Transcriptional Analysis of Mixed-Culture Fermentation of Lachancea thermotolerans and Saccharomyces cerevisiae for Natural Fruity Sour Beer
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Beer Fermentation Experiment
2.3. Analysis of Quantity Change of Different Yeast Species in Double-Yeast Mixed Fermentation System
2.4. Analytical Determinations
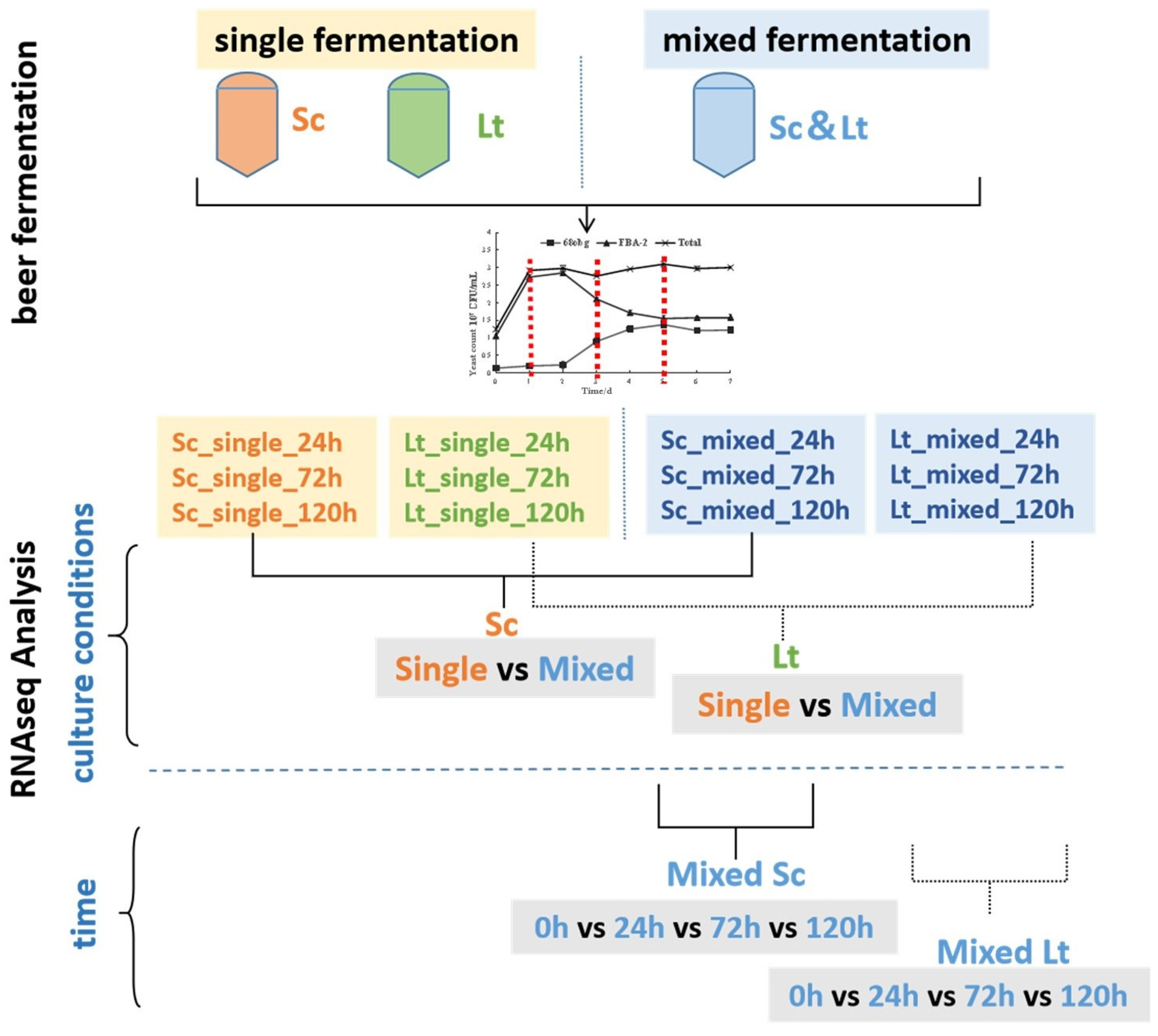
2.5. RNA Extraction Samples and RNA-Seq Analysis
2.6. Statistical Analysis
3. Results and Analysis
3.1. Non-Saccharomyces Yeast to Saccharomyces cerevisiae in Mixed-Culture Beer Fermentation
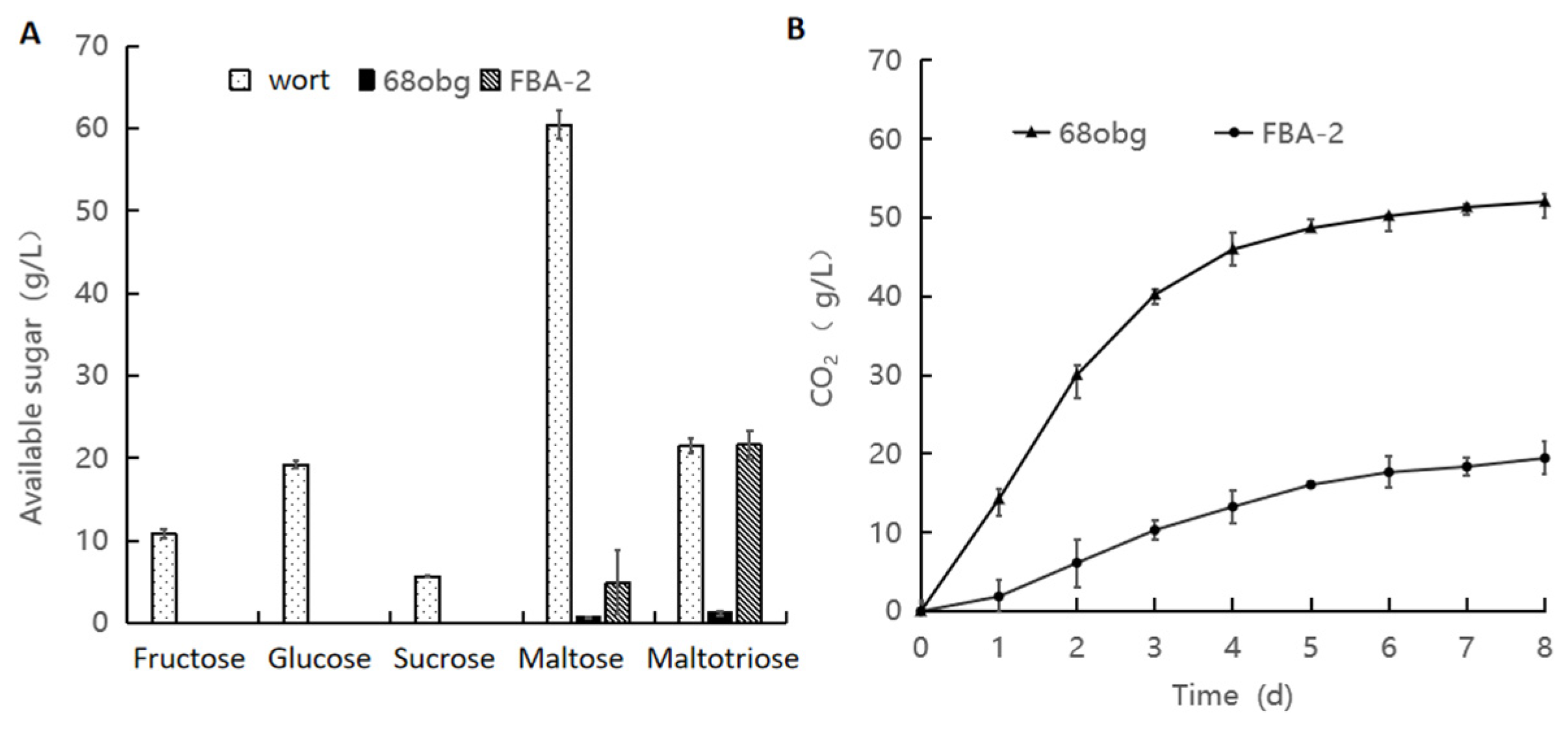
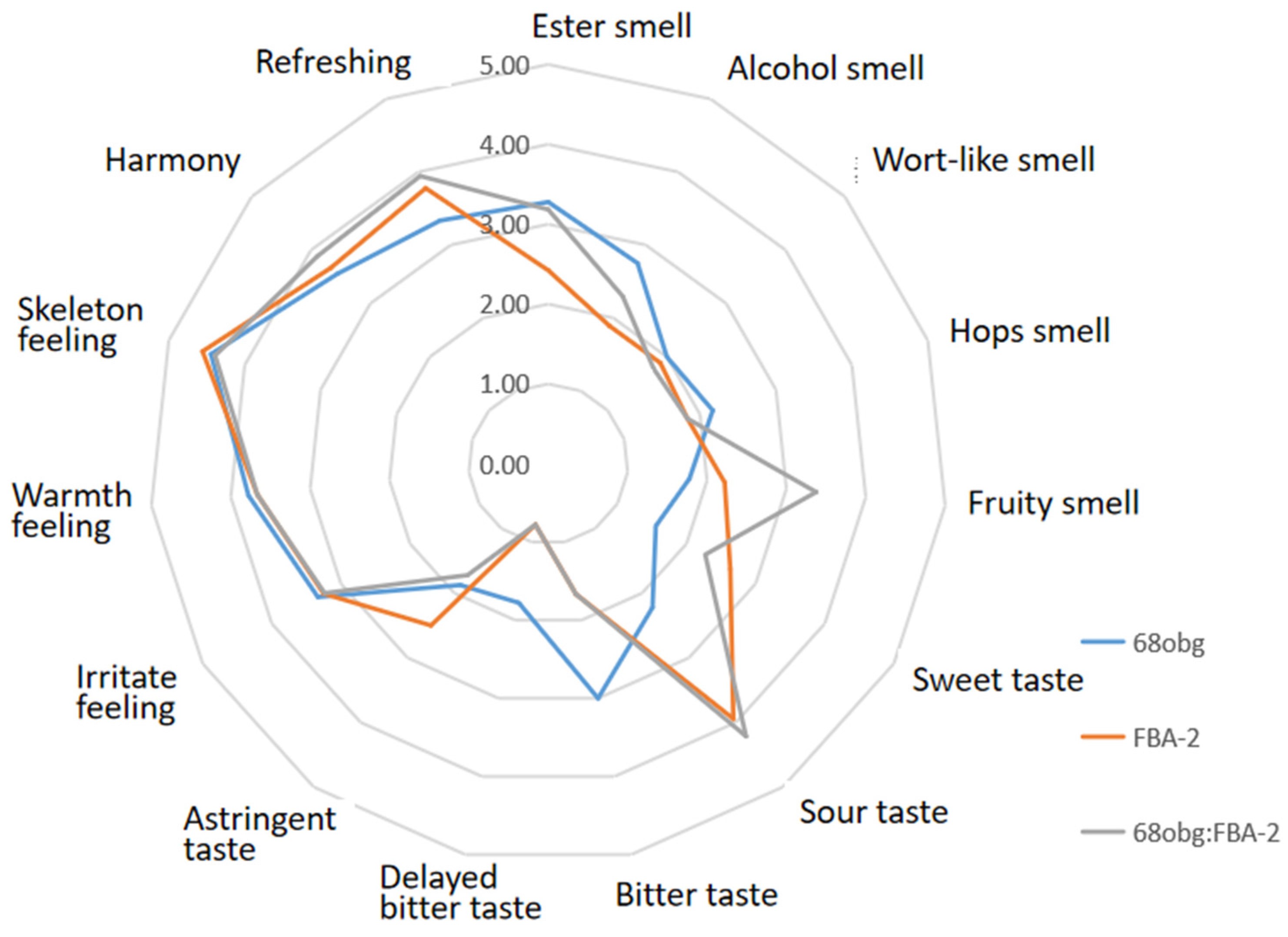
3.2. Wort with L. thermotolerans and S. cerevisiae Fermentation to Produce New Fruity Sour Beer
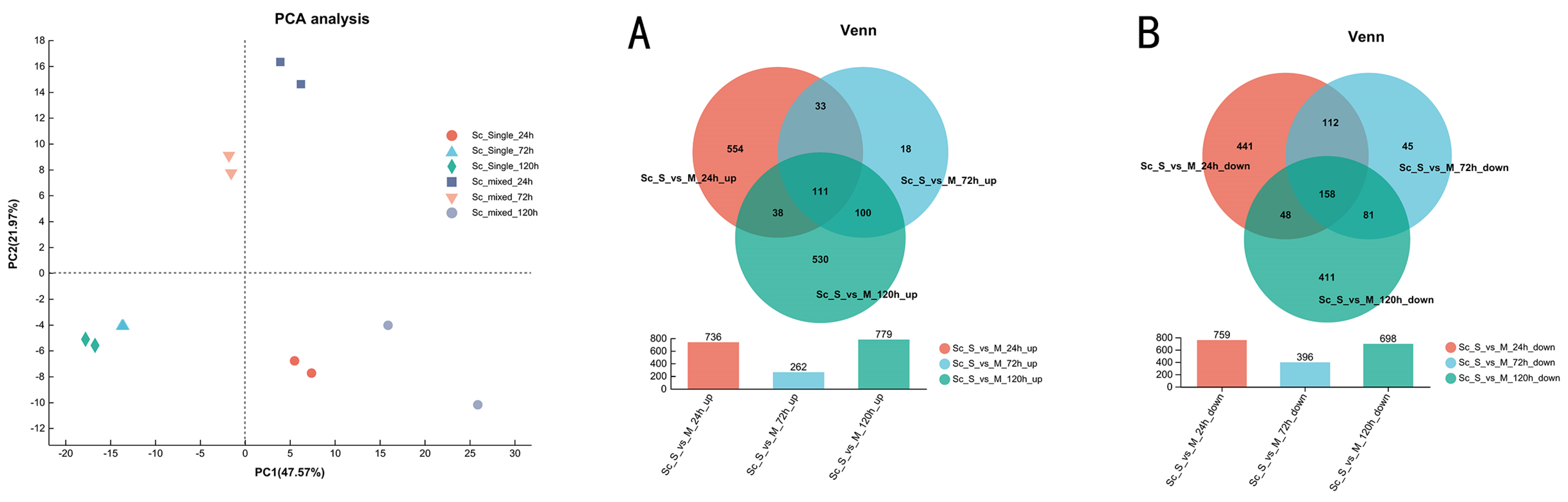
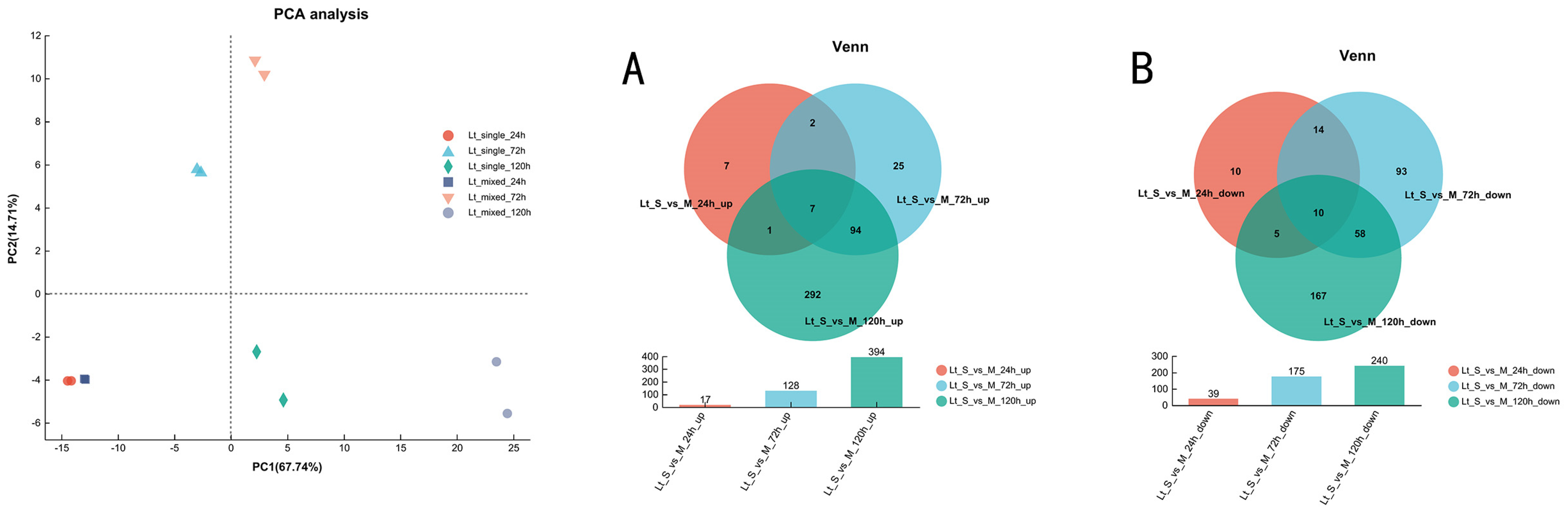
3.3. Overview of Transcriptional Response in Single- and Mixed-Culture Fermentations
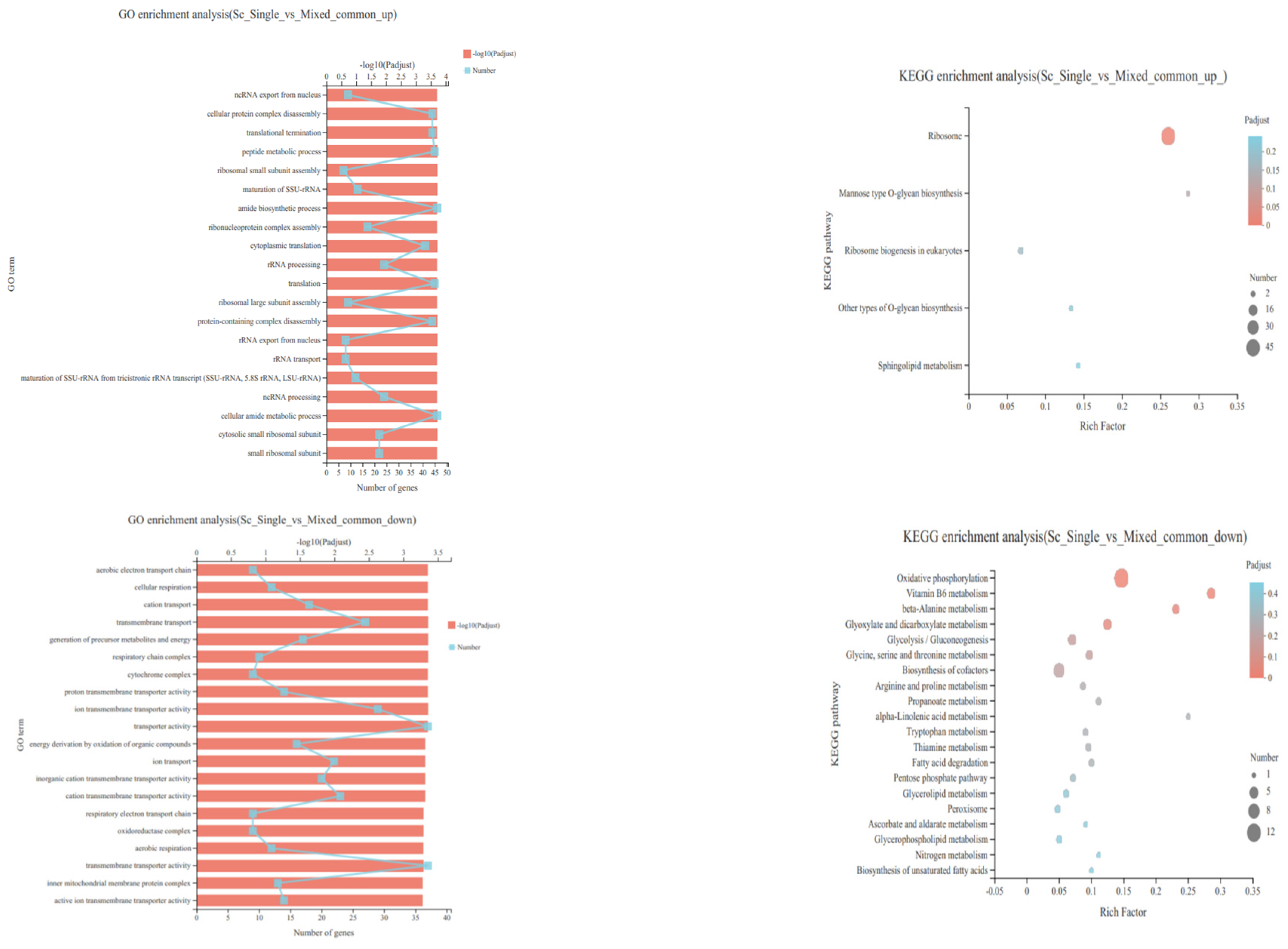
3.4. Transcriptional Profiling of S. cerevisiae in Single- and Mixed-Culture Fermentations
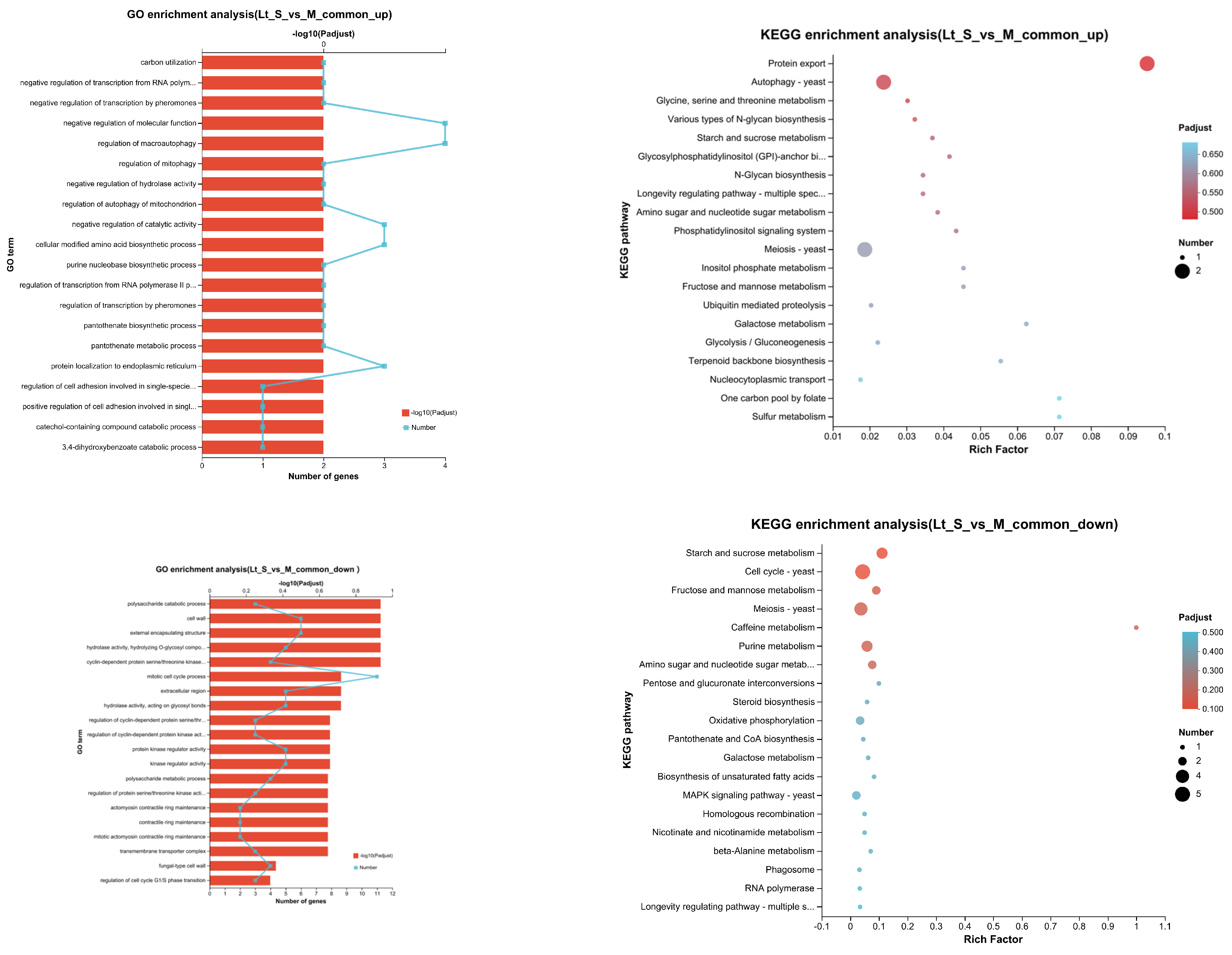
3.5. Transcriptional Profiling of L. thermotolerans in Single- and Mixed-Culture Fermentations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ciani, M.; Comitini, F.; Mannazzu, I.; Domizio, P. Controlled mixed culture fermentation: A new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res. 2010, 10, 123–133. [Google Scholar] [CrossRef]
