1. Introduction
The Mediterranean region, encompassing countries such as Spain, Italy, Greece, Turkey, and Portugal, accounts for over 80% of global olive oil production [
1]. In these countries, the long-standing tradition of olive cultivation has driven innovation not only in production processes but also in the valorization of olive byproducts, such as olive pomace (OP) and olive mill wastewater (OMW). Due to land-use restrictions and rising environmental concerns, circular economy strategies have gained momentum in Europe, promoting the reuse of agro-industrial residues. For example, Spain’s Order TED/92/2022 reclassified OP and OMW as a byproduct, enabling its use in energy generation, animal feed, and cosmetic and pharmaceutical manufacturing [
2]. These advances have been catalyzed by both regulatory initiatives and increasing consumer demand for sustainable eco-friendly practices [
3].
With an olive oil production of 40,000 tons per year, Argentina is the leading producer and exporter of olive oil in Latin America [
4,
5]; however, in contrast to Europe, it exhibits limited progress in the valorization of olive oil industry byproducts. The absence of regulatory incentives further discourages industrial stakeholders from investing in sustainable waste management practices. These limitations restrict the recovery and reuse of OP and OMW, hindering the adoption of circular economy approaches within the sector. Although Argentina holds significant potential for expansion, particularly through the reconversion and optimization of existing plantations [
6], such growth will inevitably result in increased volumes of solid and liquid residues. This scenario underscores the urgent need to develop environmentally sound and economically viable strategies for the management and valorization of these byproducts.
Given that the predominant extraction technology used in Argentina is the two-phase continuous centrifugation system, with oil yields ranging from 10% to 20%, a considerable volume of byproducts is generated during processing [
7]. Olive pomace is characterized by high acidity and elevated organic load, quantified by its chemical oxygen demand (COD) and biological oxygen demand (BOD), and a substantial concentration of polyphenolic compounds. While only about 2% of the phenolics present in olives are retained in the extracted oil, the vast majority accumulates in the pomace, contributing significantly to its toxicity [
1]. These physicochemical characteristics, combined with OP’s strong odor and pasty consistency, make it particularly difficult to handle, transport, and dispose of safely [
8]. Its direct release into the environment poses considerable ecotoxicological risks. Additionally, OP contains phytotoxic compounds, especially phenolic degradation products formed through microbial activity, that can severely inhibit seed germination, plant development, and the viability of soil microbial communities [
9].
Current strategies for managing OP include dehydration followed by residual oil extraction with hexane, after which the defatted pomace is used as boiler fuel [
1], or direct application to agricultural soils, composting, and recovery of phenolic compounds. While these approaches provide basic waste handling, they offer limited value addition and fail to fully exploit the resource potential of OP. Composting, one of the most common biological treatments, typically requires co-substrates to balance the C/N ratio and entails long processing periods, with maturation times extending up to 150–180 days [
10]. Although effective in reducing phenolic compounds, the lengthy treatment duration is a significant drawback. Similarly, the field application of OP has shown some agronomic benefits, but concerns over soil salinity and the need for long-term monitoring persist [
11,
12]. The application of low thermal treatments [
13] and natural deep eutectic solvents [
14] has proven effective for extracting hydrophilic phenols; however, these processes often involve multiple stages, controlled heating, and significant water consumption. These practices underscore the need for more efficient and innovative biological detoxification strategies that can shorten the processing times and increase the valorization outcomes.
Biological detoxification through solid-state fermentation (SSF) offers a sustainable, low-cost, and water-efficient strategy aligned with circular bioeconomy principles [
3]. Fungal species, particularly
Aspergillus sp., have demonstrated strong biodegradation potential for phenolic-rich lignocellulosic residues such as OP [
15]. Their GRAS status and ability to secrete a broad range of extracellular enzymes (e.g., cellulases, xylanases, pectinases, tannases) make them suitable agents for bioconversion processes [
16,
17]. In addition,
Aspergillus species thrive in low-water activity environments typical of SSF, making them particularly suitable for this technology [
16].
In high-production regions like Argentina, OP accumulation has become an environmental challenge, underscoring the need for scalable and efficient valorization alternatives. Within the circular economy framework, which promotes reuse, recycling, and recovery, agro-industrial residues such as OP emerge as valuable feedstocks for biotechnological applications [
18,
19]. In this context, SSF enables not only detoxification but also the generation of value-added bioproducts, such as hydrolytic enzymes, thus supporting sustainable and regionally adapted solutions for the olive oil industry.
While previous studies have applied SSF to OP, many rely on fungal–bacterial consortia, pretreated substrates, or extended composting times, often under laboratory conditions and without considerations of scalability [
10,
15]. In contrast, this study explores a dual-scale SSF system, from exploratory Petri dish trials to a fixed-bed bioreactor (FBR), using a native
Aspergillus sp. strain. Furthermore, mathematical modeling was applied to describe the detoxification and enzyme production kinetics, providing a predictive framework for future process intensification.
To enhance the fermentation performance and improve the substrate properties, grape pomace (GP), another agro-industrial residue abundantly available in the same region, was incorporated as a co-substrate. GP contributed to the nutritional balance of the medium, provided structurally diverse carbon sources, and acted as a bulking agent, improving the porosity and water retention, critical factors in SSF systems [
20]. In addition, the polyphenolic and sugar content of GP may have stimulated fungal enzyme production through induction mechanisms, as previously observed in mixed-residue fermentations [
21]. Despite its potential, GP has rarely been used in combination with OP under SSF conditions, representing an innovative aspect of this study.
In addition, a key challenge in SSF lies in monitoring the fungal growth and metabolic activity within solid heterogeneous matrices. Unlike submerged systems, direct biomass quantification is difficult, requiring indirect monitoring approaches [
22]. Among these, the carbon dioxide production rate (CPR) is widely used due to its linear correlation with substrate degradation and microbial respiration. This method provides relevant insights into biomass formation and enzymatic activity [
23]. Dry weight loss has also been reported as a simple and cost-effective indicator for evaluating fermentation performance and guiding bioprocess optimization [
24,
25,
26].
This study presents a fungal-based SSF strategy for the valorization of OP, aiming at both the detoxification and the production of industrially relevant enzymes. The approach is grounded in the use of a native
Aspergillus sp., isolated directly from OP samples obtained in the local olive oil production region. The use of a locally adapted strain offers ecological and operational advantages, such as improved tolerance to substrate-specific inhibitors and enhanced metabolic compatibility with the lignocellulosic matrix [
15]. The combination of OP and GP thus enabled the co-valorization of two abundant underutilized byproducts within a single integrated process.
Therefore, the objective of this study was to develop and evaluate a feasible SSF process using a native Aspergillus sp. and a mixed substrate of OP and GP, with the aim of achieving biological detoxification of OP alongside the production of hydrolytic enzymes, as a contribution to regional circular bioeconomy strategies. Unlike previous studies, this work investigates a dual-scale SSF system and integrates mathematical modeling to describe the weight loss, enzyme production, and detoxification kinetics in the FBR, offering valuable insights into the scalability and optimization of SSF-based biovalorization platforms.
2. Materials and Methods
The experimental design was structured in two sequential stages. In the first stage, a Petri dish assay was conducted to assess the fungal growth and preliminary detoxification capacity, using color loss, pH change, and weight reduction as indicators. In the second stage, the process was scaled up to an FBR, where substrate biotransformation was monitored through dry and wet weight loss (DWL and WWL), total phenolic content reduction, and enzymatic activity (cellulase, xylanase, endo-polygalacturonase, and tannase). Additionally, mathematical modeling of DWL and WWL and detoxification and enzymatic kinetics was performed to describe the process dynamics, providing insights rarely addressed in SSF studies with agro-industrial residues.
2.1. Byproduct Sampling and Conditioning
The OP was obtained from a local oil mill in Carpintería, Pocito, San Juan, during the 2023 olive processing season, from a two-phase oil extraction system. The GP was collected during 2023 harvest season from the temporary storage area for agro-industrial residues at a winery facility located in San Martín, San Juan. Both byproducts were placed in sterile 2 L bags and transported to the laboratory under refrigerated and aseptic conditions. Upon arrival, the samples were homogenized and subdivided into 100 g aliquots, which were stored at −20 °C to prevent undesired fermentation.
The OP and GP samples intended for the SSF experiments were dried in a convection oven (Biobase Group, model BOV-T25F, Jinan, China) at 60 ± 5 °C, until a constant weight was achieved. The dried OP and GP were then stored in labeled and sealed plastic bags until further use. Prior to the experiments, the samples were ground using a laboratory blade mill (Damai HC-100, 850 W) and sieved through a Tyler tower, using mesh sizes No. 6 (3.35 mm) and No. 4 (4.75 mm). The dried retained fraction (portion with a size between 3.35 mm and 4.75 mm) was used and will hereafter be referred to as DOP and DGP, respectively. Samples were stored in labeled and sealed plastic bags at room temperature until use.
