Identification of Microflora in a Biological Brewer’s Wort Acidification Process Run Continuously for 20 Years
Abstract
1. Introduction
2. Materials and Methods
2.1. Media and Growth Conditions
2.2. Preparing Cryogenic Cultures
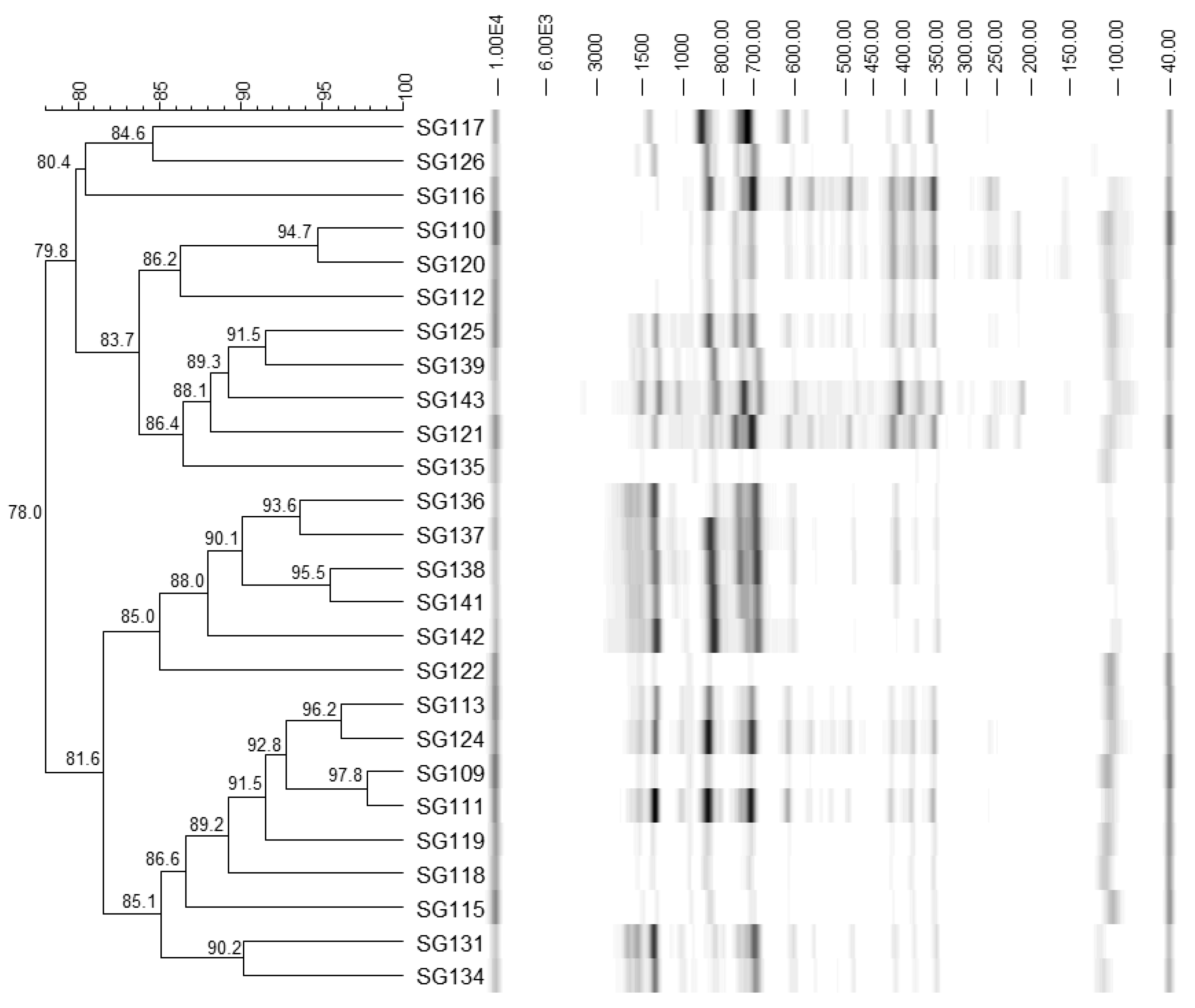
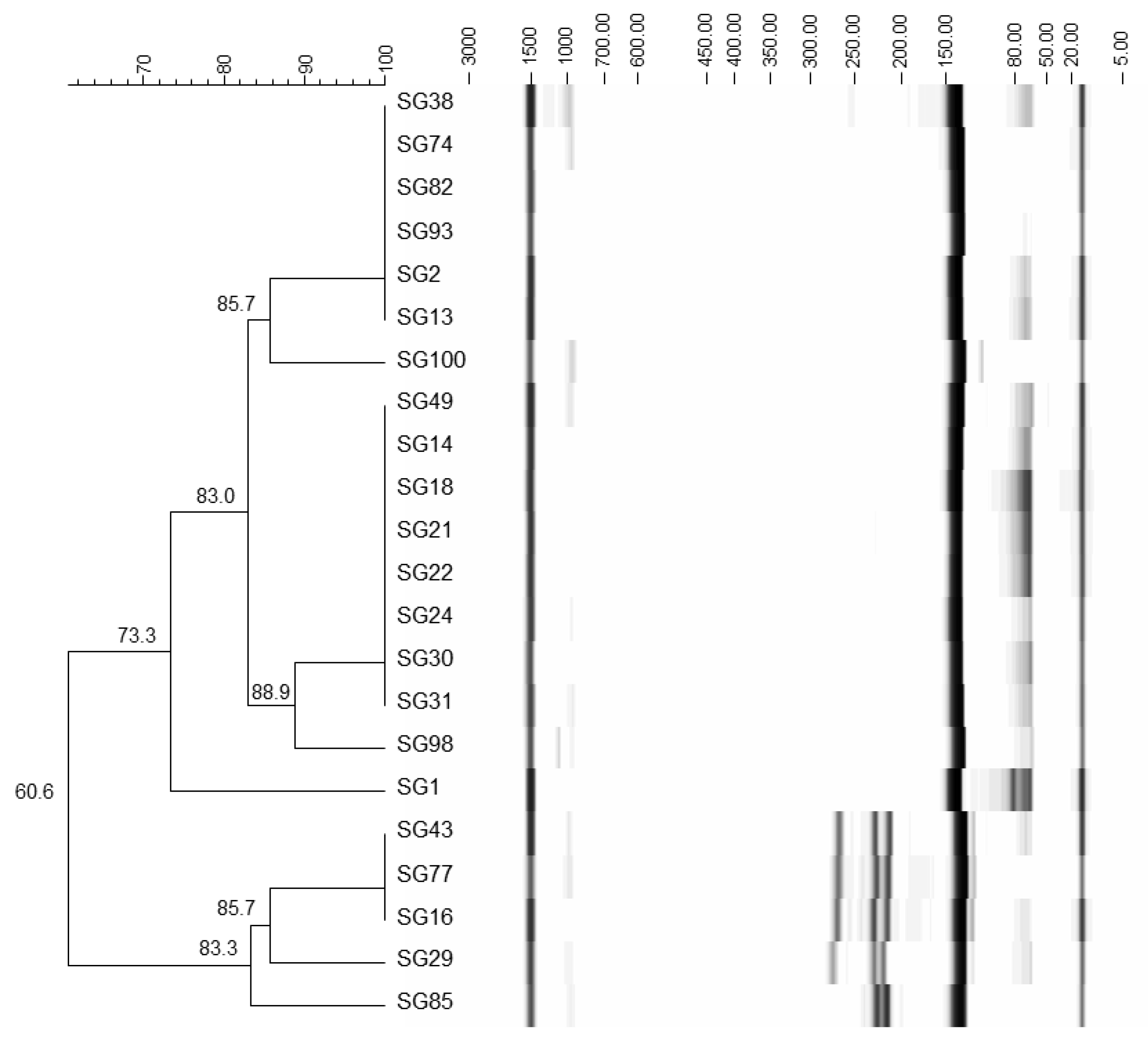
2.3. PCR and Fingerprinting
2.4. Sequencing of 16S and 26S rDNA
2.5. Real-Time PCR
2.6. Phenolic Off-Flavor Test
3. Results
3.1. Identification of Bacteria
3.2. Identification of Yeasts
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Back, W. Biologische Säuerung. Monatsschr. Brauwiss. 1988, 41, 152–156. [Google Scholar]
- Lindner, P. Über ein neues in Malzmaischen vorkommendes Milchsäure bildendes Ferment. Wochenschr. Brau. 1887, 4, 437–440. [Google Scholar]
- Leichmann, G. Über die im Brennereiprozeß bei der Bereitung von Kunsthefe auftretende spontane Milchsäuregarung (Bac. Delbrücki). Zentralbl. Bakteriol. 1896, 2, 281–285. [Google Scholar]
- Francke, O. Herstellung säuerlich schmeckender, insbesondere milchsauerer Biere. Deutsches Reichspatent Nr. 180726, 5 April 1906. [Google Scholar]
- Oliver-Daumen, B. Biological acidification in the brewing process—Part 1: Review of the literature. Brauwelt Int. 1988, 256–264. [Google Scholar]
- Grützmacher, J. Biologische Säuerung in der Brauerei. Brauwelt 1991, 40, 1762–1769. [Google Scholar]
- Back, W. Ausgewählte Kapitel der Brauereitechnologie; Hans Carl: Nürnberg, Germany, 2005. [Google Scholar]
- Lowe, D.P.; Ulmer, H.M.; Barta, R.C.; Goode, D.L.; Arendt, E.K. Biological Acidification of a Mash Containing 20% Barley Using Lactobacillus amylovorus FST 1.1: Its Effects on Wort and Beer Quality. J. Am. Soc. Brew. Chem. 2005, 63, 96–106. [Google Scholar] [CrossRef]
- Lowe, D.P.; Arendt, E.K. The Use and Effects of Lactic Acid Bacteria in Malting and Brewing with Their Relationships to Antifungal Activity, Mycotoxins and Gushing: A Review. J. Inst. Brew. 2004, 110, 163–180. [Google Scholar] [CrossRef]
