Lactococcus lactis in Dairy Fermentation—Health-Promoting and Probiotic Properties
Abstract
1. Introduction
2. Lactic Acid Bacteria
2.1. Lactococcus lactis
2.2. L. lactis Application in the Dairy Industry
2.3. Probiotic Features of L. lactis
3. Safety Assessment of L. lactis
3.1. Virulence Assessment
3.2. Antibiotic Resistance
3.3. Hemolytic Activity
3.4. Biogenic Amines
4. Bioactive Compounds Produced by L. lactis
4.1. Gamma-Aminobutyric Acid
4.2. Exopolysaccharides (EPSs)
4.3. Lactic Acid and Citrate
4.4. Bacteriocins
4.5. Bioactive Peptides
4.6. Vitamins
4.7. Conjugated Linoleic Acid (CLA)
4.8. Enzymes
5. Bioinformatic Tool Application for L. lactis Evaluation
5.1. Prediction of Potential Antimicrobial Resistance and the Detection of Antimicrobial Resistance Genes (ARGs)
5.2. Metabolic Pathway Prediction
5.3. Bacteriocin and Other Bioactive Compound Prediction and Identification
5.4. Genetic Element Annotation and Identification
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Petrescu, D.C.; Vermeir, I.; Petrescu-Mag, R.M. Consumer Understanding of Food Quality, Healthiness, and Environmental Impact: A Cross-National Perspective. Int. J. Environ. Res. Public Health 2020, 17, 169. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ren, M.; Duo, L.; Li, J.; Wang, S.; Sun, Y.; Li, M.; Ren, W.; Hou, Q.; Yu, J.; et al. Fermentation Characteristics of Lactococcus lactis subsp. lactis Isolated From Naturally Fermented Dairy Products and Screening of Potential Starter Isolates. Front. Microbiol. 2020, 11, 1794. [Google Scholar] [CrossRef] [PubMed]
- Yerlikaya, O. Probiotic Potential and Biochemical and Technological Properties of Lactococcus lactis ssp. lactis Strains Isolated from Raw Milk and Kefir Grains. J. Dairy Sci. 2019, 102, 124–134. [Google Scholar] [CrossRef]
- Yadav, M.; Shukla, P. Efficient Engineered Probiotics Using Synthetic Biology Approaches: A Review. Biotechnol. Appl. Biochem. 2020, 67, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.T.; Lu, P.; Parrella, J.A.; Leggette, H.R. Consumer Acceptance toward Functional Foods: A Scoping Review. Int. J. Environ. Res. Public Health 2022, 19, 1217. [Google Scholar] [CrossRef] [PubMed]
- Linares, D.M.; Gómez, C.; Renes, E.; Fresno, J.M.; Tornadijo, M.E.; Ross, R.P.; Stanton, C. Lactic Acid Bacteria and Bifidobacteria with Potential to Design Natural Biofunctional Health-Promoting Dairy Foods. Front. Microbiol. 2017, 8, 846. [Google Scholar] [CrossRef]
- Farnworth, E.R.; Champagne, C.P. Chapter 20—Production of Probiotic Cultures and Their Incorporation into Foods. In Probiotics, Prebiotics, and Synbiotics; Watson, R.R., Preedy, V.R., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 303–318. ISBN 978-0-12-802189-7. [Google Scholar]
- Olicón-Hernández, D.R.; Guerra-Sánchez, G.; Porta, C.J.; Santoyo-Tepole, F.; Hernández-Cortez, C.; Tapia-García, E.Y.; Chávez-Camarillo, G.M. Fundaments and Concepts on Screening of Microorganisms for Biotechnological Applications. Mini Review. Curr Microbiol 2022, 79, 373. [Google Scholar] [CrossRef]
- Carnevali, P.; Ciati, R.; Leporati, A.; Paese, M. Liquid Sourdough Fermentation: Industrial Application Perspectives. Food Microbiol. 2007, 24, 150–154. [Google Scholar] [CrossRef]