For fungal isolation, OP samples were thawed at low temperatures to prevent undesired spontaneous fermentations.
2.2. Fungal Isolation, Preservation, and Inoculum Propagation
To isolate native fungal strains from OP, a surface sterilization protocol was applied to minimize external contamination and ensure that the recovered microorganisms were naturally associated with the OP [
27]. Small solid fragments of OP were selected. Each fragment was washed with sterile distilled water, immersed in 1% sodium hypochlorite for 10 min, and rinsed twice with sterile distilled water.
Three small solid fragments obtained previously, were put directly at different locations on the surface of Petri dishes containing 20 mL of Potato Dextrose Agar (PDA), supplemented with 5 g·L
−1 of DOP and chloramphenicol (0.25 g·L
−1) [
28]. This culture medium will hereinafter be designated as PDA-DOP. Then, the Petri dishes were incubated at 25 °C for 5–7 days.
The emerging fungal colonies were subcultured using a T-streak method to obtain pure isolates. These purified strains were subsequently examined under the microscope for preliminary morphological identification, including the hyphal structure, conidiophores, and conidia characteristics. Fungal isolates were then grouped based on their macroscopic morphological features, such as the colony color, mycelial texture, and spore formation. To further characterize these traits, microscopic examinations were performed on fresh microcultures grown on PDA-DOP, following the slide culture method described in [
29]. Lactophenol Cotton Blue staining was applied [
30] to allow detailed visualization of individual fungal structures, facilitating the accurate observation of morphological features critical for taxonomic identification [
31]. Selected isolates were preserved on PDA-DOP slants and stored at 4 °C for short-term maintenance.
For inoculum propagation, the selected isolate was subculture on PDA-DOP and incubated at 25 °C for 5–7 days to promote sporulation. Conidias were harvested from the surface by adding a 0.1% (v/v) Tween 80 sterile solution, followed by agitation at 160–180 rpm using an orbital shaker (DLAB, SK-O330-Pro) for 20 min. The resulting spore stock suspension was counted using a Neubauer chamber, and serial dilutions were prepared to obtain the desired inoculum concentration for SSF assays.
2.3. Culture Medium for SSF
Based on the previously characterized physicochemical properties of OP by our research group [
32], a 2:1 (
w/
w) mixture of DOP and DGP was selected as the fermentation substrate. This formulation was designed to mitigate the potential antimicrobial effects of polyphenolic compounds present in OP, which contains approximately 98% of the total polyphenols found in the olive fruit and could adversely affect the fermentation efficiency [
8]. Furthermore, considering the semisolid consistency of OP, GP was incorporated as a bulking agent to improve the substrate porosity and moisture retention, both critical for adequate aeration and microbial proliferation [
20,
21].
Then, the culture medium used in this work for SSF was prepared by mixing DOP and DGP in a 2:1 (
w/
w) ratio. Distilled water was added to this mixture to adjust the moisture content to 60% (wet basis). The amount of water required was calculated using the following Equation (1):
where gH
2O is the amount of water added to the culture medium [g or mL], %H is the moisture content of the culture medium [g·100 g
−1], and g dry solid is the amount of dry matter in the culture medium [g].
The initial pH of the mixture composed of DOP and DGP (2:1
w/
w) was typically in the range of 4.5–5.2, depending on the batch variability. However, the pH was adjusted to 4.0 to reduce the variability and create a consistent initial condition across replicates and scales. This value was selected based on previous reports indicating that
Aspergillus species, particularly
A. niger, exhibit optimal enzyme production and growth in the acidic pH range of 3.5 to 5.5 during SSF [
33,
34]. Furthermore, phenolic compounds present in OP are known to be more soluble and reactive at slightly acidic pH, which may facilitate their biotransformation [
35]. In this study, fermentation experiments were conducted by setting the initial pH, without applying any pH correction during SSF. This decision was based on the inherent complexity of adjusting pH in solid-state systems, where real-time control is technically challenging.
This specific substrate mixture, composed of olive pomace and grape pomace in a 2:1 ratio, was designated DOGP1 for subsequent reference.
For SSF in Petri dishes, three sterile glass jars containing 100 g (wet basis) of DOGP1 each were autoclaved at 121 °C for 20 min and allowed to cool at room temperature. The substrate was then inoculated with a spore stock suspension at a concentration of 1 × 107 conidia per gram of dry solid substrate (conidia·g DSS−1). The contents were thoroughly mixed by repeated inversion and using a sterile glass rod. Subsequently, 20 g of the inoculated DOGP1 was pouted into fifteen pre-weighed and labeled sterile Petri dishes.
For SSF in FBR, 2 kg (wet basis) of DOGP1 medium was placed into a sterilization-grade polypropylene bag and autoclaved at 121 °C for 20 min. After cooling, the substrate was then inoculated with the spore stock suspension at the same concentration (1 × 107 conidia·gDSS−1). The mixture was homogenized by repeated inversion and transferred to a sterile pre-weighed FBR tray.
The pH of DOGP1 was re-measured after the sterilization process and was found to remain close to the initially adjusted value, typically ranging between 3.9 and 4.1, indicating minimal alteration due to thermal processing.
2.4. Solid-State Fermentation Processes
2.4.1. SSF in Petri Dishes Scale
The objective of this experiment was to monitor the SSF through simple tests, in order to quickly obtain preliminary results on the effect of the fungus isolated from the OP on the culture medium used.
Fifteen Petri dishes were incubated for 7 days in a microbial culture chamber at a constant temperature of 25 °C. Then, samples were taken in triplicate at predetermined times (0, 24, 48, 72, and 168 h), substrate weights were recorded to monitor the weight loss, and the samples were kept in refrigerated conditions until analyzed.
Sampling Strategy
Due to the inherent heterogeneity of SSF systems, marked by spatial variability in moisture, oxygen availability, microbial activity, and metabolite concentrations, obtaining representative samples is essential for reliable analytical results [
36]. To address this, a composite sampling strategy was implemented. At each sampling point, five subsamples (~1 g wet substrate) were collected from distinct regions of the culture dish: one from the center and one from each quadrant. These subsamples were combined to form a 5 g composite sample, manually homogenized under aseptic conditions and either analyzed immediately or stored at –20 °C. This approach minimizes the effects of local variability and enhances result reproducibility, consistent with established SSF sampling guidelines [
37].
Analysis of Samples
The moisture content was determined by analyzing 2 g of the composite sample using a halogen moisture analyzer (OHAUS MB35). Based on the resulting data, the DWL was calculated [mg·g
−1], and it was determined as follows:
where idw [g] and fdw [g] represent the initial and final dry weights of the sample, respectively. In addition, the WWL was calculated [mg·g
−1] as follows:
where iww [g] and fww [g] are the initial and final wet weights of the sample, respectively.
These parameters, DWL and WWL, were used to evaluate the mass reduction during SSF.
The remaining 3 g of the sample was mixed with distilled water at a 1:4 ratio and agitated in an orbital shaker (DLAB, SK-O330-Pro, Beijing, China) at 250 rpm for 30 min. The mixture was then filtered and centrifuged at 5000 rpm for 5 min. The resulting supernatant (aqueous extract) was used for pH determination with a digital pH meter (ADWA AD800, Szeged, Hungary) and for monitoring the dark color intensity [
38]. The absorbance was measured at 395 nm using a microplate spectrophotometer (Thermo Scientific Multiskan FC, Hillsboro, OR, USA) at various time points throughout the SSF process. The results were expressed as the percentage of decolorization:
where
A0 is the initial absorbance at time zero, and
At is the absorbance at time t. This parameter was used as an indirect indicator of the transformation or degradation of chromophoric compounds, primarily phenolic constituents, within the substrate matrix.
All experiments were conducted in triplicate.