- Vaughan, A.; O’Sullivan, T.; Sinderen, D. Enhancing the Microbiological Stability of Malt and Beer—A Review. J. Inst. Brew. 2005, 111, 355–371. [Google Scholar] [CrossRef]
- Bohak, I.; Back, W.; Richter, L.; Ehrmann, M.; Ludwig, W.; Schleifer, K.H. Lactobacillus amylolyticus sp. nov., Isolated from Beer Malt and Beer Wort. Syst. Appl. Microbiol. 1998, 21, 360–364. [Google Scholar] [CrossRef]
- Goranov, B.; Denkova, Z.; Kostov, G.; Denkova, R. Possibilities for the biological acidification of mash in the production of wort: Kinetics of lactic acid production in a free cell culture of Lactobacillus delbrueckii ssp. bulgaricus M3. Presented at 50 Years FoodRDI. Food Technologies and Health, International Scientific-Practical Conference, Plovdiv, Bulgaria, 8 November 2012. [Google Scholar]
- Boulton, C.; Quain, D. Brewing Yeast and Fermentation; Blackwell Science; Iowa State University Press: Malden, MA, USA; Ames, IA, USA, 2001. [Google Scholar]
- Versalovic, J.; Schneider, M.; de Bruijn, F.J.; Lupski, J.R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 1994, 5, 25–40. [Google Scholar]
- Hutzler, M. Getränkerelevante Hefen—Identifizierung und Differenzierung. Wie Können Hefen Praxisrelevant Unterschieden Werden, und wie Können Identifizierungsergebnisse Technologisch Bewertet Werden? Südwestdt. Verlag für Hochschulschriften: Saarbrücken, Germany, 2010; ISBN-13: 9783838114828. [Google Scholar]
- Zhang, H.; You, C.; Ren, J.; Xu, D.; Han, M.; Liao, W. A simple one-step PCR walking method and its application of bacterial rRNA for sequencing identification. Curr. Microbiol. 2014, 68, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Ehrmann, M.A.; Müller, M.R.A.; Vogel, R.F. Molecular analysis of sourdough reveals Lactobacillus mindensis sp. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Nossa, C.W.; Oberdorf, W.E.; Yang, L.; Aas, J.A.; Paster, B.J.; Desantis, T.Z.; Brodie, E.L.; Malamud, D.; Poles, M.A.; Pei, Z. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J. Gastroenterol. 2010, 16, 4135–4144. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 2003, 3, 417–432. [Google Scholar] [CrossRef]