- Comitini, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [CrossRef]
- Hansen, E.H.; Nissen, P.; Sommer, P.; Nielsen, J.C.; Arneborg, N. The effect of oxygen on the survival of non-Saccharomyces yeasts during mixed culture fermentations of grape juice with Saccharomyces cerevisiae. J. Appl. Microbiol. 2001, 91, 541–547. [Google Scholar] [CrossRef]
- Quirós, M.; Rojas, V.; Gonzalez, R.; Morales, P. Selection of non-Saccharomyces yeast strains for reducing alcohol levels in wine by sugar respiration. Int. J. Food Microbiol. 2014, 181, 85–91. [Google Scholar] [CrossRef]
- Shekhawat, K.; Bauer, F.F.; Setati, M.E. Impact of oxygenation on the performance of three non-Saccharomyces yeasts in co-fermentation with Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2017, 101, 2479–2491. [Google Scholar] [CrossRef]
- Ciani, M.; Beco, L.; Comitini, F. Fermentation behavior and metabolic interactions of multistarter wine yeast fermentations. Int. J. Food Microbiol. 2006, 108, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; Capece, A.; Comitini, F.; Canonico, L.; Siesto, G.; Romano, P. Yeast interactions in inoculated wine fermentation. Front. Microbiol. 2016, 7, 555. [Google Scholar] [CrossRef]