2.4.2. SSF in Fixed-Bed Bioreactor
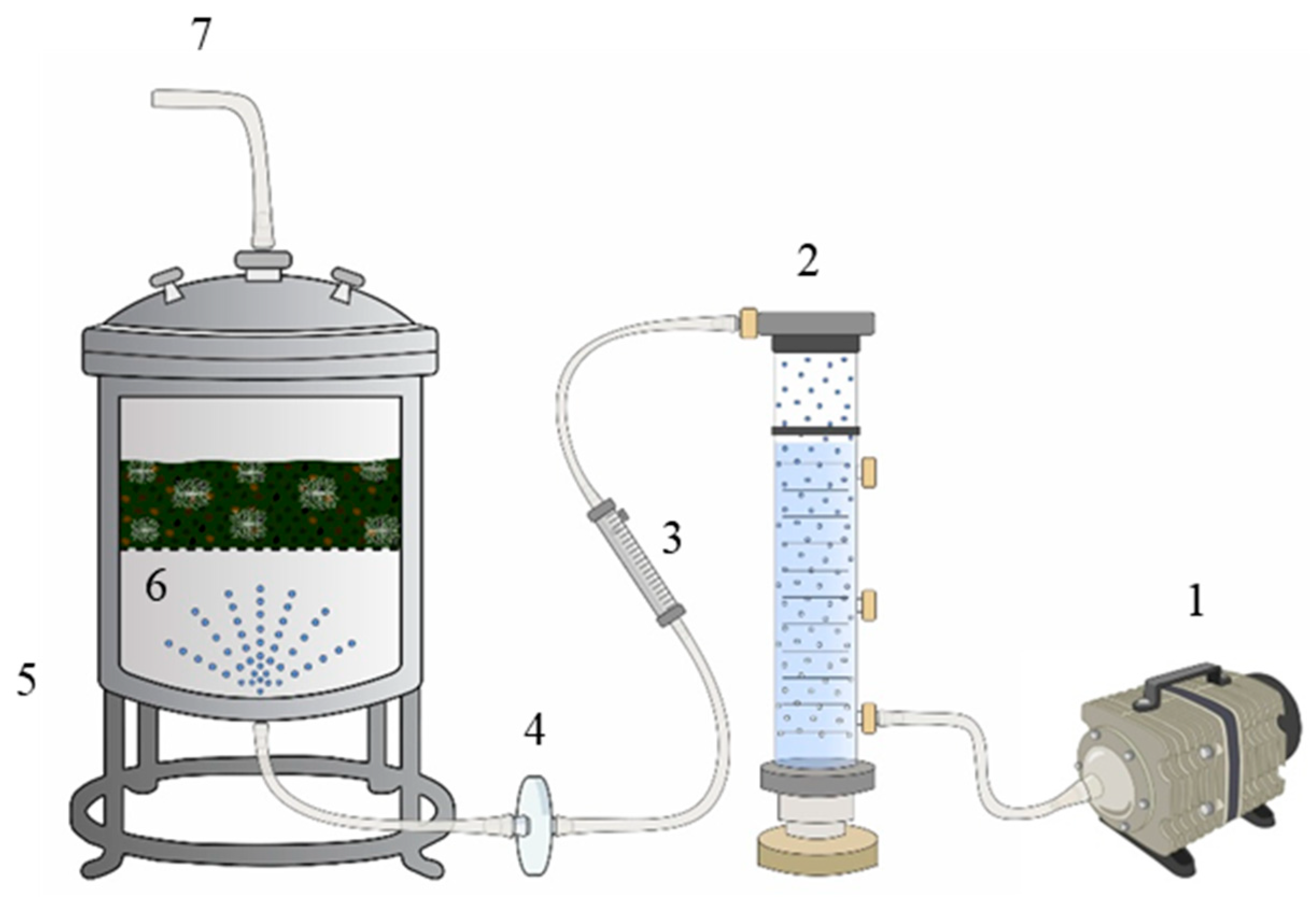
Bioreactor Setup and Operation
The FBR used in this assay is detailed in
Figure 1.
Prior to SSF operation, all system components, including the perforated tray, air filter, tubing, and humidification column, were sterilized by autoclaving at 121 °C for 20 min. After sterilization, the system was assembled in a controlled-environment cultivation room and positioned on a laboratory bench. The inlet and outlet tubing were connected, and humidified air, previously passed through a water-filled column, was continuously supplied at a flow rate of 2 L·min−1, regulated by a rotameter. The outlet was fitted with a gas filter to retain the fermentation-derived emissions.
Subsequently, the inoculated substrate (as described in
Section 2.3) was loaded into the reactor, which was then hermetically sealed. The start time of the fermentation (day and hour) was recorded.
Fermentation was carried out for 7 days at 25 °C. Monitoring was performed at 0, 8, 26, 32, 48, 56, 72, 80, 96, 104, 119, and 165 h. The final weight of the perforated tray was recorded at the end of fermentation.
The airflow rate selected for the FBR was designed to ensure that the solid substrate remained adequately moist throughout the fermentation, a key requirement for effective SSF, without reaching forced aeration conditions. In this system, the bed is surrounded by a stream of humidified air, which maintains the substrate–fungus matrix at optimal moisture levels while also allowing the passive release of fermentation gases, primarily carbon dioxide produced by fungal metabolism, without generating excessive airflow that could dry out the medium or disrupt the microbial activity.
Sampling Strategy
To obtain representative samples from the FBR, a composite sampling strategy was applied [
37]. At each sampling point, six subsamples of approximately 2 g of wet substrate were collected from distinct spatial positions: one from the central surface, one from each of the four corners (near the surface), and one from the mid-depth center (approximately 2.5 cm deep). Where feasible, additional subsamples were collected from deeper layers (up to 5 cm depth) at different lateral positions to account for potential vertical gradients. All subsamples were aseptically collected and immediately combined to form a single 12 g composite sample per time point. The composite samples were homogenized under aseptic conditions and either processed immediately or stored at −20 °C for further analysis.
Sample Processing
The samples were used for the following analyses: moisture content, DWL, and WWL, determined according to the procedures described in
Section 2.4.1.
The remaining sample material was allocated for extract preparation:
Total polyphenolic (TP) extraction: a 5 g of wet substrate was mixed with 20 mL of a methanol–water solution (60:40
v/
v), corresponding to a solid–liquid ratio of 1:4. The mixture was shaken at 250 rpm for 30 min [
39].
Enzyme extraction: An equivalent amount of wet substrate (5 g) was mixed with 10 mL of a 0.01% Tween 80 aqueous solution, maintaining a solid–liquid ratio of 1:2. The mixture was agitated at 250 rpm under the same conditions [
40]. The original method used distilled water for enzyme extraction; however, it was slightly modified by incorporating Tween 80, a non-ionic surfactant that improves enzyme recovery by reducing the surface tension and promoting the desorption of enzymes bound to biomass or substrate particles [
41].
Both extracts were filtered and centrifuged at 5000 rpm for 5 min. The supernatants were stored at −20 °C until further analysis.
Analysis of Samples
The total polyphenol content was determined using the total polyphenol method with the Folin–Ciocalteu reagent [
39]. To minimize potential interference, extract samples were diluted (1:10) prior to the reaction, and the Folin–Ciocalteu assay was conducted under buffered conditions with sodium carbonate, ensuring a neutral to slightly alkaline pH for proper chromogenic development. Gallic acid was used as the standard reagent for the calibration curve. The absorbance was measured at 590 nm after 60 min of reaction (Thermo Scientific Multiskan FC). This was reported as
where A
0 corresponds to the initial absorbance at t = 0 h, and Aₜ corresponds to the absorbance at time t. This parameter was used to quantify the reduction in polyphenol content throughout the SSF process, serving as an indicator of the detoxification of the solid substrate by fungal enzymes.
Experimental data for % TP reduction, acquired from samples collected throughout SSF, were used to fit a kinetic curve. This curve was subsequently fitted to a first-order model (FO). This model has found wide application across various scientific disciplines, including population statistics, ecology, laser physics, economics, and in the modeling of tumor growth dynamics [
42], and the mathematical expression is
where TP (t) represents the accumulated percentage reduction in polyphenol content as a function of time [mg·100 g
−1], A is the maximum asymptotic reduction value of polyphenols [mg·100 g
−1], and k is the kinetic reduction constant (h
−1), indicating the rate at which the asymptotic value is approached. This model assumes that the degradation or transformation of polyphenols follows first-order kinetics, where the rate of change is proportional to the remaining number of reactive compounds. A good fit to this model allows for a straightforward and comparative quantification of the detoxification efficiency under different fermentation treatments or conditions.
Enzymatic Activity. The quantification of enzymatic activities was performed through the measurement of colorimetric reaction products, following the method described in [
43], as detailed below.
Cellulose: The colorimetric method was based on the release of reducing sugars from carboxymethyl cellulose (CMC). The assay was carried out in sodium citrate buffer (0.05 M, pH 4.8) as the reaction medium, with CMC (20 g·L−1) as the substrate and a glucose standard solution for calibration curve preparation. The reducing sugars were quantified using the 3,5-dinitrosalicylic acid reagent (DNS) at 540 nm. Enzymatic activity was expressed as glucose units released per minute under standard conditions.
Xylanase: The colorimetric assay was based on the release of reducing sugars from xylan (10.0 g·L−1 4-O-methyl glucuronoxylan) as the substrate. The assay was conducted in phosphate–citrate buffer (0.025 M Na2HPO4-0.05 M citric acid, pH 5.3). A D-xylose solution prepared in 0.05 M citrate buffer was used for the standard curve. The reducing sugars were detected using the DNS reagent at 540 nm. Enzymatic activity was expressed as units of xylose released per minute under standard conditions.
