Enzymes for Wine Fermentation: Current and Perspective Applications
Abstract
1. Introduction
2. Pectinases
2.1. Commercial Pectinases
- Polygalacturonase (homogalacturonan-hydrolase) (PG): hydrolytic depolymerization of the polygalacturonic acid chain. One can differentiate enzymes that cleave either single galacturonic acid units from the chain end (exo-activity, exoPG, EC 3.2.1.67), or in the middle of the chain (endo-activity, endoPG, EC 3.2.1.15).
- Pectinlyase/pectate lyase (EC 4.2.2.2 and 4.2.2.9): nonhydrolytic cleavage of the polygalacturonic acid chain.
- Pectinesterase (EC 3.1.1.11): hydrolytic cleavage of methanol from the d-galacturonic acid chain, causing drastic viscosity reduction in the liquid portion of the mash and better must flow.
- Acetylesterase (EC 3.1.1.6): cleaves acetyl residues from d-galacturonic acid with release of acetic acid. By this way the interfering acetyl residues at the connecting points of the side chains of the “hairy regions” are removed which facilitates further enzymatic degradation.
2.1.1. Effect on Juice Extraction, Clarification and Filtration
2.1.2. Effect on Color Extraction
2.1.3. Immobilization
2.2. Yeast Pectinases
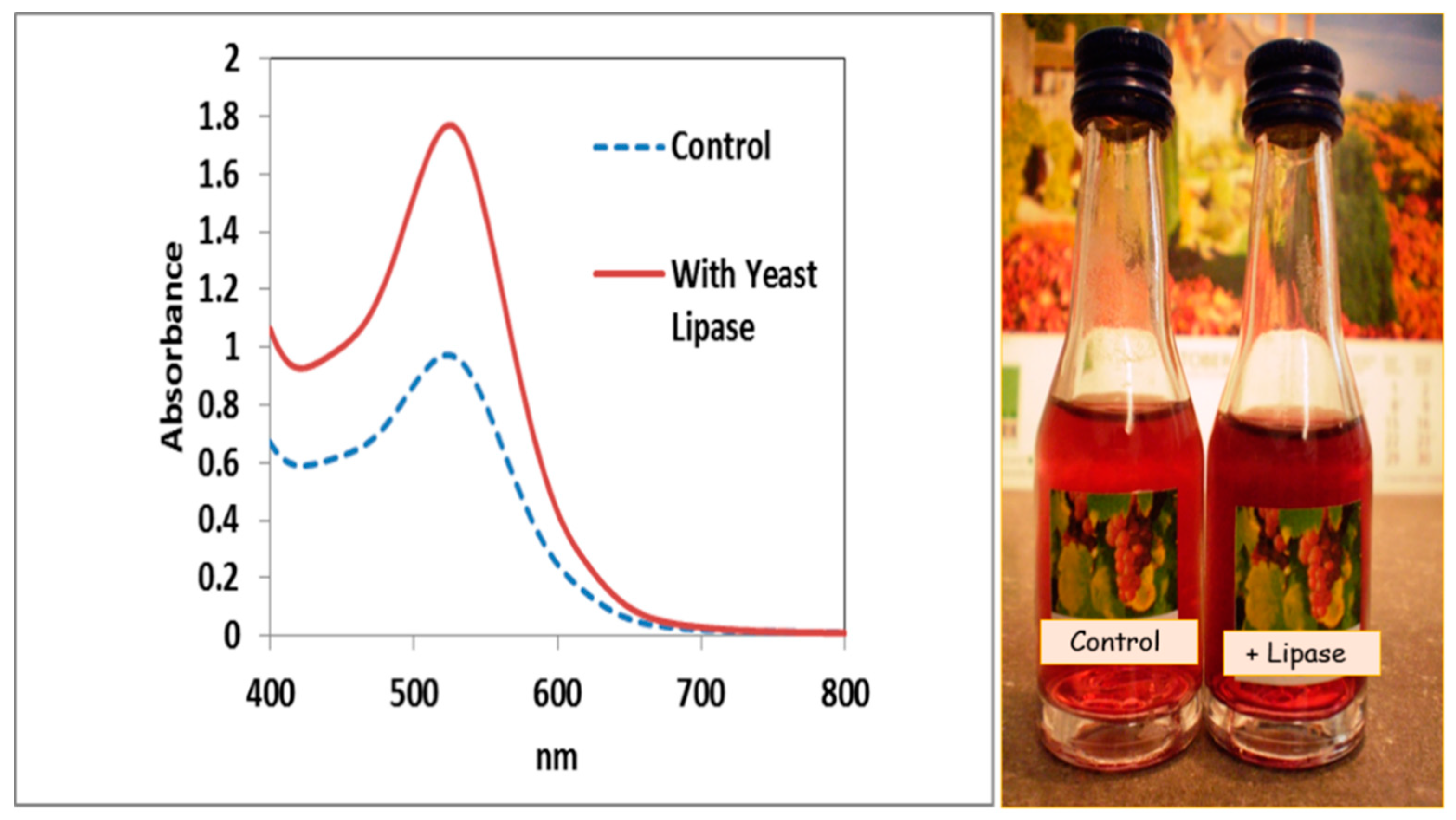
3. Lipases
4. Glucanases
4.1. Commercial Glucanases
4.2. Microbial Glucanases
5. Glycosidases
6. Esterhydrolases and -Synthetases
7. Proteinases
7.1. Proteases from Fungal and Plant Sources
7.2. Microbial Proteases
8. Phenoloxidases
9. Urease
10. Lysozyme
11. Legislative Regulations
12. Conclusions
Conflicts of Interest
References
- Mojsov, K. Use of enzymes in wine making: A review. Int. J. Market. Technol. 2013, 3, 112–127. [Google Scholar]
- Mojsov, K.; Andronikov, D.; Janevski, A.; Jordeva, S.; Zezova, S. Enzymes and wine—The enhanced quality and yield. Adv. Technol. 2015, 4, 94–100. [Google Scholar] [CrossRef]
- Ugliano, M. Enzymes in winemaking. In Wine Chemistry and Biochemistry; Morena-Arribas, M.V., Polo, M.C., Eds.; Springer: New York, NY, USA, 2009; pp. 103–126. [Google Scholar]
- Van Rensburg, P.; Pretorius, I.S. Enzymes in winemaking: Harnessing natural catalysts for efficient biotransformations: A review. S. Afr. J. Enol. Vitic. 2000, 21, 52–73. [Google Scholar]
- Hüfner, E.; Haßelbeck, G. Application of microbial enzymes during winemaking. In Biology of Microorganisms on Grapes, in Must and in Wine, 2nd ed.; König, H., Unden, G., Fröhlich, J., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 635–658. [Google Scholar]
- Mojsov, K.; Ziberoski, J.; Bozinovic, Z. The effect of pectolytic enzyme treatments on red grapes mash of Vranec on grape juice yields. Perspect. Innov. Econ. Bus. 2011, 7, 84–86. [Google Scholar] [CrossRef]
- Mojsov, K.; Ziberoski, J.; Bozinovic, Z.; Petreska, M. A comparison of effects of three commercial pectolytic enzyme preparations in red winemaking. Int. J. Pure Appl. Sci. Technol. 2010, 1, 127–136. [Google Scholar]
- Mojsov, K.; Ziberoski, I.; Bozinovic, Z.; Petreska, M. Effects of pectolytic enzyme treatments on white grape mashs of Smedervka on grape juice yields and volume of lees. In Proceedings of the 46th Croatian and 6th International Symposium on Agriculture, Opatija, Croatia, 14–18 February 2011; pp. 968–971. [Google Scholar]
- Cosme, F.; Jordão, A.M. Grape phenolic composition and antioxidant capacity. In Wine—Phenolic Composition, Classification and Health Benefits; El Rayess, Y., Ed.; Nova Publishers: New York, NY, USA, 2014; pp. 1–40. [Google Scholar]
- Sommer, S.; Cohen, S.D. Comparison of different extraction methods to predict anthocyanin concentration and color characteristics of red wines. Fermentation 2018, 4, 39. [Google Scholar] [CrossRef]
- Reyes, N.; Rivas Ruiz, I.; Dominguez-Espinosa, R.; Solis, S. Influence of immobilization parameters on endopolygalacturonase productivity by hybrid Aspergillus sp. HL entrapped in calcium aliginate. Biochem. Eng. J. 2006, 32, 43–48. [Google Scholar] [CrossRef]
- Sario, K.; Demir, N.; Acar, J.; Mutlu, M. The use of commercial pectinase in the fruit industry, part 2: Determination of the kinetic behaviour of immobilized commercial pectinase. J. Food Eng. 2001, 47, 271–274. [Google Scholar]
- Romero, C.; Sanchez, S.; Manjon, S.; Iborra, J.L. Optimization of the pectinesterase/endo-d-polygalacturonase immobilization process. Enzym. Microb. Technol. 1989, 11, 837–843. [Google Scholar] [CrossRef]
- Lozano, P.; Manjon, A.; Iborra, J.L.; Canovas, M.; Romojaro, F. Kinetic and operational study of a cross-flow reactor with immobilized pectolytic enzymes. Enzym. Microb. Technol. 1990, 12, 499–505. [Google Scholar] [CrossRef]