- Moenne, M.I.; Saa, P.; Felipe Laurie, V.; Ricardo Pérez-Correa, J.; Agosin, E. Oxygen incorporation and dissolution during industrial-scale red wine fermentations. Food Bioprocess Technol. 2014, 9, 2627–2636. [Google Scholar] [CrossRef]
- Boulton, R.B.; Singleton, V.L.; Bisson, L.F.; Kunkee, R.E. Principles and Practices of Winemaking; Springer-Verlag Inc.: New York, NY, USA, 1996. [Google Scholar]
- Gobbi, M.; Comitini, F.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Lachancea thermotolerans and Saccharomyces cerevisiae in simultaneous and sequential co-fermentation: A strategy to enhance acidity and improve the overall quality of wine. Food Microbiol. 2013, 33, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Sadoudi, M.; Tourdot-Marechal, R.; Rousseaux, S.; Steyer, D.; Gallardo-Chacon, J.J.; Ballester, J.; Vichi, S.; Guerin-Schneider, R.; Caixach, J.; Alexandre, H. Yeast-yeast interactions revealed by aromatic profile analysis of sauvignon blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol. 2012, 32, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Soden, A.; Francis, I.L.; Oakey, H.; Henschke, P.A. Effect of co-fermentation with Candida stellata and Saccharomyces cerevisiae on the aroma and composition of chardonnay wine. Aust. J. Grape Wine Res. 2000, 6, 21–30. [Google Scholar] [CrossRef]