Endo-polygalacturonase: The colorimetric assay was based on the release of D-galacturonic acid from polygalacturonic acid (PGA) as the substrate (2.0 g·L−1), prepared in either acetate buffer (20 mM, pH 5.0) or citrate–phosphate buffer (50 mM citric acid-25 mM Na2HPO4, pH 3.0), both maintained at 4 °C. The reducing sugars were detected using the DNS reagent at 540 nm. The standard curve was prepared with a D-galacturonic acid. Enzymatic activity was expressed as units of galacturonic acid released per minute under standard conditions.
Tannase activity was determined by incubating 50 μL of fermentation extract with 1 mL of tannic acid solution (0.3 mM) in 0.1 M acetate buffer (pH 5) at 30 °C for 30 min. The reaction was stopped with 0.2 mL of 2 M HCl. The gallic acid released during hydrolysis was quantified using the rhodanine colorimetric method. After adding rhodanine and KOH to the reaction mixture, the absorbance was measured at 520 nm. Enzymatic activity was expressed as μmol of gallic acid released per mL per minute. The standard curve was prepared with gallic acid [
44].
All experiments were conducted in triplicate.
3. Kinetic Modeling of SSF in Fixed-Bed Reactor
In general terms, the microbial metabolism that occurs during SSF can be represented by the following global equation:
During SSF, filamentous fungi degrade complex carbon sources (C.S), e.g., polysaccharides and lignocellulose, via extracellular enzymes, generate simpler compounds used in catabolic pathways for energy. Under aerobic conditions, most metabolic energy is obtained through respiration, involving oxygen (O
2) consumption and carbon dioxide (CO
2) release. Consequently: (a). C.S from the substrate are transformed into fungal biomass (BIOMASS) and CO
2; (b). nitrogen sources (N.S) are assimilated for biomass synthesis (BIOMASS) through anabolic processes; (c). valuable metabolites such as organic acids, hydrolytic enzymes, pigments, and antimicrobials (BIOPRODUCT), and (d). water (H
2O) and heat (∆H
R) are produced [
45].
Monitoring of the SSF process was carried out in an FBR by measuring both the DWL and WWL, as previously detailed. This decision was based on previous studies [
21], which demonstrated an almost linear correlation between the DWL and CO
2 production during SSF. These parameters were selected as indirect indicators of microbial activity and metabolic progress throughout the biotransformation. A description of the SSF based on the water mass balance can be expressed as follows [
46]:
where WWL is the weight loss before drying the solid matter [mg·g
−1], DWL is the weight loss after drying the solid matter [mg·g
−1], ΔO
2 is the oxygen consumed [mg·g
−1], ΔCO
2 is the carbon dioxide evolved [mg·g
−1], ΔP represents the volatile products [mg·g
−1], W
MET is the water produced due to carbohydrate metabolism [mg·g
−1], W
HYD is the water consumed for substrate hydrolysis [mg·g
−1], and W
REL is the water released from the reactor [mg.g
−1]. Then, combining Equations (8) and (9), we have
The contributions of the variables W
HYD and W
MET to weight loss were significantly lower than W
REL, indicating that they did not significantly contribute to the weight loss and could be eliminated from Equation (10). Consequently, Equation (10) can be rearranged to form Equation (11), as follows:
This indicates that both weight loss parameters will follow a similar trend, with the discrepancy attributable solely to a constant value representing water released from the reactor during the SSF.
Experimental data for the WWL and DWL, acquired from samples collected throughout the SSF, were used to develop kinetic curves. These curves were subsequently fitted to a logistic model. This model has found wide application across various scientific disciplines, including population statistics, ecology, laser physics, economics, and in the modeling of tumor growth dynamics [
42], and the mathematical expression is
where WL(t) is the rate of change in weight loss over time (DWL or WWL, [mg·g
−1·h
−1]), a is the maximum value of WL (DWL or WWL, [mg·g
−1]), b is the growth rate [h
−1], t is the time [h] and, c is the value of midpoint of the time [h].
Data Analysis
All graphs were constructed using the mean values from triplicate experiments. Kinetic data were fitted to mathematical models using Matlab R2015a. The statistical data of the estimated parameters were also reported.
4. Results and Discussion
4.1. Morphological Fungal Identification
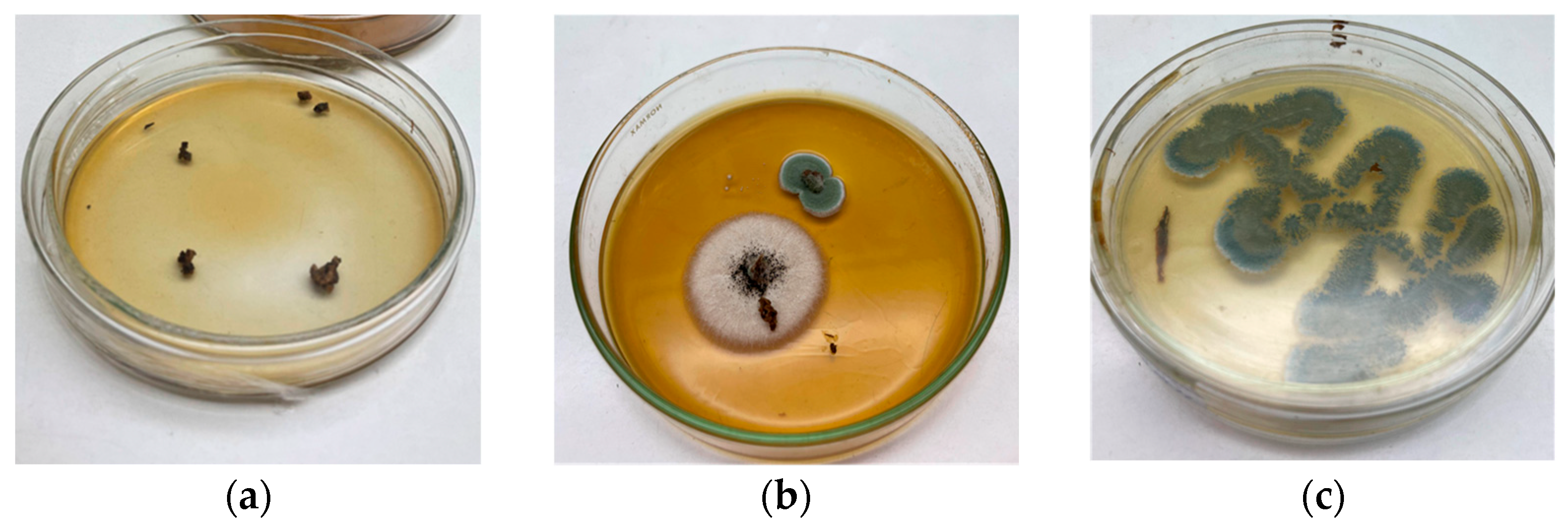
Among the isolates, two morphologically distinct colonies were observed as shown in
Figure 2.
The observation shown in
Figure 2b was carried out five days after inoculating OP fragments into the PDA-DOP culture medium. Based on this, a macroscopic evaluation was conducted, highlighting the following:
The larger central fungal colony exhibited the following macroscopic characteristics: color: cottony white at the periphery, with a darker center (possibly a sclerotium or sporulation zone); texture: dense and filamentous, with a woolly or cotton-like appearance typical of early growth stages; elevation: slightly raised in the center; edge: regular and well defined.
The smaller upper fungal colony, in contrast, presented the following features: color: bluish-green with a white margin; texture: velvety to powdery, typical of sporulating species; elevation: flat; edge: circular and well defined, with a clear halo at the periphery; size: considerably smaller, suggesting slower growth or an earlier developmental stage.
The green fungal colony was selected for subculturing using the T-streak method to obtain a pure isolate (
Figure 2c). Although it exhibited smaller radial growth, it reached sporulation rapidly, suggesting a more efficient nutrient uptake and metabolic activity. This behavior may be associated with the secretion of a potent enzymatic complex capable of quickly degrading the substrate, followed by sporulation triggered by nutrient depletion.
Filamentous fungi regulate nutrient acquisition through the production of extracellular enzymes that depolymerize complex substrates into assimilable compounds. This strategy enables rapid exploitation of the environment but often results in accelerated nutrient exhaustion, which in turn induces developmental transitions such as sporulation [
47]. As further noted by [
48], fungal secretion systems are tightly linked to morphological responses in fluctuating environments. Therefore, the early sporulation observed in this isolate likely reflects an efficient enzymatic system coupled to a rapid lifecycle adaptation typical of filamentous fungi.
