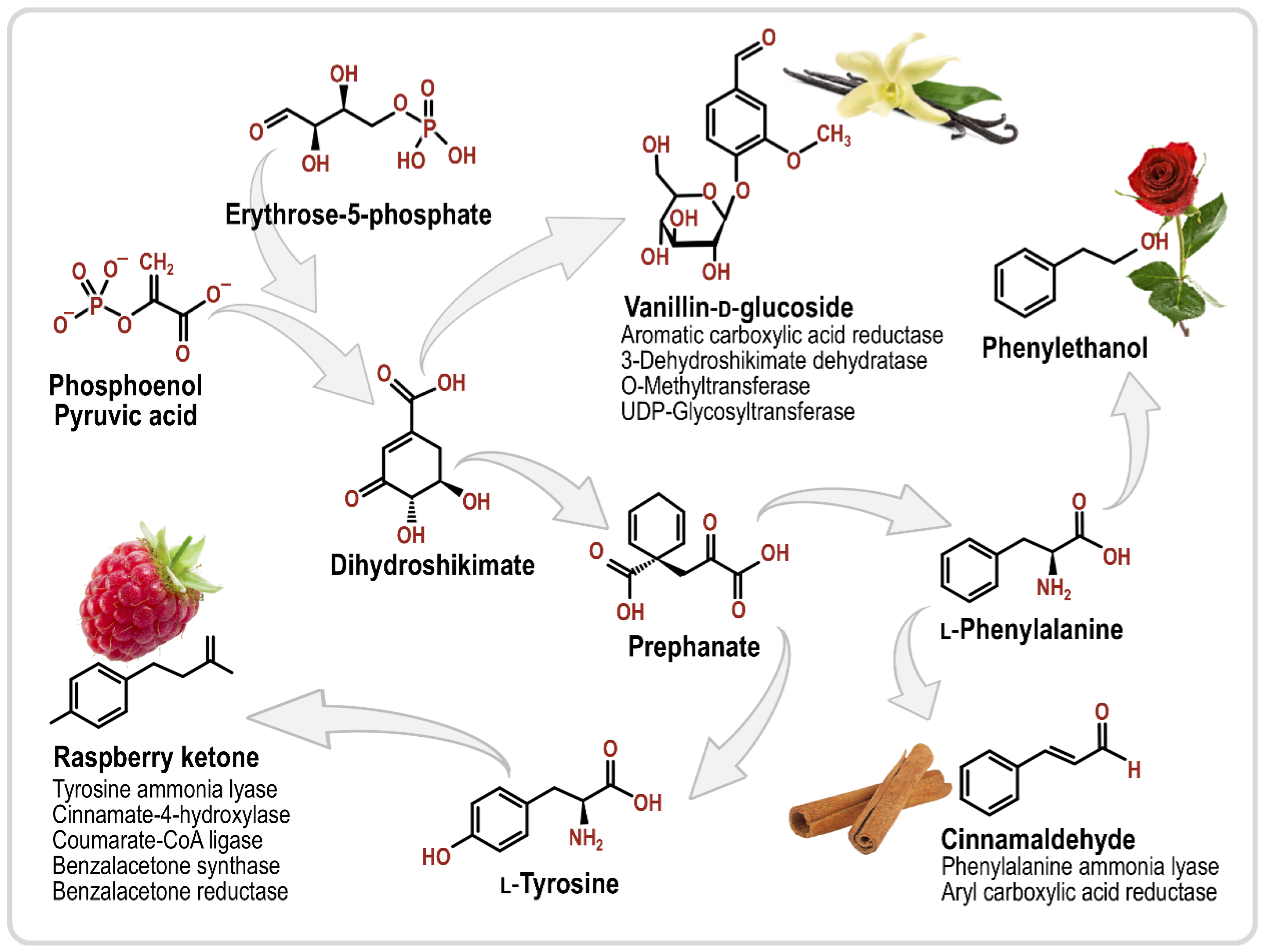
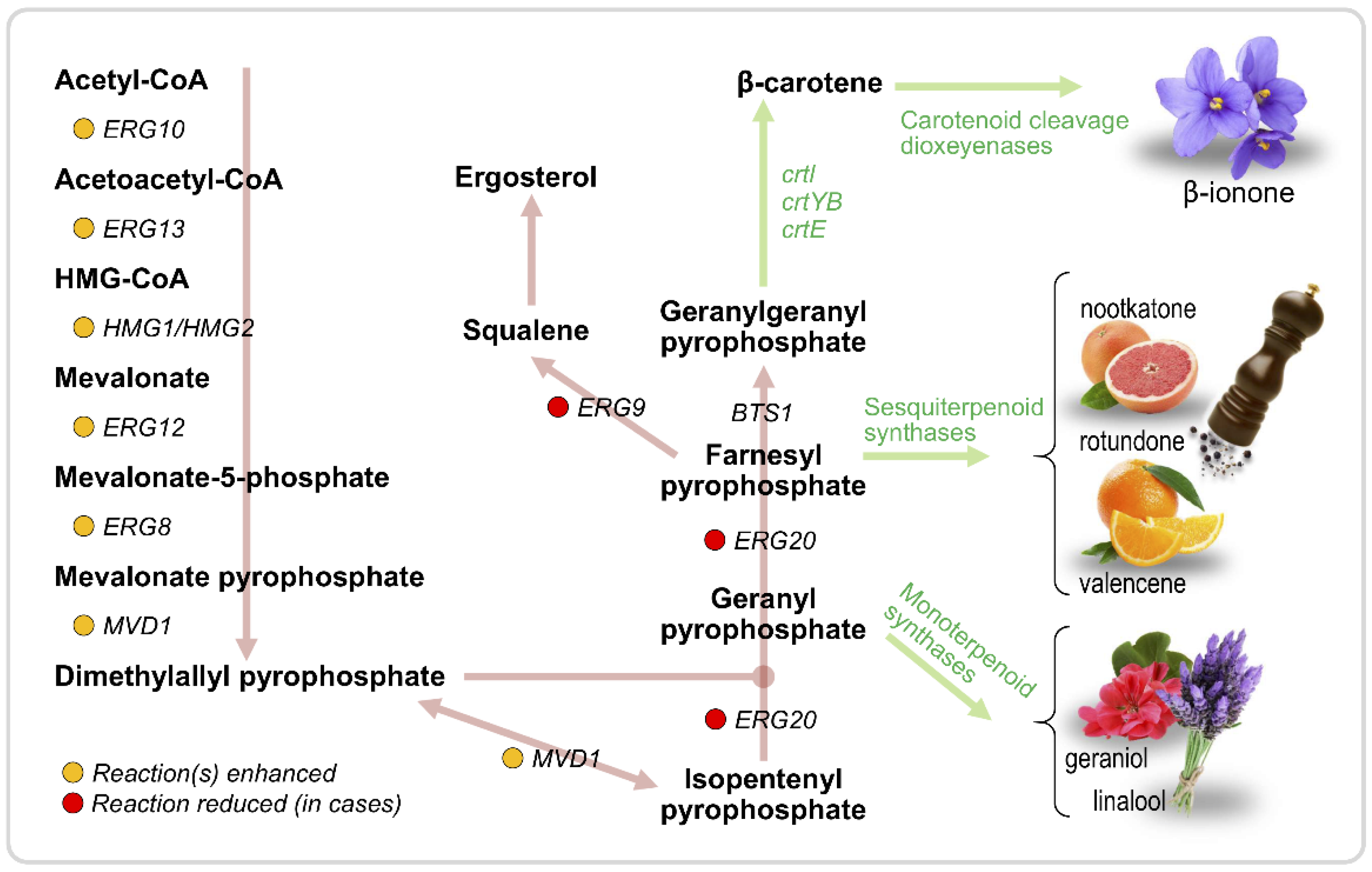

| Aroma Compound | Molecule(s) Sensed | Description | Reference |
|---|---|---|---|
| Shikimate pathway | |||
| β-phenylethanol | Aromatic amino acids | Allosteric transcription factor sensor. Transcriptional regulation LacZ reporter gene by the aromatic amino acid responsive ARO9 promoter. Increased β-galactosidase activity correlated with elevated β-phenylethanol levels. | [73] |
| Precursor | Betaxanthin | Enzyme-coupled sensor. Highly yeast-active heterologous l-tyrosine hydroxylases were identified, based on increased betaxanthin fluorescence intensities in yeast expressing the plant DOPA dioxygenase. | [70] |
| p-Coumaric acid | p-Coumaric acid | Exogenous bacterial sensor. Droplet sorting of encapsulated p-coumaric acid producing yeast cells and p-coumaric acid sensing E. coli cells, to select producers based on bacterial YFP-fluorescence output. | [80] |
| Precursor | Muconic acid | Heterologous allosteric transcription factor. Used an Acinetobacter sp. transcriptional regulator to drive GFP expressing in the presence of muconic acid. | [77] |
| Mevalonate pathway | |||
| Precursor | Malonyl-CoA | Recombinant allosteric transcription factor sensor. Used a bacterial FapR transcription factor and FapO operator pair to identify strains from a genome-wide overexpression library that produce high levels of malonyl-CoA. | [75,76] |