- Nissen, P.; Nielsen, D.; Arneborg, N. Viable Saccharomyces cerevisiae cells at high concentrations cause early growth arrest of non-Saccharomyces yeasts in mixed cultures by a cell-cell contact- mediated mechanism. Yeast 2003, 20, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Nevado, F.; Albergaria, H.; Hogg, T.; Girio, F. Cellular death of two non-Saccharomyces wine-related yeasts during mixed fermentations with Saccharomyces cerevisiae. Int. J. Food Microbiol. 2006, 108, 336–345. [Google Scholar] [PubMed]
- Albergaria, H.; Francisco, D.; Gori, K.; Arneborg, N.; Girio, F. Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl. Microbiol. Biotechnol. 2010, 86, 965–972. [Google Scholar] [CrossRef]
- Rossouw, D.; Meiring, S.; Bauer, F.F. Modifying Saccharomyces cerevisiae adhesion properties regulates yeast ecosystem dynamics. mSphere 2018, 3, e00383-18. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.E.; Brown, T.A.; Trumpower, B.L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990, 18, 3091–3092. [Google Scholar] [CrossRef] [PubMed]
- HannonLab. Fastx-Toolkit. 2010. Available online: http://hannonlab.cshl.edu/fastx_toolkit/index.html (accessed on 10 May 2016).
- Morgan, M.; Anders, S.; Lawrence, M.; Aboyoun, P.; Pagès, H.; Gentleman, R. ShortRead: A Bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 2009, 25, 2607–2608. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, I.