- Brandl, A. Entwicklung und Optimierung von PCR-Methoden zur Detektion und Identifizierung von Brauereirelevanten Mikroorganismen zur Routine-Anwendung in Brauereien. Ph.D. Thesis, TU München, Freising, Germany, 2006. [Google Scholar]
- Salinas, F.; Garrido, D.; Ganga, A.; Veliz, G.; Martínez, C. Taqman real-time PCR for the detection and enumeration of Saccharomyces cerevisiae in wine. Food Microbial. 2009, 26, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Meier-Dörnberg, T.; Hutzler, M.; Michel, M.; Methner, F.-J.; Jacob, F. The Importance of a Comparative Characterization of Saccharomyces Cerevisiae and Saccharomyces Pastorianus Strains for Brewing. Fermentation 2017, 3, 41. [Google Scholar] [CrossRef]
- Stackebrand, E.; GOEBEL, B.M. Taxonomic Note: A Place for DNA-DNA Reassociation and 16S rRNA Sequence Analysis in the Present Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Thurston, P.A.; Tubb, R.S. Screening yeast strains for their ability to produce phenolic off-flavours: A simple method for determining phenols in wort and beer. J. Inst. Brew. 1981, 87, 177–179. [Google Scholar] [CrossRef]
- Gallone, B.; Steensels, J.; Prahl, T.; Soriaga, L.; Saels, V.; Herrera-Malaver, B.; Merlevede, A.; Roncoroni, M.; Voordeckers, K.; Miraglia, L.; et al. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 2016, 166, 1397–1410. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, M.; Pontes, A.; Almeida, P.; Barbosa, R.; Serra, M.; Libkind, D.; Hutzler, M.; Goncalves, P.; Sampaio, J.P. Distinct domestication trajectories in top-fermenting beer yeasts and wine yeasts. Curr. Biol. 2016, 26, 2750–2761. [Google Scholar] [CrossRef] [PubMed]