- Manini, F.; Casiraghi, M.C.; Poutanen, K.; Brasca, M.; Erba, D.; Plumed-Ferrer, C. Characterization of Lactic Acid Bacteria Isolated from Wheat Bran Sourdough. LWT-Food Sci. Technol. 2016, 66, 275–283. [Google Scholar] [CrossRef]
- de Souza, E.L.; de Oliveira, K.Á.; de Oliveira, M.E. Influence of Lactic Acid Bacteria Metabolites on Physical and Chemical Food Properties. Curr. Opin. Food Sci. 2023, 49, 100981. [Google Scholar] [CrossRef]
- Ribeiro, S.C.; O’Connor, P.M.; Ross, R.P.; Stanton, C.; Silva, C.C.G. An Anti-Listerial Lactococcus lactis Strain Isolated from Azorean Pico Cheese Produces Lacticin 481. Int. Dairy J. 2016, 63, 18–28. [Google Scholar] [CrossRef]
- Arena, M.P.; Capozzi, V.; Russo, P.; Drider, D.; Spano, G.; Fiocco, D. Immunobiosis and Probiosis: Antimicrobial Activity of Lactic Acid Bacteria with a Focus on Their Antiviral and Antifungal Properties. Appl. Microbiol. Biotechnol. 2018, 102, 9949–9958. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Nero, L.A.; Todorov, S.D. Safety Profiles of Beneficial Lactic Acid Bacteria Isolated from Dairy Systems. Braz. J. Microbiol. 2020, 51, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Bojanic Rasovic, M.; Mayrhofer, S.; Martinovic, A.; Dürr, K.; Domig, K.J. Lactococci of Local Origin as Potential Starter Cultures for Traditional Montenegrin Cheese Production. Food Technol. Biotechnol. 2017, 55, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Xie, X.; Du, T.; Jiang, X.; Miao, W.; Wang, T. Lactococcus lactis, a Bacterium with Probiotic Functions and Pathogenicity. World J. Microbiol. Biotechnol. 2023, 39, 325. [Google Scholar] [CrossRef] [PubMed]
- Cano-Lozano, J.A.; Villamil Diaz, L.M.; Melo Bolivar, J.F.; Hume, M.E.; Ruiz Pardo, R.Y. Probiotics in Tilapia (Oreochromis Niloticus) Culture: Potential Probiotic Lactococcus lactis Culture Conditions. J. Biosci. Bioeng. 2022, 133, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Bottacini, F.; van Sinderen, D. Towards the Diversification of Lactococcal Starter and Non-Starter Species in Mesophilic Dairy Culture Systems. Microb. Biotechnol. 2023, 16, 1745–1754. [Google Scholar] [CrossRef] [PubMed]
- Castellone, V.; Bancalari, E.; Rubert, J.; Gatti, M.; Neviani, E.; Bottari, B. Eating Fermented: Health Benefits of LAB-Fermented Foods. Foods 2021, 10, 2639. [Google Scholar] [CrossRef]
- Mokoena, M.P.; Omatola, C.A.; Olaniran, A.O. Applications of Lactic Acid Bacteria and Their Bacteriocins against Food Spoilage Microorganisms and Foodborne Pathogens. Molecules 2021, 26, 7055. [Google Scholar] [CrossRef]
- Burgain, J.; Scher, J.; Francius, G.; Borges, F.; Corgneau, M.; Revol-Junelles, A.M.; Cailliez-Grimal, C.; Gaiani, C. Lactic Acid Bacteria in Dairy Food: Surface Characterization and Interactions with Food Matrix Components. Adv. Colloid Interface Sci. 2014, 213, 21–35. [Google Scholar] [CrossRef]
- Grosu-Tudor, S.-S.; Stancu, M.-M.; Pelinescu, D.; Zamfir, M. Characterization of Some Bacteriocins Produced by Lactic Acid Bacteria Isolated from Fermented Foods. World J. Microbiol. Biotechnol. 2014, 30, 2459–2469. [Google Scholar] [CrossRef] [PubMed]