- Ramirez, H.L.; Gómez Brizuela, L.; Úbeda Iranzo, J.; Areval-Villena, M.; Briones Pérez, A.I. Pectinase immobilization on a chitosan-coated chitin support. J. Food Proc. Eng. 2016, 39, 97–104. [Google Scholar] [CrossRef]
- Charoenchai, C.; Fleet, G.H.; Henschke, P.A.; Todd, B.E.N. Screening of non-Saccharomyces wine yeasts for the presence of extracellular hydrolytic enzymes. Aust. J. Grape Wine Res. 1997, 3, 2–8. [Google Scholar] [CrossRef]
- Fernández, M.; Úbeda, J.F.; Briones, A.I. Typing of non-Saccharomyces yeasts with enzymatic activities of interest in winemaking. Int. J. Food Microbiol. 2000, 59, 29–36. [Google Scholar] [CrossRef]
- Strauss, M.L.A.; Jolly, N.P.; Lambrechts, M.G.; van Rensburg, P. Screening for the production of extracellular hydrolytic enzymes by non-Saccharomyces wine yeasts. J. Appl. Microbiol. 2001, 91, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Claus, H. Microbial Enzymes: Relevance for Winemaking. In Biology of Microorganisms on Grapes, in Must and in Wine, 2nd ed.; König, H., Unden, G., Fröhlich, J., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 315–338. [Google Scholar]
- Mateo, J.J.; Maicas, S. Application of Non-Saccharomyces yeasts to winemaking process. Fermentation 2016, 2, 14. [Google Scholar] [CrossRef]
- Merín, M.G.; Mendoza, L.M.; Farías, M.E.; de Ambrosini, V.I.M. Isolation and selection of yeast from wine grape ecosystem secreting cold-active pectinolytic activity. Int. J. Food Microbiol. 2011, 147, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Sahay, S.; Hanid, B.; Singh, P.; Ranjan, K.; Chauhan, D.; Rana, R.S.; Chaurse, V.K. Evaluation of pectinolytic activities for oenological uses from psychrotrophic yeasts. Lett. Appl. Microbiol. 2013, 57, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Merín, M.G.; de Ambrosini, V.I.M. Highly cold-active pectinases under wine-like conditions from non-Saccharomyces yeasts for enzymatic production during winemaking. Lett. Appl. Microbiol. 2015, 60, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Gallander, J.F.; Peng, A.C. Lipid and fatty acid composition of different wine grapes. Am. J. Enol. Vitic. 1980, 31, 24–27. [Google Scholar]
- Pueyo, E.; Martinez-Rodriquez, A.; Polo, M.C.; Santa-Maria, G.; Bartomé, B. Release of lipids during yeast autolysis in model wine. J. Agric. Food Chem. 2000, 48, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Fragopoulou, E.; Antonopoulou, S.; Demopoulos, C.A. Biologically active lipids with antiatherogenic properties from white wine and must. J. Agric. Food Chem. 2002, 50, 2684–2694. [Google Scholar] [CrossRef] [PubMed]
- Vakhlu, J.; Kour, A. Yeast lipases: Enzyme purification, biochemical properties and gene cloning. Electron. J. Biotechnol. 2006, 9. [Google Scholar] [CrossRef]
- Matthews, A.; Grbin, P.R.; Jiranek, V. A survey of lactic acid bacteria for enzymes of interest in oenology. Aust. J. Grape Wine Res. 20026, 12, 235–244. [Google Scholar] [CrossRef]
- Madrigal, T.; Maicas, S.; Tolosa, J.J.M. Glucose and ethanol tolerant enzymes produced by Pichia (Wickerhamomyces) isolates from enological ecosystems. Am. J. Enol. Vitic. 2013, 64, 126–133. [Google Scholar] [CrossRef]
- Molnárova, J.; Vadkertiová, R.; Stratilová, E. Extracellular enzymatic activities and physiological profiles of yeasts colonizing fruit trees. J. Basic Microbiol. 2014, 51, S74–S84. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulou, M.; Lonvauf-Funel, A.; Dols-Lafargue, M. Polysaccharide production by grapes must and wine microorganisms. In Biology of Microorganisms on Grapes, in Must and in Wine, 2nd ed.; König, H., Unden, G., Fröhlich, J., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 241–258. [Google Scholar]
- Pozo-Bayón, M.A.; Monagas, M.; Bartolomé, B.; Moreno-Arribas, M.V. Wine features related to safety and consumer health: An integrated perspective. Crit. Rev. Food Sci. Nutr. 2012, 52, 31–54. [Google Scholar] [CrossRef] [PubMed]
- Campos, F.M.; Couto, J.A.; Hogg, T. Utilisation of natural and by-products to improve wine safety. In Wine Safety, Consumer Preference, and Human Health; Morena-Arribas, M.V., Bartolomé Sualdea, B., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 27–49. [Google Scholar]
- Marchal, R.; Jeandet, P. Use of enological additives for colloids and tartrate salt stabilization in white wines and for improvement of sparkling wine foaming properties. In Wine Chemistry and Biochemistry; Morena-Arribas, M.V., Polo, M.C., Eds.; Springer: New York, NY, USA, 2010; pp. 127–158. [Google Scholar]
- Venturi, F.; Andrich, G.; Quartacci, M.F.; Sanmartin, C.; Andrich, L.; Zinnai, A. A kinetic method to identify the optimum temperature for glucanase activity. S. Afr. J. Enol. Vitic. 2013, 34, 281–286. [Google Scholar] [CrossRef]