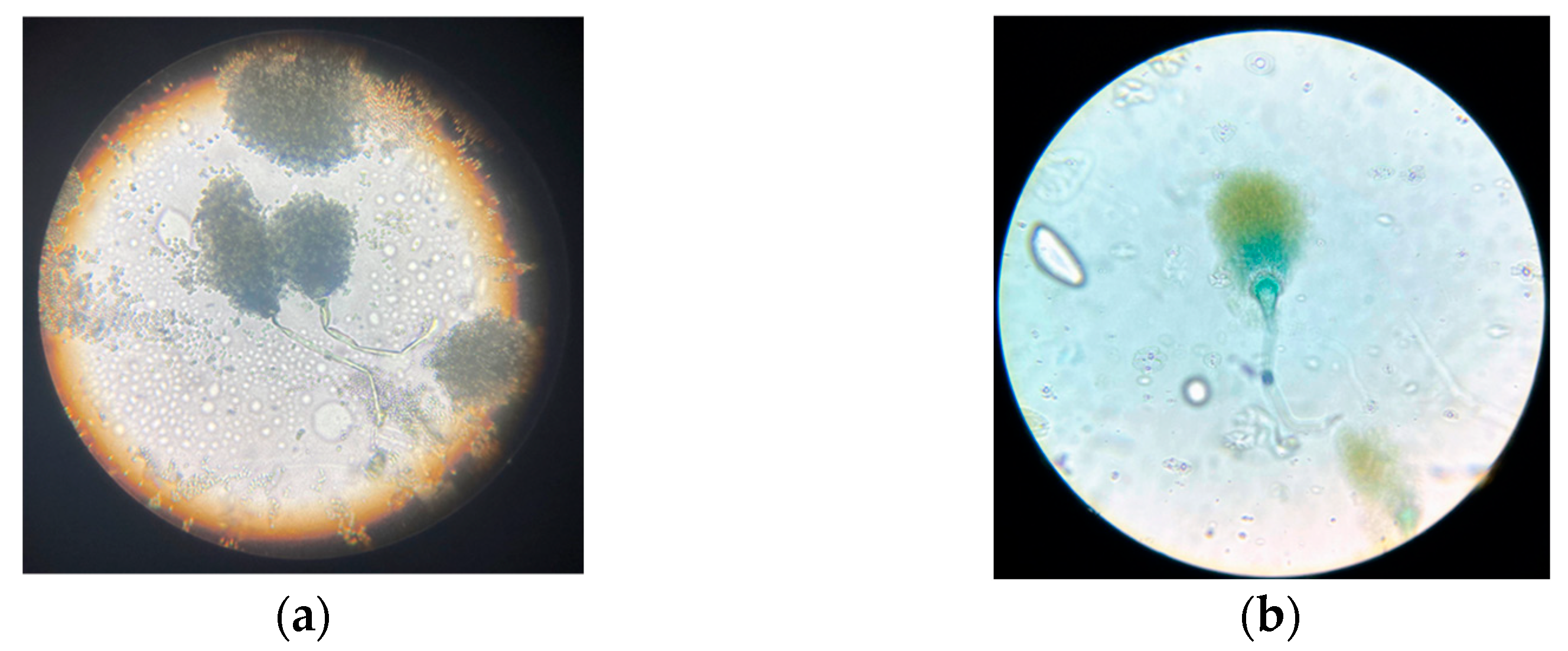
These features initially suggested that the isolate might belong to the genus
Penicillium, which is commonly associated with lignocellulosic residues, although the microscopic examination under lactophenol blue staining provided clearer insights into its identity, as shown in
Figure 3.
The fungus displayed a smooth, upright, and non-septate conidiophore terminating in a well-defined globose vesicle, from which phialides radiated outward to form a compact conidial head. This morphology is typical of the genus
Aspergillus, particularly species such as
A. niger or
A. flavus [
49,
50], and is inconsistent with the brush-like conidiophore branching pattern characteristic of
Penicillium. This reclassification highlights the importance of integrating multiple levels of analysis for accurate fungal identification. While macroscopic traits can provide an initial hypothesis, detailed microscopic analysis, especially with specific stains, enables clearer discrimination between morphologically similar genera.
The presence of
Aspergillus in OP was reported in other works [
28], consistent with its ecological versatility and ability to colonize substrates rich in phenolic compounds and with low water activity, conditions typical of this agro-industrial byproduct [
29]. Moreover, many
Aspergillus species are known producers of extracellular enzymes (e.g., tannases, cellulases, xylanases), indicating their biotechnological potential in waste valorization and bioremediation processes [
51].
Altogether, these findings reinforce the value of OP not only as a substrate to be treated but also as a source of robust microorganisms suitable for applied bioprocesses, aligning with circular economy strategies. Although molecular characterization (e.g., ITS or β-tubulin sequencing) was not included in the present study, such analysis is currently underway and will be included in future work to complement the morphological data.
Based on these morphological traits, the first isolate (olive-green colony) was selected for subsequent SSF experiments. The selected isolation is named Aspergillus sp.
4.2. SSF in Petri Dishes Scale
To evaluate the capacity of
Aspergillus sp. to grow on the DOGP1 substrate, a laboratory-scale SSF was carried out in Petri dishes over a 7-day period. This preliminary assay aimed to assess the substrate degradation by monitoring variations in moisture content, pH, color intensity, DWL, and WWL. The results of this evaluation are summarized in
Table 1.
In SSF, variations in moisture content during fungal growth are indicative of metabolic activity. The moisture content remained relatively stable during the initial days (0 to 72 h) but showed a significant decrease by the end of the process (168 h). This trend may indicate that the system maintained adequate moisture levels during the early stages but experienced cumulative losses over time due to evaporation and/or metabolic consumption. However, the value of 36.20% observed on day 7 (168 h) could be limiting for microbial activity [
52,
53]. Although the culture medium was initially formulated at 60% humidity (wet basis), post-sterilization measurements indicated a slightly reduced initial value (~48.6%), which remained within the optimal range (40–70%) reported for SSF systems [
54,
55]. This moisture level was sufficient to support fungal growth and enzymatic activity, although some reduction in performance was observed after 7 days, possibly due to cumulative moisture loss.
As indicators of substrate transformation and microbial activity, a progressive increase in DWL was observed from 24 h onward, reaching a peak of 200 mg·g
−1 at 72 h. Subsequently, the DWL slightly decreased and stabilized at 149 mg·g
−1 by the end of the process at 168 h. In contrast, the WWL showed a continuous upward trend over time, with moderate changes during the first 48 h (15–42 mg·g
−1), followed by a marked increase from 72 h onward, reaching 218 mg·g
−1 at 168 h. These weight losses reflect the dynamic interactions between fungal metabolism and the substrate. The DWL primarily indicates substrate degradation and biomass formation, whereas the WWL captures both mass loss and moisture dynamics [
46]. The observed trends support the occurrence of active microbial colonization and substrate bioconversion during the SSF.
The pH of the aqueous extracts showed a marked increase after 48–72 h of incubation, which is consistent with the transition from active mycelial growth to the onset of sporulation. The sporulation phase in filamentous fungi is often associated with shifts in carbon and nitrogen metabolism, as reported in [
56]; during SSF with
Aspergillus sp., using wheat bran and soybean meal as substrates for xylanase production, a progressive increase in pH was observed between 24 and 72 h of fermentation. This trend was attributed to the depletion of easily available carbon sources and the subsequent consumption of organic acids. Furthermore, the breakdown of nitrogenous compounds and proteins can lead to the release of ammonium ions, both of which contribute to an increase in pH [
57].
The decolorization of the substrate progressively increased throughout the SSF process. A slight reduction in color intensity was observed after 24 h, becoming more evident at 48 h (39.68 ± 5.10%) and reaching 55.90 ± 4.62% at 72 h. The maximum decolorization (68.42 ± 4.30%) was recorded at 168 h. This trend suggests an effective transformation of chromophore compounds in OP, likely through adsorption, modification, or enzymatic degradation by the fungal biomass. It is known that during olive oil processing, only a minimal fraction of the phenolic compounds is recovered, with approximately 98% remaining in byproducts such as OP [
58]. However, partial detoxification also confirmed that a substantial portion of phenolic compounds remained undegraded. It is important to note that the polyphenolic compounds naturally present in OP and GP, such as hydroxytyrosol, tyrosol, caffeic acid, ferulic acid, and flavonoids such as luteolin and apigenin [
14,
59] are known to exhibit color variations ranging from pale yellow to brown. These color changes can occur not only due to microbial enzymatic degradation but also as a result of spontaneous oxidation and polymerization reactions during sterilization or exposure to air. Therefore, the observed decolorization may result from a combination of microbial activity and oxidative processes intrinsic to the phenolic matrix.