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with highthroughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Barbosa, C.; Mendes-Faia, A.; Lage, P.; Mira, N.P.; Mendes-Ferreira, A. Genomic expression program of Saccharomyces cerevisiae along a mixed-culture wine fermentation with Hanseniaspora guilliermondii. Microb. Cell Factories 2015, 14, 124. [Google Scholar] [CrossRef] [PubMed]
- Bron, P.A.; Wels, M.; Bongers, R.S.; van Bokhorst-van, V.; Wiersma, A.; Overmars, L.; Marco, M.L.; Kleerebezem, M. Transcriptomes reveal genetic signatures underlying physiological variations imposed by different fermentation conditions in Lactobacillus plantarum. PLoS ONE 2012, 7, e38720. [Google Scholar] [CrossRef] [PubMed]
- de Groot, M.J.L.; Daran-Lapujade, P.; van Breukelen, B.; Knijnenburg, T.H.; Pronk, J.T.; Slijper, M.; Heck, A.J.R. Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post-transcriptional regulation of key cellular processes. Microbiology 2007, 153, 3864–3878. [Google Scholar] [CrossRef] [PubMed]
- Gasch, A.P.; Spellma, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef] [PubMed]
- Koskenniemi, K.; Laakso, K.; Koponen, J.; Kankainen, M.; Greco, D.; Auvinen, P.; Savijoki, K.; Nyman, T.A.; Surakka, A.; Salusjarvi, T.; et al. Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol. Cell. Proteom. 2011, 10, M110.002741. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Pajarillo, E.A.; Kim, M.J.; Chae, J.P.; Kang, D.K. Proteomic and transcriptional analysis of Lactobacillus johnsonii PF01 during bile salt exposure by iTRAQ shotgun proteomics and quantitative RT-PCR. J. Proteome Res. 2013, 12, 432–443. [Google Scholar] [CrossRef] [PubMed]
- Mostert, T.T.; Divol, B. Investigating the proteins released by yeasts in synthetic wine fermentations. Int. J. Food Microbiol. 2014, 171, 108–118. [Google Scholar] [CrossRef]
- Rossouw, D.; Du Toit, M.; Bauer, F.F. The impact of co-inoculation with Oenococcus oeni on the transcriptome of Saccharomyces cerevisiae and on the flavour-active metabolite profiles during fermentation in synthetic must. Food Microbiol. 2012, 29, 121–131. [Google Scholar] [CrossRef]
- Baumann, K.; Dato, L.; Graf, A.A.B.; Frascotti, G.; Dragositis, M.; Porro, D.; Mattanovich, D.; Ferrer, P.; Branduardi, P. The impact of oxygen on the transcriptome of recombinant S. cerevisiae and P. pastoris–a comparative analysis. BMC Genom. 2011, 12, 218. [Google Scholar] [CrossRef]
- Hodgins-Davis, A.; Adomas, A.B.; Warringer, J.; Townsend, J.P. Abundant geneby-environment interactions in gene expression reaction norms to copper within Saccharomyces cerevisiae. Genome Biol. Evol. 2012, 11, 1061–1079. [Google Scholar] [CrossRef]