- Styger, G.; Prior, B.; Bauer, F.F. Wine flavor and aroma. J. Ind. Microbiol. 2011, 38, 1145–1159. [Google Scholar] [CrossRef] [PubMed]
- Ugliano, M.; Henschke, P.A. Yeasts and wine flavour. In Wine Chemistry and Biochemistry; Morena-Arribas, M.V., Polo, M.V., Eds.; Springer: New York, NY, USA, 2010; pp. 313–392. [Google Scholar]
- Hjelmeland, A.K.; Ebeler, S.E. Glycosidically bound volatile aroma compounds in grapes and wine: A review. Am. J. Enol. Vitic. 2015, 66, 1–10. [Google Scholar] [CrossRef]
- Mateo, J.J.; DiStefano, R. Description of the beta-glucosidase activity of wine yeasts. Food Microbiol. 1997, 14, 583–591. [Google Scholar] [CrossRef]
- Maicas, S.; Mateo, J.J. Microbial glycosidases for wine production. Beverages 2016, 2, 20. [Google Scholar] [CrossRef]
- Perez-Martin, F.; Sesena, S.; Miguel Izquierdo, P.; Martin, R.; Llanos Palop, M. Screening for glycosidase activities of lactic acid bacteria as a biotechnological tool in oenology. World J. Microbiol. Biotechnol. 2012, 28, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Michlmayer, H.; Nauer, S.; Brandes, W.; Schumann, C.; Kulbe, K.D.; del Hierro, A.M.; Eder, R. Release of wine monoterpenes from natural precursors by glycosidases from Oenococcus oeni. Food Chem. 2012, 135, 80–87. [Google Scholar] [CrossRef]
- Iranzo, J.F.U.; Pérez, A.I.B.; Cañas, P.M.I. Study of oenological characteristics and enzymatic activities of wine yeasts. Food Microbiol. 1998, 15, 399–406. [Google Scholar] [CrossRef]
- Zoecklein, B.; Marcy, J.; Williams, S.; Jasinski, Y. Effect of native yeasts and selected strains of Saccharomyces cerevisiae on glycosyl glucose, potential volatile terpenes, and selected aglycons of white Riesling (Vitis vinfera L.) wines. J. Food Comp. Anal. 1997, 10, 55–65. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Xu, Y.; Li, J. Evaluating potential applications of indigenous yeasts and their β-glucosidases. J. Inst. Brew. 2015, 121, 642–648. [Google Scholar] [CrossRef]
- Delcroix, A.; Gunata, Z.; Sapis, L.C.; Salmon, J.M.; Baynone, C. Glycosidase activities of three enological yeast strains during winemaking: Effect on the terpenol content of Muscat wine. Am. J. Enol. Vitic. 1994, 45, 291–296. [Google Scholar]
- Sabel, A.; Martens, S.; Petri, A.; König, H.; Claus, H. Wickerhamomyces anomalus AS1: A new strain with potential to improve wine aroma. Anal. Microbiol. 2014, 64, 483–491. [Google Scholar] [CrossRef]
- Rodríguez, M.E.; Lopes, C.A.; van Broock, M.; Valles, S.; Ramón, D.; Caballero, A.C. Screening and typing of Patagonian wine yeasts for glycosidase activities. J. Appl. Microbiol. 2004, 96, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Gonzales-Pombo, O.; Farina, L.; Carreau, F.; Batista-Viera, F.; Brena, B.M. A novel extracellular beta-glucosidase from Issatschenkia terricola: Immobilization and application for aroma enhancement of white Muscat wine. Process Biochem. 2011, 46, 385–389. [Google Scholar] [CrossRef]
- Hu, K.; Zhu, X.L.; Mu, H.; Ma, Y.; Ullah, N.; Tao, Y.S. A novel extracellular glycosidase activity from Rhodotorula mucilaginosa: Its application potential in wine aroma enhancement. Lett. Appl. Microbiol. 2016, 62, 169–176. [Google Scholar] [CrossRef] [PubMed]
- López, M.C.; Mateo, J.J.; Maicas, S. Screening of β-glucosidase and β-xylosidase activities in four non-Saccharomyces yeast isolates. J. Food Sci. 2015, 80, C1696–C1704. [Google Scholar] [CrossRef] [PubMed]
- Schwentke, J.; Sabel, A.; Petri, A.; König, H.; Claus, H. The wine yeast Wickerhamomyces anomalus AS1 secretes a multifunctional exo-β-1,3 glucanase with implications for winemaking. Yeast 2014, 31, 349–359. [Google Scholar] [CrossRef] [PubMed]
- El Rayess, Y. Wine: Phenolic Composition, Classification and Health Benefits; Nova Publishers: New York, NY, USA, 2014; ISBN 978-1-63321-048-6. [Google Scholar]
- Padilla, B.; Gil, J.V.; Manzanares, P. Past and future of Non-Saccharomyces yeasts: From spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front. Microbiol. 2016, 7, 111. [Google Scholar] [CrossRef] [PubMed]
- Saerens, S.M.G.; Delvaux, F.; Verstrepen, K.J.; van Dijck, P.; Thevelein, J.M.; Delvaux, F.R. Parameters affecting ethyl ester production by Saccharomyces cerevisiae during fermentation. Appl. Environ. Microbiol. 2008, 74, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Maicas, S.; Gil, J.V.; Pardo, I.; Ferrer, S. Improvement of volatile composition of wines by controlled addition of malolactic bacteria. Food Res. Int. 1999, 32, 491–496. [Google Scholar] [CrossRef]