Notably, while other studies have achieved complete polyphenol degradation using microbial consortia in complex systems like abandoned evaporation ponds [
15], the present study demonstrates a high level of detoxification using a single fungal isolate. This underscores the strong bioconversion potential of the
Aspergillus sp. strain isolated from OP and reinforces its suitability as a robust biocatalyst for SSF processes. The effectiveness of this monoculture approach not only simplifies the process but also offers advantages in scalability, control, and cost, aligning well with circular economy principles and industrial applications [
60].
In this context, the correlation between the increased decolorization, elevated pH values, and substrate weight loss suggests an active oxidative metabolism, driven by the synergistic action of hydrolytic and oxidative enzyme systems secreted by the fungus during substrate colonization and transformation. It is important to note that no uninoculated control was included in the SSF assays performed in this study. Although the observed changes in weight loss, pH, decolorization, and phenolic reduction were consistent with active fungal metabolism, future experiments will incorporate abiotic controls to isolate and quantify the specific contributions of microbial activity from physicochemical or oxidative changes occurring independently in the substrate.
Finally, to confirm the fungal growth and substrate transformation, the results also revealed key limitations and opportunities for process improvement. The marked moisture loss after seven days suggests the need for strategies that enhance water retention or enable controlled humidification of the solid matrix to sustain microbial activity. Furthermore, the observed decolorization points to potential phenolic compound degradation, warranting further investigation through quantitative analysis and enzymatic profiling. To address these aspects and assess scalability, the next section presents a second fermentation trial in a 2 kg FBR, with enhanced monitoring of moisture dynamics and metabolic performance.
4.3. SSF in FBR Scale
4.3.1. Detoxiphication and Weight Loss
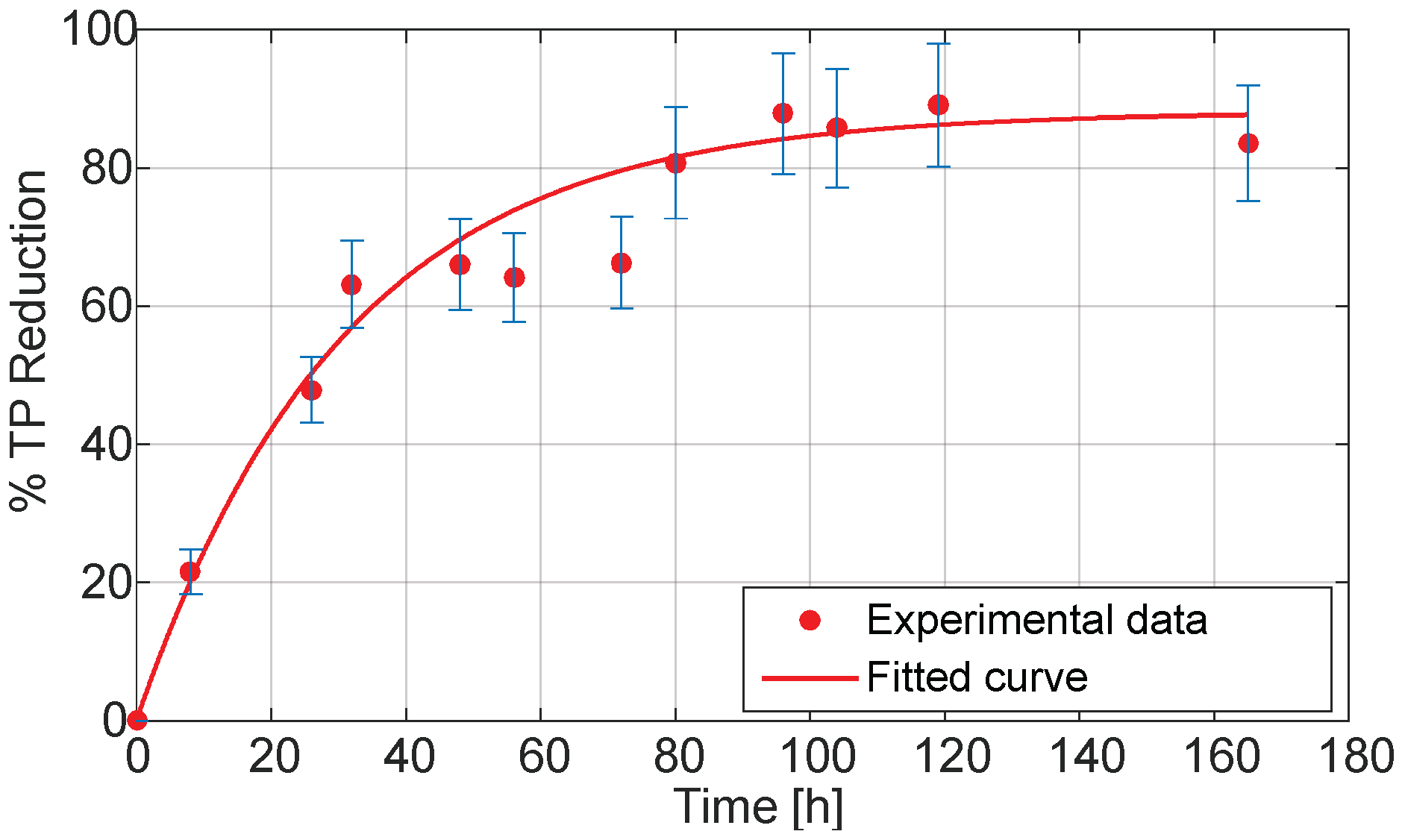
The TP reduction curve (
Figure 4) demonstrates that
Aspergillus sp. achieved a progressive and sustained decrease in phenolic content during the SSF of olive pomace (OP), reaching a maximum reduction of 88.10% at 168 h. The data showed a characteristic asymptotic behavior, with a rapid detoxification phase in the first 72 h, followed by a plateau indicative of substrate saturation or enzymatic limitation.
To describe this behavior, a first-order (FO) kinetic model was applied, yielding a rate constant (k) of 0.0325 h
−1 and an excellent fit to the experimental data (R
2 = 94.71%), as shown in
Table 2.
The FO model, commonly used to represent degradation processes, assumes that the detoxification rate is proportional to the concentration of phenolics remaining. This assumption appears valid for the current system, as the microbial and enzymatic activity likely declined as phenolics were depleted or transformed into less reactive compounds.
The detoxification percentage obtained in this study is among the highest reported for OP treated by fungal SSF. For example, Ref. [
15] reported polyphenol degradation efficiencies ranging from 40% to 100% using fungal–bacterial consortia applied to olive mill waste sludge. In contrast, the current study achieved comparable detoxification levels using a single
Aspergillus isolate, highlighting the biotechnological potential of locally adapted fungal strains. This approach reduces the complexity and cost, facilitating the process scalability and reproducibility in industrial settings.
In similar SSF-based studies, composting with mixed inocula or natural microbiota required 150–180 days to achieve significant detoxification [
10], while in our case, the majority of phenolic reduction occurred within 96 h, underscoring the efficiency of this fungal strain under optimized conditions.
Importantly, a comparative analysis between the Petri dish trials and the FBR system revealed a more pronounced removal of polyphenolic compounds in the FBR. This difference is evident, as shown in
Figure 4 and
Table 1, where the reduction percentages achieved under both conditions are presented, highlighting the improved detoxification performance at larger scale. This suggests that the FBR configuration may offer improved conditions for mass transfer, aeration, or enzymatic activity, thereby enhancing the biodegradation efficiency. These findings are promising and provide a solid foundation for future scale-up studies, positioning FBR-based SSF as a viable bioprocess for industrial detoxification of OP.
Nonetheless, the plateau observed after 120 h suggests that not all phenolic compounds were metabolized. This may be due to the recalcitrant nature of certain polyphenolic structures or the inhibition of enzymatic activity by the accumulation of fermentation products. As previously proposed [
14], the integration of a pre-treatment stage, such as selective extraction using natural deep eutectic solvents (NADES), could improve the overall phenolic removal and enable valorization of the recovered bioactives, thereby increasing the circularity of the process.