| Precursor | Isopentenyl diphosphate | Synthetic transcription factor to allow feedback-regulated evolution of phenotype. Higher intracellular IPP concentrations resulted in increased GAL10 transcription, generating an evolvable growth phenotype on galactose | [74] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Wyk, N.; Kroukamp, H.; Pretorius, I.S. The Smell of Synthetic Biology: Engineering Strategies for Aroma Compound Production in Yeast. Fermentation 2018, 4, 54. https://doi.org/10.3390/fermentation4030054
Van Wyk N, Kroukamp H, Pretorius IS. The Smell of Synthetic Biology: Engineering Strategies for Aroma Compound Production in Yeast. Fermentation. 2018; 4(3):54. https://doi.org/10.3390/fermentation4030054
Chicago/Turabian StyleVan Wyk, Niël, Heinrich Kroukamp, and Isak S. Pretorius. 2018. "The Smell of Synthetic Biology: Engineering Strategies for Aroma Compound Production in Yeast" Fermentation 4, no. 3: 54. https://doi.org/10.3390/fermentation4030054
APA StyleVan Wyk, N., Kroukamp, H., & Pretorius, I. S. (2018). The Smell of Synthetic Biology: Engineering Strategies for Aroma Compound Production in Yeast. Fermentation, 4(3), 54. https://doi.org/10.3390/fermentation4030054