- Matthews, A.; Grbin, P.R.; Jiranek, V. Biochemical characterization of the esterase activities of wine lacti acid bacteria. Appl. Microbiol. Biotechnol. 2007, 77, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Fia, G.; Oliver, V.; Cavaglioni, A.; Canuti, V.; Zanoni, B. Side activities of commercial enzyme preparations and their influence on hydroxycinnamic acids, volatile compounds and nitrogenous components of white wine. Aust. J. Grape Wine Res. 2016, 22, 366–375. [Google Scholar] [CrossRef]
- Van Sluyter, S.C.; McRae, J.M.; Falconer, R.J.; Smith, P.A.; Bacic, A.; Waters, E.J.; Marangon, M. Wine protein haze: Mechanisms of formation and advances in prevention. J. Agric. Food Chem. 2015, 63, 4020–4030. [Google Scholar] [CrossRef] [PubMed]
- Weber, P.; Kratzin, H.; Brockow, K.; Ring, J.; Steinhart, H.; Paschke, A. Lysozyme in wine: A risk evaluation for consumers allergic to hen’s egg. Mol. Nutr. Food Res. 2009, 53, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Peñas, E.; di Lorenzo, C.; Uberti, F. Allergenic proteins in enology: A review on technological applications and safety aspects. Molecules 2015, 20, 13144–13174. [Google Scholar] [CrossRef] [PubMed]
- Rizzi, C.; Mainente, F.; Pasini, G.; Simonato, B. Hidden exogenous proteins in wine: Problems, methods of detection and related legislation—A review. Czech J. Food Sci. 2016, 34, 93–104. [Google Scholar] [CrossRef]
- Selitrennikoff, C.P. Antifungal proteins. Appl. Environ. Microbiol. 2001, 67, 2883–2894. [Google Scholar] [CrossRef] [PubMed]
- Jaeckels, N.; Tenzer, S.; Rosch, A.; Scholten, G.; Decker, H.; Fronk, P. β-Glucosidase removal due to bentonite fining during wine making. Eur. Food Res. Technol. 2015, 241, 253–262. [Google Scholar] [CrossRef]
- Esti, M.; Benucci, I.; Lombardelli, C.; Liburdi, K.; Garzillo, A.M.V. Papain from papaya (Carica papaya L.) fruit and latex: Preliminary characterization in alcoholic-acidic buffer for wine application. Food Bioprod. Process. 2013, 91, 595–598. [Google Scholar] [CrossRef]
- Benucci, I.; Esti, M.; Liburdi, K. Effect of free and immobilised stem bromelain on protein haze in white wine. Aust. J. Grape Wine Res. 2014, 20, 347–352. [Google Scholar] [CrossRef]
- Manrangon, M.; van Sluyter, S.C.; Robinson, E.M.C.; Muhlack, R.A.; Holt, H.E.; Haynes, P.A.; Godden, P.W.; Smith, P.A.; Waters, E.J. Degradation of white wine haze proteins by Aspergillopepsin I and II during flash pasteurization. Food Chem. 2012, 135, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Van Sluyter, S.C.; Warnock, N.I.; Schmidt, S.; Anderson, P.; van Kan, J.A.L.; Bacic, A.; Waters, E.J. Aspartic acid protease from Botrytis cinerea removes haze-forming proteins during white winemaking. J. Agric. Food Chem. 2013, 61, 9705–9711. [Google Scholar] [CrossRef] [PubMed]
- Dizy, M.; Bisson, L.F. Proteolytic activity of yeast strains during grape juice fermentation. Am. J. Enol. Vitic. 2000, 51, 155–167. [Google Scholar]
- Younes, B.; Cilindre, C.; Villaume, S.; Parmentier, M.; Jeandet, P.; Vasserot, Y. Evidence for an extracellular and proteolytic activity secreted by living cells of Saccharomyces cerevisiae PIR1: Impact on grape proteins. J. Agric. Food Chem. 2011, 59, 6239–6246. [Google Scholar] [CrossRef] [PubMed]
- Younes, B.; Cilindre, C.; Jeandet, P.; Vasserot, Y. Enzymatic hydrolysis of thermos-sensitive grape proteins by a yeast protease as revealed by a proteomic approach. Food Res. Int. 2013, 54, 1298–1301. [Google Scholar] [CrossRef]
- Song, L.; Chen, Y.; Du, Y.; Wang, X.; Guo, X.; Dong, J.; Xiao, D. Saccharomyces cerevisiae proteinase A excretion and wine making. World J. Microbiol. Biotechnol. 2017, 33, 210. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.A.; Choi, E.S.; Hong, W.K.; Kim, J.Y.; Ko, S.M.; Sohn, J.H.; Rhee, S.K. Proteolytic stability of recombinant human serum albumin secreted in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2000, 53, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Schlander, M.; Distler, U.; Tenzer, S.; Thines, E.; Claus, H. Purification and properties of yeast proteases secreted by Wickerhamomyces anomalus 227 and Metschnikowia pulcherrima 446 during growth in a white grape juice. Fermentation 2017, 3, 2. [Google Scholar] [CrossRef]
- Theron, L.W.; Bely, M.; Divol, B. Characterisation of the enzymatic properties of MpAPr1, an aspartic protease secreted by the wine yeast Metschikowia pulcherrima. J. Sci. Food Agric. 2017, 97, 3584–3593. [Google Scholar] [CrossRef] [PubMed]
- Theron, L.W.; Bely, M.; Divol, B. Monitoring the impact of an aspartic protease (MpApr1) on grape proteins and wine properties. Appl. Microbiol. Biotechnol. 2018, 102, 5173–5183. [Google Scholar] [CrossRef] [PubMed]