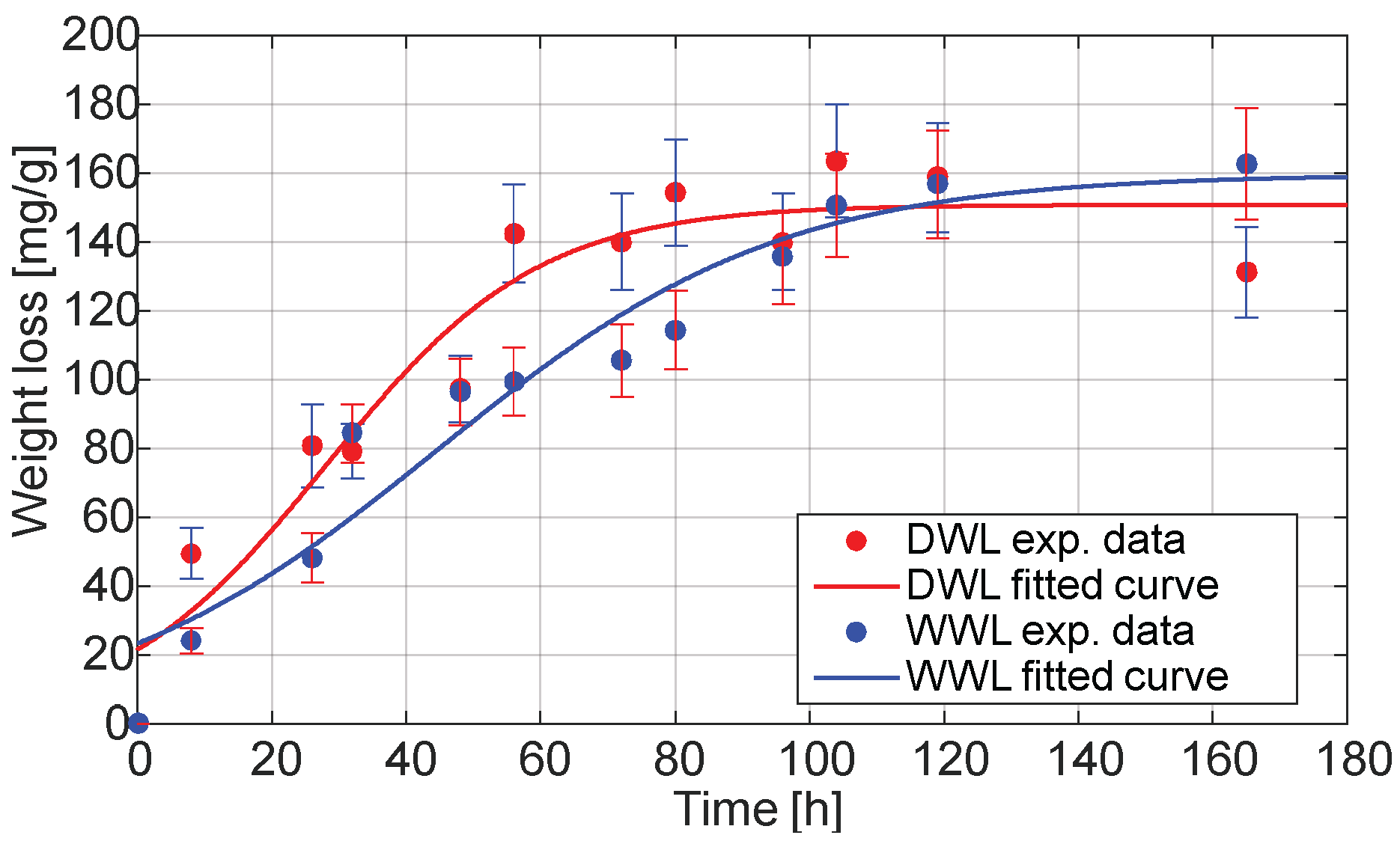
Figure 5 displays the DWL and WWL of the substrate over the 168 h SSF process. These parameters serve as indirect indicators of the substrate degradation and fungal metabolic activity and are frequently used in SSF systems where direct biomass quantification is challenging.
Both the DWL and WWL followed a sigmoidal trend, characteristic of logistic kinetics. The DWL showed a more rapid initial increase, reaching a maximum value of 150.7 mg·g−1 with a high kinetic coefficient (b = 0.0635). In contrast, the WWL progressed more gradually, peaking at 159.5 mg·g−1 with a slower growth rate (b = 0.0394), suggesting a lag in total moisture reduction compared to dry matter consumption.
The logistic model provided a good fit for both datasets (see
Table 2), with R
2 values of 91.87% for DWL and 94.04% for WWL, confirming that the weight loss follows a typical microbial growth-associated dynamic. However, higher RMSE values in the DWL model indicate greater variability in the dry weight measurements, likely due to localized differences in aeration, humidity gradients, and fungal colonization within the FBR. These factors are known to introduce heterogeneity in SSF systems, particularly in solid matrices with non-uniform porosity or moisture distribution.
The DWL curve reflects the true extent of substrate consumption, while the WWL includes both dry matter degradation and moisture dynamics, including evaporation, metabolic water retention, and partial absorption by the mycelium. The hysteresis observed between both curves, particularly in the 48–96 h window, may reflect an imbalance between the substrate degradation and moisture release, a phenomenon previously described in SSF systems [
36]. This discrepancy also emphasizes the importance of composite sampling and moisture control to ensure reproducibility and process consistency in upscaled systems. The observed hysteresis underscores the relevance of simultaneously monitoring both the dry and wet weight loss to accurately capture the progression of biotransformation and the associated physicochemical changes during SSF.
Overall, these results validate the use of the DWL and WWL as effective performance indicators in SSF with Aspergillus sp. and highlight the potential of this configuration for process intensification. The efficient substrate degradation achieved under non-forced aeration further supports the viability of fixed-bed SSF systems for large-scale applications.
Notably, this type of kinetic response has not been previously reported in similar studies, and the identification of such hysteresis adds valuable insight into the internal dynamics of solid-state systems. It also suggests that future scale-up and modeling efforts should incorporate moisture-phase behavior as a critical factor for optimizing SSF performance.
4.3.2. Enzymes Production
The secretion of extracellular enzymes by filamentous fungi under SSF conditions is a well-documented phenomenon, widely reported in the literature [
61,
62]. These organisms are particularly efficient in producing a broad spectrum of hydrolytic and oxidative enzymes, such as cellulases, xylanases, tannase, and laccases, which contribute to the degradation and transformation of complex organic substrates [
51]. This enzymatic versatility is one of the main advantages of SSF and underlies the capacity of filamentous fungi to adapt to and valorize agro-industrial residues. These biochemical processes not only reflect detoxification activity but also play a crucial role in substrate degradation and fungal development.
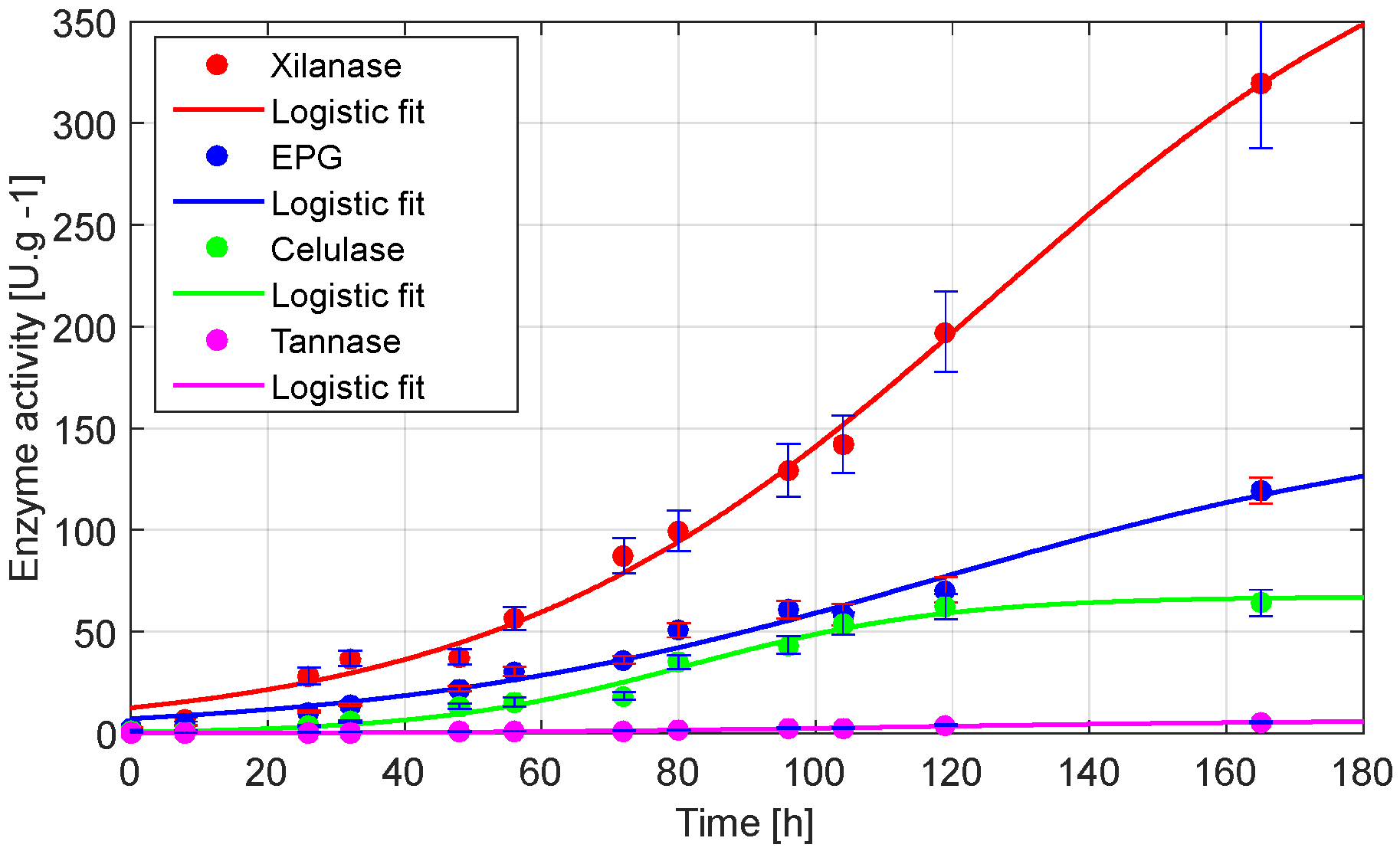
Figure 6 shows the enzymatic profiles obtained during the SSF of OP and GP by the native
Aspergillus sp. revealed differential kinetic behaviors and production capacities for cellulase, xylanase, EPG, and tannase:
Table 3 presents the parameters of the Logistic model and the associated statistical indicators describing these kinetics.