- Casaburi, A.; Di Martino, V.; Ferranti, P.; Picariello, L.; Villani, F. Technological Properties and Bacteriocins Production by Lactobacillus curvatus 54M16 and Its Use as Starter Culture for Fermented Sausage Manufacture. Food Control 2016, 59, 31–45. [Google Scholar] [CrossRef]
- Lahiri, D.; Nag, M.; Dutta, B.; Sarkar, T.; Pati, S.; Basu, D.; Abdul Kari, Z.; Wei, L.S.; Smaoui, S.; Wen Goh, K.; et al. Bacteriocin: A Natural Approach for Food Safety and Food Security. Front. Bioeng. Biotechnol. 2022, 10, 1005918. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, E.; LeBlanc, J.G.; Pérez, G. Technological Properties of Lactic Acid Bacteria Isolated from Raw Cereal Material. LWT 2016, 70, 185–191. [Google Scholar] [CrossRef]
- Lahiri, D.; Nag, M.; Sarkar, T.; Ray, R.R.; Shariati, M.A.; Rebezov, M.; Bangar, S.P.; Lorenzo, J.M.; Domínguez, R. Lactic Acid Bacteria (LAB): Autochthonous and Probiotic Microbes for Meat Preservation and Fortification. Foods 2022, 11, 2792. [Google Scholar] [CrossRef] [PubMed]
- Sadiq, S.; Imran, M.; Hassan, M.N.; Iqbal, M.; Zafar, Y.; Hafeez, F.Y. Potential of Bacteriocinogenic Lactococcus lactis subsp. lactis Inhabiting Low pH Vegetables to Produce Nisin Variants. LWT-Food Sci. Technol. 2014, 59, 204–210. [Google Scholar] [CrossRef]
- Ünlü, G.; Nielsen, B.; Ionita, C. Production of Antilisterial Bacteriocins from Lactic Acid Bacteria in Dairy-Based Media: A Comparative Study. Probiotics Antimicro. Prot. 2015, 7, 259–274. [Google Scholar] [CrossRef]
- Tavares, L.M.; de Jesus, L.C.L.; da Silva, T.F.; Barroso, F.A.L.; Batista, V.L.; Coelho-Rocha, N.D.; Azevedo, V.; Drumond, M.M.; Mancha-Agresti, P. Novel Strategies for Efficient Production and Delivery of Live Biotherapeutics and Biotechnological Uses of Lactococcus lactis: The Lactic Acid Bacterium Model. Front. Bioeng. Biotechnol. 2020, 8, 517166. [Google Scholar] [CrossRef]
- Martín, R.; Langella, P. Emerging Health Concepts in the Probiotics Field: Streamlining the Definitions. Front. Microbiol. 2019, 10, 1047. [Google Scholar] [CrossRef]
- Cavanagh, D.; Fitzgerald, G.F.; McAuliffe, O. From Field to Fermentation: The Origins of Lactococcus lactis and Its Domestication to the Dairy Environment. Food Microbiol. 2015, 47, 45–61. [Google Scholar] [CrossRef]
- Nicolescu, C.M.; Bumbac, M.; Buruleanu, C.L.; Popescu, E.C.; Stanescu, S.G.; Georgescu, A.A.; Toma, S.M. Biopolymers Produced by Lactic Acid Bacteria: Characterization and Food Application. Polymers 2023, 15, 1539. [Google Scholar] [CrossRef] [PubMed]
- Abdul Hakim, B.N.; Xuan, N.J.; Oslan, S.N.H. A Comprehensive Review of Bioactive Compounds from Lactic Acid Bacteria: Potential Functions as Functional Food in Dietetics and the Food Industry. Foods 2023, 12, 2850. [Google Scholar] [CrossRef] [PubMed]
- Passerini, D.; Beltramo, C.; Coddeville, M.; Quentin, Y.; Ritzenthaler, P.; Daveran-Mingot, M.-L.; Bourgeois, P.L. Genes but Not Genomes Reveal Bacterial Domestication of Lactococcus lactis. PLoS ONE 2010, 5, e15306. [Google Scholar] [CrossRef] [PubMed]