- Folio, P.; Ritt, J.F.; Alexandre, H.; Remize, F. Characterization of EprA, a major extracellular protein of Oenococcus oeni with protease activity. Int. J. Food Microbiol. 2008, 127, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Reid, V.J.; Theron, L.W.; du Toit, M.; Divol, B. Identification and partial characterization of extracellular aspartic protease genes from Metschnikowia pulcherrima IWBT Y1123 and Candida apicola IWBT Y1384. Appl. Environ. Microbiol. 2012, 78, 6838–6849. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.J.; Maicas, S.; Thießen, C. Biotechnological characterisation of extracellular proteases produced by enological Hanseniaspora isolates. Int. J. Food Sci. Technol. 2015, 50, 218–225. [Google Scholar] [CrossRef]
- Lagace, L.S.; Bisson, L.F. Survey of yeast acid proteases for effectiveness of wine haze reduction. Am. J. Enol. Vitic. 1990, 41, 147–155. [Google Scholar]
- Chasseriaud, L.; Miot-Sertier, C.; Coulon, J.; Iturmendi, N.; Moine, V.; Albertin, W.; Bely, M. A new method for monitoring the extracellular proteolytic activity of wine yeasts during alcoholic fermentation of grape must. J. Microbiol. Methods 2015, 119, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Claus, H.; Sabel, A.; König, H. Wine phenols and laccase: An ambivalent relationship. In Wine, Phenolic Composition, Classification and Health Benefits; Rayess, Y.E., Ed.; Nova Publishers: New York, NY, USA, 2014; pp. 155–185. [Google Scholar]
- Claus, H.; Decker, H. Bacterial tyrosinases. Syst. Appl. Microbiol. 2006, 29, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Fronk, P.; Hartmann, H.; Bauer, M.; Solem, E.; Jaenicke, E.; Tenzer, S.; Decker, H. Polyphenoloxidase from Riesling and Dornfelder wine grapes (Vitis vinfera) is a tyrosinase. Food Chem. 2015, 183, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Riebel, M.; Sabel, A.; Claus, H.; Fronk, P.; Xia, N.; Li, H.; König, H.; Decker, H. Influence of laccase and tyrosinase on the antioxidant capacity of selected phenolic compounds on human cell lines. Molecules 2015, 20, 17194–17207. [Google Scholar] [CrossRef] [PubMed]
- Riebel, M.; Sabel, A.; Claus, H.; Xia, N.; Li, H.; König, H.; Decker, H.; Fronk, P. Antioxidant capacity of phenolic compounds on human cell lines as affected by grape-tyrosinase and Botrytis-laccase oxidation. Food Chem. 2017, 229, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Sabel, A.; Bredefeld, S.; Schlander, M.; Claus, H. Wine phenolic compounds: Antimicrobial properties against yeasts, lactic acid and acetic acid bacteria. Beverages 2017, 3, 29. [Google Scholar] [CrossRef]
- Zinnai, A.; Venturi, F.; Sanmartin, C.; Quartacci, M.F.; Andrich, G. Chemical and laccase catalysed oxidation of gallic acid: Determination of kinetic parameters. Res. J. Biotechnol. 2013, 6, 62–65. [Google Scholar]
- Servili, M.; de Stefano, G.; Piacquadio, P.; Sciancalepore, V. A novel method for removing phenols from grape must. Am. J. Enol. Vitic. 2000, 51, 357–361. [Google Scholar]
- Minussi, R.C.; Rossi, M.; Bolgna, L.; Rotilio, D.; Pastore, G.M.; Durán, N. Phenols removal in musts; strategy for wine stabilization by laccase. J. Mol. Catal. B Enzym. 2007, 45, 102–107. [Google Scholar] [CrossRef]
- Maier, G.; Dietrich, H.; Wucherpfennig, K. Winemaking without SO2-with the aid of enzymes? Weinwirtschaft-Technik 1990, 126, 18–22. [Google Scholar]
- Brenna, O.; Bianchi, E. Immobilized laccase for phenolic removal in must and wine. Biotechnol. Lett. 1994, 16, 35–40. [Google Scholar] [CrossRef]
- Lettera, V.; Pezzella, C.; Cicatiello, P.; Piscitelli, A.; Giacobelli, V.G.; Galano, E.; Amoresano, A.; Sannia, G. Efficient immobilization of a fungal laccase and its exploitation in fruit juice clarification. Food Chem. 2016, 196, 1272–1278. [Google Scholar] [CrossRef] [PubMed]
- Lustrato, G.; De Leonardis, A.; Macciola, V.; Ranalli, G. Preliminary lab scale of advanced techniques as new tools to reduce ethylphenols content in synthetic wine. Agro Food Ind. Hi-Tech 2015, 26, 51–54. [Google Scholar]
- Smit, A.A.; du Toit, W.J.; du Toit, M. Biogenic amines in wine: Understanding the headache. S. Afr. J. Enol. Vitic. 2008, 29, 109–127. [Google Scholar] [CrossRef]
- Guo, Y.Y.; Yang, Y.P.; Peng, Q.; Han, Y. Biogenic amines in wine: A review. Int. J. Food Sci. Technol. 2015, 50, 1523–1532. [Google Scholar] [CrossRef]
- Preti, R.; Vieri, S.; Vinci, G. Biogenic amine profiles and antioxidant properties of Italian red wines from different price categories. J. Food Comp. Anal. 2016, 46, 7–14. [Google Scholar] [CrossRef]
- Sebastian, P.; Herr, P.; Fischer, U.; König, H. Molecular identification of lactic acid bacteria occurring in must and wine. S. Afr. J. Enol. Vitic. 2011, 32, 300–309. [Google Scholar] [CrossRef]