The logistic model provided an excellent fit to the experimental data for all enzymes, with R2 values ranging from 98.08% to 99.45%, indicating that the enzyme synthesis followed a sigmoidal pattern typical of microbial metabolic activity in SSF systems.
Xylanase exhibited the highest enzymatic yield, reaching a maximum activity (a) of 424.6 U·g
−1, with a slower growth rate (b = 0.0278 h
−1) and a late inflection point (c = 125.1 h), indicating prolonged and sustained production. The xylanase yield is comparable to that reported in other studies, such as the use of
Aspergillus niger on the solid residue remaining after sugar recovery from white grape pomace, which achieved 579 U·g
−1 of xylanase [
63]. However, significantly higher yields have been reported—for instance,
Aspergillus niger CECT 2700 produced 1400.80 U·g
−1 of xylanase when grown on pretreated brewery spent grain [
64]. These results highlight the marked differences in xylanase production depending on whether the solid residue used for fermentation undergoes a pretreatment step.
The EPG activity followed a similar kinetic pattern to xylanase, with a = 153.6 U·g
−1, b = 0.0252 h
−1, and c = 118.4 h. The delayed peak aligns with the progressive degradation of pectic polysaccharides, which are typically less accessible and require sequential enzymatic activity for depolymerization. In comparison, another study using barley bran mixed with sugar beet pulp as substrates for polygalacturonase production by
Aspergillus niger reported a maximum enzyme yield after 7 days, achieving over 900 U·g
−1 under optimized conditions [
65]. In a different study, various solid substrates (bean testa, mango peels, plantain peels, and their 1:1:1 mixtures) were evaluated using different
Aspergillus species. The polygalacturnase production ranged from 0.0377 to 141.0095 U·g
−1, with
Aspergillus tamarii,
Aspergillus terreus, and
Aspergillus piperis showing the highest activities on mango peels. These findings strongly suggest that the nature and composition of the substrate play a crucial role in determining the yield of this type of enzyme.
Cellulase reached a peak activity of 67.43 U·g
−1, with a higher growth rate constant (b = 0.0528 h
−1) and an earlier inflection point (c = 81.6 h), indicating rapid initial production followed by stabilization. This behavior may be associated with the fast degradation of cellulose fractions in the OP–GP mixture, which provide easily accessible carbon sources for fungal growth. The cellulase yield obtained in this study is higher than that reported in other works: for example, 6.23 U·g
−1 was obtained using the same pretreated substrate as used for xylanase production [
64]. In this case, the pretreatment did not enhance cellulase production. In another study, wheat bran and groundnut shell were used as native lignocellulosic substrates with
Aspergillus niger to compare cellulolytic enzyme production under submerged fermentation (SmF) and SSF. Higher enzyme yields were achieved in SSF, with wheat bran proving to be a more effective solid matrix than groundnut shell [
66].
Finally, tannase exhibited the lowest absolute activity (6.72 U·g
−1), yet showed the best model fit, with an R
2 of 99.27% and RMSE of 0.154. However, it also displayed slower production kinetics (b = 0.0309 h
−1; c = 113.4 h). In one study, leafy vegetables such as mango, jamun, and coffee leaves, as well as agro-industrial residues like coffee husks, rice husks, and wheat bran, were used as substrates for tannase production by
Aspergillus sp. GM4 under solid-state fermentation, yielding values ranging from 0.13 to 1.44 U·mg
−1 [
67]. Another study focused on optimizing SSF conditions for green tea processing residues using
Aspergillus niger TBG 28A, achieving a high enzymatic yield of 246.82 U·g
−1 [
68]. These results also highlight the direct influence of the tannin content in the fermented substrate on tannase production yields. In this context, the limited activity of tannase observed in this study may be attributed to the fact that tannins were supplied exclusively by the GP fraction of the substrate mixture, potentially restricting their availability for enzyme induction and expression.
In an SSF study using OP mixed with winery waste as solid substrates and
A. niger as the biotransforming agent [
69] reported xylanase and cellulase production values of 74.2 ± 6.3 U·g
−1 and 38.2 ± 2.23 U·g
−1, respectively. In another study, based on a fungal collection containing a diversity of strains isolated from olive oil industry residues capable of producing various enzymes [
28],
A. niger produced 9.92 U·g
−1 of cellulase and 6.22 U·g
−1 of tannase. These values are notably lower than those obtained in the present work, highlighting the enhanced enzymatic potential of the
Aspergillus sp. strain isolated and evaluated here.
These findings not only confirm the enzymatic versatility of the isolated Aspergillus strain but also validate the use of logistic modeling to characterize enzyme kinetics in SSF systems. The high productivity, particularly for xylanase and EPG, underscores the biotechnological value of this strain for lignocellulosic valorization. Additionally, the use of a single, non-genetically modified, and locally isolated microorganism offers operational advantages over consortia, such as simplified process control and reduced contamination risk, further enhancing the feasibility of scale-up for industrial applications.
4.4. Limitations and Challenges of the Process
This study presents promising results for the bioconversion of olive pomace residues via solid-state fermentation using Aspergillus sp. However, several limitations must be acknowledged. First, no abiotic control was included in the experimental design, making it difficult to fully distinguish the contribution of the microbial metabolism from spontaneous substrate changes. Second, the water retention capacity of the substrate was not measured, which could have provided better insight into the moisture dynamics throughout the process.
Additionally, the use of olive oil production residues, such as OP and GP, poses intrinsic challenges. These materials are highly heterogeneous, seasonally variable, and often contain high concentrations of antimicrobial polyphenols and fatty acids, which may inhibit microbial activity. Their dense structure also complicates aeration and homogenization during SSF. These factors should be carefully considered in future process optimization and scale-up efforts, particularly in non-sterile industrial environments.
5. Conclusions
This study demonstrates the feasibility and effectiveness of SSF using a native Aspergillus sp. for the simultaneous detoxification of polyphenol-rich OP and GP and the production of industrially relevant hydrolytic enzymes. The fungal isolate achieved a high percentage of polyphenol reduction within a relatively short fermentation period, accompanied by consistent weight loss and significant enzymatic activity, underscoring its metabolic efficiency under SSF conditions.
One of the key contributions of this work is the demonstration that a single native fungal strain, without the need for microbial consortia or genetic modification, can drive complex biotransformations in lignocellulosic phenolic-rich substrates. This supports circular economy strategies by converting agro-industrial residues into value-added bioproducts through low-cost and scalable biotechnological approaches.
The two-phase experimental design, integrating Petri dish assays with FBR trials, provides robust evidence of the process scalability and lays a strong foundation for further optimization. The successful application of first-order and logistic kinetic models to describe detoxification and bioconversion dynamics offers valuable predictive tools for process control and scale-up. Additionally, the FBR system proved to be a cost-effective and operationally simple platform for process intensification, enabling reproducible results and real-time monitoring of SSF performance.
Nevertheless, certain limitations should be acknowledged. No uninoculated controls were included in this study, which will be addressed in future work to better distinguish microbial contributions from abiotic changes. Furthermore, while hydrolytic enzyme activities were characterized, the potential role of oxidative enzymes, particularly laccases, remains unexplored.
Future research should focus on longer fermentation times, oxidative enzymatic pathways, and the validation of the process in pilot-scale or continuous-flow systems. These steps will be essential for evaluating the full industrial potential and broader applicability of the proposed bioprocess in sustainable waste management and bioproduct generation.