- Rivero, D.; Berná, L.; Stefanini, I.; Baruffini, E.; Bergerat, A.; Csikász-Nagy, A.; Filippo, C.; Cavalieri, D. Hsp12p and PAU genes are involved in ecological interactions between natural yeast strains. Environ. Microbiol. 2015, 17, 3069–3081. [Google Scholar] [CrossRef] [PubMed]
- Henschke, P.; Jiranek, V. Yeasts-metabolism of nitrogen compounds. In Wine Microbiology and Biotechnology; Fleet, G.H., Ed.; Harwood Academic: Chur, Switzerland, 1993; pp. 77–164. [Google Scholar]

| Organic Acid mg/L | Yeast Strains | |||||
|---|---|---|---|---|---|---|
| K. marxianus 1695 | K. marxianus 1042 | L. thermotolerans 1548 | L. thermotolerans FBA-2 | B. bruxellens WDB24 | S. cerevisiae 68obg | |
| Lactic acid | 539.6 ± 2.31 d | 966.83 ± 6.13 c | 1517.57 ± 6.75 b | 2573.18 ± 3.66 a | 254.66 ± 4.18 e | 156.37 ± 3.63 f |
| Acetic acid | 168.12 ± 3.81 d | 239.23 ± 0.45 b | 226.6 ± 1.42 c | 121.92 ± 2.14 f | 324.17 ± 2.43 a | 104.31 ± 1.59 e |
| Formic acid | 3.75 ± 0.44 cd | 4.3 ± 0.10 c | 4.15 ± 0.07 cd | 11.54 ± 0.57 a | 8.87 ± 0.52 b | 3.64 ± 0.33 d |
| Pyruvic acid | 106.68 ± 4.16 d | 132.15 ± 0.78 c | 98.59 ± 2.52 e | 145.94 ± 1.95 b | 130.74 ± 1.78 c | 157.38 ± 2.73 a |
| Succinic acid | 249.27 ± 4.43 c | 249.83 ± 1.69 c | 312.49 ± 0.50 b | 209.69 ± 2.33 d | 212.37 ± 3.29 d | 513.41 ± 10.21 a |
| Oxalic acid | 26.22 ± 0.45 c | 29.52 ± 1.22 b | 22.7 ± 1.25 d | 37.33 ± 0.92 a | 24.86 ± 0.84 cd | 29.54 ± 2.71 b |