- Röcken, W.; Marg, C. Nachweis von Fremdhefen in der obergärigen Brauerei. Vergleich verschiedener Nährböden. Monatsschr. Brauwiss. 1983, 1983, 276–279. [Google Scholar]
| Primer | Sequence 5′ → 3′ | Reference |
|---|---|---|
| GTG5 | GTGGTGGTGGTGGTG | [14] |
| IGS2-314-fp | CGGGTAACCCAGTTCCTCACT GTAGCATATATTTCTTGTGTGAGAAAGGT | [15] |
| IGS2-314-rp | ||
| 16S-27f | AGAGTTTGATCM(C/A)TGGCTCAG TACGGY(C/T)TACCTTGTTACGACTT | [16] |
| 1492r | ||
| 612r | GTAAGGTTY(C/T)TNCGCGT | [17] |
| 926r | CCGTCAATTCM(C/A)TTTRAGT | [18] |
| NL1 | GCATATCAATAAGCGGAGGAAAAG GGTCCGTGTTTCAAGACGG | [19] |
| NL4 |
| Strain | Sequencing Primer | Accession Number (Length in bp) | NCBI Identification | Identities of the Target Microorganism (%) | Real-Time PCR Identification |
|---|---|---|---|---|---|
| SG1 | NL4 | KJ506733.1 (609) | S. cerevisiae | 99 | S. cerevisiae |
| SG14 | NL4 | CP006467.1 (1893211) | S. cerevisiae | 99 | S. cerevisiae |
| SG16 | NL4 | JQ672609.1 (601) | S. cerevisiae | 100 | S. cerevisiae |
| SG22 | NL4 | JQ277730.1 (9076) | S. cerevisiae | 99 | S. cerevisiae |
| SG82 | NL4 | JQ672587.1 (587) | S. cerevisiae | 99 | S. cerevisiae |
| SG85 | NL4 | JQ968592.1 (573) | S. cerevisiae | 100 | S. cerevisiae |
| SG93 | NL4 | JX103179.1 (620) | S. cerevisiae | 99 | S. cerevisiae |
| SG100 | NL4 | JX103179.1 (620) | S. cerevisiae | 100 | S. cerevisiae |
| SG109 | 1492r | Y17361.1 (1544) | L. amylolyticus | 99 | L. amylolyticus |
| NR_075048.1 (1575) | L. amylovorus | 99 | |||
| SG115 | 1492r | NR_117069.1 (1443) | L. amylolyticus | 99 | L. amylolyticus |
| NR_075048.1 (1575) | L. amylovorus | 99 | |||
| SG126 | 1492r | NR_029352.1 (1544) | L. amylolyticus | 99 | L. amylolyticus |
| EF120375.1 (1550) | L. amylovorus | 99 | |||
| SG139 | 1492r | Y17361.1 (1558) | L. amylolyticus | 99 | L. amylolyticus |
| NR_075048.1 (1575) | L. amylovorus | 99 |
| Yeast Strain | OG-COXII | SCTM | UG-LRE1 | UG300 | Sbp |
|---|---|---|---|---|---|
| S. cerevisiae SG1 | + | + | − | − | − |