- Christ, E.; König, H.; Pfeiffer, P. Bacterial formation of biogenic amines in grape juice: Influence of culture conditions. Deutsche Lebensmittel-Rundschau 2012, 108, 73–78. [Google Scholar]
- Henríquez-Aedo, K.; Durán, D.; Garcia, A.; Hengst, M.B.; Aranda, M. Identification of biogenic amines-producing lactic acid bacteria isolated from spontaneous malolactic fermentation of Chilean red wines. LWT Food Sci. Technol. 2016, 68, 183–189. [Google Scholar] [CrossRef]
- Moreno-Arribas, M.V.; Polo, M.C. (Eds.) Amino acids and biogenic amines. In Wine Chemistry and Biochemistry; Springer: New York, NY, USA, 2010; pp. 163–189. [Google Scholar]
- Kushnereva, E.V. Formation of biogenic amines in wine production. Appl. Biochem. Microbiol. 2015, 51, 108–112. [Google Scholar] [CrossRef]
- Yagodina, O.V.; Nikol’skaya, E.B.; Khovanskikh, A.E.; Kormilitsyn, B.N. Amine oxidases of microorganisms. J. Evol. Biochem. Physiol. 2002, 38, 251–258. [Google Scholar] [CrossRef]
- Klinman, J.P. The multi-functional topa-quinone copper amine oxidases. Biochim. Biophys. Acta 2003, 1647, 131–137. [Google Scholar] [CrossRef]
- Corpillo, D.; Valetti, F.; Giuffrida, M.G.; Conti, A.; Rossi, A.; Finazzi-Agrò, A.; Giunta, C. Induction and characterization of a novel amine oxidase from the yeast Kluyveromyces marxianus. Yeast 2003, 20, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Bäumlisberger, M.; Moellecken, U.; König, H.; Claus, H. The potential of the yeast Debaryomyces hansenii H525 to degrade biogenic amines in food. Microorganisms 2015, 3, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Callejón, S.; Sendra, R.; Ferrer, S.; Pardo, I. Identification of a novel enzymatic activity from lactic acid bacteria able to degrade biogenic amines in wine. Appl. Microbiol. Biotechnol. 2014, 98, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Callejón, S.; Sendra, R.; Ferrer, S.; Pardo, I. Cloning and characterization of a new laccase from Lactobacillus plantarum J16 CECT 8944 catalyzing biogenic amine degradation. Appl. Microbiol. Biotechnol. 2016, 100, 3113–3124. [Google Scholar] [CrossRef] [PubMed]
- Lonvaud-Funel, A. Undesirable compounds and spoilage microorganisms in wine. In Wine Safety, Consumer Preference, and Human Health; Morena-Arribas, M.V., Bartolomé Sualdea, B., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 3–26. [Google Scholar]
- König, H.; Fröhlich, J. Lactic acid bacteria. In Biology of Microorganisms on Grapes, in Must and in Wine, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2017. [Google Scholar]
- Liburdi, K.; Benucci, I.; Esti, M. Lysozyme in wine: An overview of current and future applications. Compr. Rev. Food Sci. Food Saf. 2014, 13, 1062–1073. [Google Scholar] [CrossRef]
- Mojsov, K.; Petreska, M.; Ziberoski, J. Risks of microbial spoilage in wine. In Proceedings of the EHEDG World Congress on Hygienic Engineering & Design, Ohrid, Macedonia, 22–24 September 2011. [Google Scholar]
- Blättel, V.; Wirth, K.; Claus, H.; Schlott, P.; Pfeiffer, P.; König, H. A lytic enzyme cocktail from Streptomyces sp. B578 for the control of lactic and acid bacteria in wine. Appl. Microbiol. Biotechnol. 2009, 83, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Sommer, S.; Wegmann-Herr, P.; Wacker, M.; Fischer, U. Influence of lysozyme addition on hydroxycinnamic acids and volatile phenols during wine fermentation. Fermentation 2018, 4, 5. [Google Scholar] [CrossRef]
- Gonzales, R.; Tronchoni, J.; Quirós, M.; Morales, P. Genetic improvement and genetically modified microorganisms. In Wine Safety, Consumer Preference, and Human Heath; Morena-Arribas, M.V., Bartolomé Sualdea, B., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 71–96. [Google Scholar]
- Pretorius, I.S. Tailoring wine yeast for the new millenium: Novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [CrossRef]
- Pretorius, I.S. Conducting wine symphonics with the aid of yeast genomics. Beverages 2016, 2, 36. [Google Scholar] [CrossRef]

| Application/Process | Enzymatic Activity | Aim |
|---|---|---|
| Enhancement of filtration/clarification of must | Pectinolytic enzymes | Degradation of viscosity (pectin) |
| Mash fermentation/heating (red wine) | Pectinase with side activities (cellulases, hemicellulases) | Hydrolysis of plant cell wall polysaccharides. Improvement of skin maceration and color extraction of grapes, quality, stability, filtration of wines |
| Late phase of fermentation (white wine) | Glycosidases | Improvement of aroma by splitting sugar residues from odorless precursors |
| Young wine | Glucanases | Lysis of yeast cell walls, release of mannoproteins |