| Fumaric acid | 2.08 ± 0.17 c | 2.59 ± 0.07 c | 2.45 ± 0.29 c | 2.68 ± 0.33 c | 3.31 ± 0.08 b | 5.36 ± 0.75 a |
| Citric acid | 195.50 ± 1.11 c | 194.17 ± 0.56 c | 212.52 ± 0.88 b | 193.26 ± 2.99 c | 188.97 ± 1.29 d | 293.41 ± 1.23 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, X.; Guo, L.; Li, Y.; Chen, X.; Song, Y.; Li, S. Transcriptional Analysis of Mixed-Culture Fermentation of Lachancea thermotolerans and Saccharomyces cerevisiae for Natural Fruity Sour Beer. Fermentation 2024, 10, 180. https://doi.org/10.3390/fermentation10040180
Fu X, Guo L, Li Y, Chen X, Song Y, Li S. Transcriptional Analysis of Mixed-Culture Fermentation of Lachancea thermotolerans and Saccharomyces cerevisiae for Natural Fruity Sour Beer. Fermentation. 2024; 10(4):180. https://doi.org/10.3390/fermentation10040180
Chicago/Turabian StyleFu, Xiaofen, Liyun Guo, Yumeng Li, Xinyu Chen, Yumei Song, and Shizhong Li. 2024. "Transcriptional Analysis of Mixed-Culture Fermentation of Lachancea thermotolerans and Saccharomyces cerevisiae for Natural Fruity Sour Beer" Fermentation 10, no. 4: 180. https://doi.org/10.3390/fermentation10040180
APA StyleFu, X., Guo, L., Li, Y., Chen, X., Song, Y., & Li, S. (2024). Transcriptional Analysis of Mixed-Culture Fermentation of Lachancea thermotolerans and Saccharomyces cerevisiae for Natural Fruity Sour Beer. Fermentation, 10(4), 180. https://doi.org/10.3390/fermentation10040180