| S. cerevisiae SG14 | + | + | − | − | − |
| S. cerevisiae SG16 | + | + | − | − | − |
| S. cerevisiae SG22 | + | + | − | − | − |
| S. cerevisiae SG82 | + | + | − | − | − |
| S. cerevisiae SG85 | + | + | − | − | − |
| S. cerevisiae SG93 | + | + | − | − | − |
| S. cerevisiae SG100 | + | + | − | − | − |
| Flavor | Precursor Cinnamic Acid, Product Styrene “Styrofoam-Like” | Precursor Ferulic Acid, 4-Vinylguaiacol “Clove-Like” | |
|---|---|---|---|
| Yeast | |||
| Frisinga-TUM 34/70® | − | − | |
| LeoBavaricus-TUM 68® | + | + | |
| S. cerevisiae SG1 | ++ | + | |
| S. cerevisiae SG14 | ++ | + | |
| S. cerevisiae SG16 | − | − | |
| S. cerevisiae SG22 | ++ | + | |
| S. cerevisiae SG24 | ++ | + | |
| S. cerevisiae SG29 | − | − | |
| S. cerevisiae SG82 | ++ | ++ | |
| S. cerevisiae SG85 | ++ | ++ | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hutzler, M.; Čmielová, J.; Frank, T.; Brandl, A.; Jacob, F.; Michel, M. Identification of Microflora in a Biological Brewer’s Wort Acidification Process Run Continuously for 20 Years. Fermentation 2018, 4, 51. https://doi.org/10.3390/fermentation4030051
Hutzler M, Čmielová J, Frank T, Brandl A, Jacob F, Michel M. Identification of Microflora in a Biological Brewer’s Wort Acidification Process Run Continuously for 20 Years. Fermentation. 2018; 4(3):51. https://doi.org/10.3390/fermentation4030051
Chicago/Turabian StyleHutzler, Mathias, Jana Čmielová, Tobias Frank, Andreas Brandl, Fritz Jacob, and Maximilian Michel. 2018. "Identification of Microflora in a Biological Brewer’s Wort Acidification Process Run Continuously for 20 Years" Fermentation 4, no. 3: 51. https://doi.org/10.3390/fermentation4030051
APA StyleHutzler, M., Čmielová, J., Frank, T., Brandl, A., Jacob, F., & Michel, M. (2018). Identification of Microflora in a Biological Brewer’s Wort Acidification Process Run Continuously for 20 Years. Fermentation, 4(3), 51. https://doi.org/10.3390/fermentation4030051