| Contaminated juice | Glucanases | Lysis of microbial exopolysacharides to improve clarification |
| Wine | Urease | Hydrolysis of yeast derived urea, preventing formation of ethyl carbamate |
| Must, wine | Lysozyme from hen egg | Control of bacterial growth |
| Must, wine | Proteases | Wine stabilization by prevention of protein haze; Reduction of bentonite demand |
| Enzyme | Remarks |
|---|---|
| Fungi (Botrytis cinerea) | |
| Glycosidases | Influence aromatic potential of infected grapes by release of volatile aroma compounds |
| Laccases | Broad specificity to phenolic compounds, cause oxidation and browning |
| Pectinases | Depolymerizing enzymes, cause degradation of plant cell walls and grape rotting |
| Cellulases | Multi-component complexes: endo-, exoglucanases and cellobiases; synergistic working, degrade plant cell walls |
| Lipases | Degrade lipids (e.g., in cell membranes) |
| Esterases | Involved in ester formation and degradation |
| Proteases | Aspartic proteases occur early in fungal infection, determine rate and extent of rotting caused by pectinases |
| Yeasts | |
| Glucosidases | Some yeasts produce β-glucosidases which are not repressed by glucose and are resistant to ethanol and low pH; positive influence on wine flavor |
| Glucanases | Occur extracellular, cell wall associated and intracellular, accelerate autolysis process and release of mannoproteins |
| Proteases | Acidic endoproteases accelerate autolysis process and degradation of grape proteins |
| Pectinases | Degrade pectin in grape cell walls |
| Lactic acid bacteria | |
| Malolactic enzymes | Convert malic acid to lactic acid |
| Esterases | Involved in ester formation and degradation |
| Glycosidases | Deliberate flavor compounds |
| Lipases | Degrade lipids |
| Lichenases, Glucanases, Cellulases, Xylanases | Degradation of polysaccharides |
| Proteases | Hydrolysis of proteins |
| Tannases | Hydrolysis of tannins (polymeric phenolic compounds) |
| Laccases | Oxidation of phenolic compounds |
| Supplier | Enzyme Preparation | Purpose of Application |
|---|---|---|
| AEB, South Africa | Pectocel L | Improvement of clarification, filtration and product yield |
| Endozym Pectoflot | Improvement of clarification, filtration and product yield | |
| Endozym Contact Pelliculaire | Enhancement of extraction and color stabilization | |
| Endozym Rouge | Enhancement of extraction and color stabilization | |
| Endozyme Active | Improvement of clarification, filtration and product yield | |
| Begerow, Germany | Siha Panzym Extract G | Enhanced extraction and release of color and aroma |
| Siha Panzym Clair Rapide G | Improvement of clarification, filtration and product yield | |
| Siha Panzym Fino G (β-Glucanase) | Improvement of clarification, filtration and sensory | |
| Siha Panzym Arome G (β-Glucosidase) | Enhanced aroma development | |
| Darleon, South Africa | Influence | Improvement of clarification, filtration and product yield |
| Enzym’ Color Plus | Enhancement of extraction and color stabilization | |
| DSM, Switzerland | Rapidase Filtration | Improvement of clarification, filtration and product yield |
| Rapidase Vino Super | Improvement of clarification, filtration and product yield | |
| Enartis, Italy | Uvazym 1000S | Clarification of white juices—facilitation of fining and filtration |
| Progress Quick | Must flotation | |
| Uvazym couleur | Enhanced extraction during short macerations | |
| Erbslöh, Germany | Trenolin bukett DF (β-Glycosidase) | Enhanced aroma development—Improvement of clarification |
| Trenolin Super DF | Improvement of clarification, filtration and product yield | |
| Trenolin Flot DF | Must flotation | |
| Trenolin 4000 DF | Enhancement of sugar yield | |
| Trenolin Filtro DF (β-Glucanase) | Improvement of clarification and filtration; Hydrolysis of Botrytis cinerea exopolysaccharide slime | |
| Trenolin Bukett DF | Enhance of color and aroma release from red grapes | |
| Laffort, France | Lafazym press | Enhanced color and tannin extraction—Facilitation of fining and filtration |
| Lafazym CL | Improvement of clarification, filtration and product yield | |
| Lafase 60 | Improvement of clarification, filtration and product yield | |
| Lafase HE | Enhancement of extraction and color stabilization | |
| Lallemand, France | Lallemand EX | Enhancement of extraction and color stabilization |
| Lallemand OE | Enhancement of extraction and color stabilization | |
| Novo Nordisk, Denmark | Novoclair FCE | Improvement of clarification, filtration and product yield |
| Vinozym EC | Enhancement of extraction and color stabilization | |
| Glucanex (β-Glucanase) | Improvement of clarification and filtration; Hydrolysis of Botrytis cinerea exopolysaccharide slime | |
| Ultrazym | Improvement of clarification and filtration | |
| Pectinex Superpress | Improvement of clarification and filtration | |
| Valley Research, USA | Crystalzyme | Rapid clarification–Color improvement–Increased complexity–Process efficiency |
| Species | Specificity | Substrate | MW(kDa) |
|---|---|---|---|
| Candida albicans | Endo-β-1,3- | L, OL, P | 49 |
| Exo-β-1,3- | L | 107 | |
| Candida hellenica | nd | G | nd |
| Candida lambica | nd | G | nd |
| Candida pulcherrima | nd | G, Li | nd |
| Candida stellata | nd | G, Li | nd |
| Candida utilis | Endo-β-1-3- | L, PNPG | 20 |
| Exo-β-1,3-1,6- | L, P, PNPG | 20 | |
| Endo-β-1,3- | L, OL | 21 | |
| Kloeckera apiculata | nd | G, Li | nd |
| Kluyveromyces phaseolosporus | Endo-β-1,3-(I) | L, OL | 180 |
| Exo-β-1,3-(II) | L, OL | 45 | |
| Exo-β-1,3-1,6-(III) | L, P | 18.5 | |
| Exo-β-1,3-1,6-(IV) | L, Ol, P | 8.7 | |
| Pichia polymorpha | Endo-β-1,3-(I) | L, OL | 47 |
| Exo-β-1,3-1,6-(II) | L, OL, P, PNPG | 40 | |
| Exo-β-1,3-(III) | L, PNPG | 30 | |
| Schizosaccharomyces pombe | Endo-β-1,3-(I) | L, OL | 160 |
| Endo-β-1-3-(II) | L, OL | 75 | |
| Schizosaccharomyces versatilis | Endo-β-1,3- | L, OL | 97 |
| Exo-β-1,3-1,6- | L, P, PNPG | 43 | |
| Wickerhamomyces anomalus AS1 | Exo-β-1,3- | L, PNPG | 47.5 |
| Species | Enzymatic Activities* | ||||
|---|---|---|---|---|---|
| β-d-Glucosidase | α-l-Arabino-Furanosidase | α-l-Rhamnosidase | β-d-Xylosidase | Carbon-Sulfur Lyase | |
| Aureobasidium pullulans | + | + | + | ||
| Brettanomyces anomalus | + | ||||
| Candida guillermondii | + | + | + | ||
| Candida molischiana | + | ||||
| Candida stellata | + | + | + | ||
| Candida utilis | + | ||||
| Candida zemplinia | + | ||||
| Debaryomyces castelli | + | ||||
| Debaryomyces hansenii | + | ||||
| Debaryomyces polymorphus | + | ||||
| Debaryomyces pseudopolymorphus | + | ||||
| Debaryomyces vannjii | + | ||||
| Hanseniaspora guillermondii | + | ||||
| Hanseniaspora osmophila | + | + | |||
| Hanseniaspora vineae | + | + | + | + | |
| Hanseniaspora uvarum | + | + | + | + | |
| Issatschenkia terricola | + | ||||
| Kluyveromyces thermotolerans | + | + | |||
| Metschnikowia pulcherrima | + | + | + | ||
| Pichia angusta | + | ||||
| Picha anomala | + | + | + | + | |
| Pichia capsulata | + | ||||
| Pichia guilliermondii | + | ||||
| Pichia kluyveri | + | ||||
| Pichia membranaefaciens | + | + | |||
| Saccharomycodes ludwigii | + | ||||
| Schizosaccharomyces pombe | + | ||||
| Sporidiobolus pararoseus | + | ||||
| Torulasporus delbrueckii | + | + | |||
| Torulasporus asahii | + | ||||
| Wickerhamomyces anomalus | + | + | + | ||
| Zygosaccharomyces bailii | + | ||||
| Species | Mode of Identification | Characterization | Reference |
|---|---|---|---|
| Candida apicola | Skim milk agar (pH 3.5), Gene Sequencing | Aspartic protease CaPR1 (39.2 kDa) | [78] |
| Candida stellata | Casein agar | nd | [18] |
| Hanseniaspora guelliermondii Hansenispora valbyenis Hanseniapora occidentalis | Casein agar and broth (pH 6.0) | nd | [79] |
| Hanseniaspora uvarum | Skim milk agar (pH 3.5) | nd | [16] |
| Casein agar | nd | [18] | |
| Kloeckera apiculata | Enzymatic; Inhibitor studies | Acid endopeptidase | [80] |
| Metschnikowia pulcherrima | Skim milk agar (pH 3.5) | nd | [16] |
| Azocasein hydrolysis during fermentation of grape must | nd | [81] | |
| Skim milk agar (pH 3.5), Sequencing of protease gene, Purification | Aspartic protease pAPR1 (40.8 kDa) | [75,78] | |
| Casein agar | nd | [18] | |
| Skim milk agar (pH 4.5), Enzymatic; Inhibitor studies, LC-MS/MS | Aspartic protease | [74] | |
| Wickerhamomyces anomalus | Skim milk agar (pH 3.5) | nd | [16] |
| Skim milk agar (pH 4.5), Enzymatic; Inhibitor studies, LC-MS/MS | Aspartic protease WaAPR1 (47 kDa) | [74] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Claus, H.; Mojsov, K. Enzymes for Wine Fermentation: Current and Perspective Applications. Fermentation 2018, 4, 52. https://doi.org/10.3390/fermentation4030052
Claus H, Mojsov K. Enzymes for Wine Fermentation: Current and Perspective Applications. Fermentation. 2018; 4(3):52. https://doi.org/10.3390/fermentation4030052
Chicago/Turabian StyleClaus, Harald, and Kiro Mojsov. 2018. "Enzymes for Wine Fermentation: Current and Perspective Applications" Fermentation 4, no. 3: 52. https://doi.org/10.3390/fermentation4030052
APA StyleClaus, H., & Mojsov, K. (2018). Enzymes for Wine Fermentation: Current and Perspective Applications. Fermentation, 4(3), 52. https://doi.org/10.3390/fermentation4030052