- Tidona, F.; Meucci, A.; Povolo, M.; Pelizzola, V.; Zago, M.; Contarini, G.; Carminati, D.; Giraffa, G. Applicability of Lactococcus hircilactis and Lactococcus laudensis as Dairy Cultures. Int. J. Food Microbiol. 2018, 271, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Li, T.T.; Tian, W.L.; Gu, C.T. Elevation of Lactococcus lactis subsp. cremoris to the Species Level as Lactococcus cremoris sp. nov. and Transfer of Lactococcus lactis subsp. tructae to Lactococcus cremoris as Lactococcus cremoris subsp. tructae comb. nov. Int. J. Syst. Evol. Microbiol. 2019, 71, 004727. [Google Scholar] [CrossRef] [PubMed]
- Samelis, J.; Giannou, E.; Pappa, E.C.; Bogović-Matijašić, B.; Lianou, A.; Parapouli, M.; Drainas, C. Behavior of Artificial Listerial Contamination in Model Greek Graviera Cheeses Manufactured with the Indigenous Nisin A-Producing Strain Lactococcus lactis subsp. cremoris M104 as Costarter Culture. J. Food Saf. 2017, 37, e12326. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. Notification That New Names of Prokaryotes, New Combinations and New Taxonomic Opinions Have Appeared in Volume 71, Part 3 of the IJSEM. Int. J. Syst. Evol. Microbiol. 2021, 71, 004812. [Google Scholar] [CrossRef]
- Bozoudi, D.; Kotzamanidis, C.; Hatzikamari, M.; Tzanetakis, N.; Menexes, G.; Litopoulou-Tzanetaki, E. A Comparison for Acid Production, Proteolysis, Autolysis and Inhibitory Properties of Lactic Acid Bacteria from Fresh and Mature Feta PDO Greek Cheese, Made at Three Different Mountainous Areas. Int. J. Food Microbiol. 2015, 200, 87–96. [Google Scholar] [CrossRef]
- Kleerebezemab, M.; Hols, P.; Hugenholtz, J. Lactic Acid Bacteria as a Cell Factory: Rerouting of Carbon Metabolism in Lactococcus lactis by Metabolic Engineering. Enzym. Microb. Technol. 2000, 26, 840–848. [Google Scholar] [CrossRef]
- Ruggirello, M.; Cocolin, L.; Dolci, P. Fate of Lactococcus lactis Starter Cultures during Late Ripening in Cheese Models. Food Microbiol. 2016, 59, 112–118. [Google Scholar] [CrossRef]
- Rattanachaikunsopon, P.; Phumkhachorn, P. Lactic Acid Bacteria: Their Antimicrobial Compounds and Their Uses in Food Production. Ann. Biol. Res. 2010, 1, 218–228. [Google Scholar]
- Alegría, A.; Delgado, S.; Roces, C.; López, B.; Mayo, B. Bacteriocins Produced by Wild Lactococcus lactis Strains Isolated from Traditional, Starter-Free Cheeses Made of Raw Milk. Int. J. Food Microbiol. 2010, 143, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Song, A.A.-L.; In, L.L.A.; Lim, S.H.E.; Rahim, R.A. A Review on Lactococcus lactis: From Food to Factory. Microb. Cell Fact. 2017, 16, 55. [Google Scholar] [CrossRef] [PubMed]
- Mileriene, J.; Serniene, L.; Kondrotiene, K.; Lauciene, L.; Kasetiene, N.; Sekmokiene, D.; Andruleviciute, V.; Malakauskas, M. Quality and Nutritional Characteristics of Traditional Curd Cheese Enriched with Thermo-Coagulated Acid Whey Protein and Indigenous Lactococcus lactis Strain. Int. J. Food Sci. Technol. 2021, 56, 2853–2863. [Google Scholar] [CrossRef]
- Mileriene, J.; Serniene, L.; Kondrotiene, K.; Santarmaki, V.; Kourkoutas, Y.; Vasiliauskaite, A.; Lauciene, L.; Malakauskas, M. Indigenous Lactococcus lactis with Probiotic Properties: Evaluation of Wet, Thermally- and Freeze-Dried Raisins as Supports for Cell Immobilization, Viability and Aromatic Profile in Fresh Curd Cheese. Foods 2022, 11, 1311. [Google Scholar] [CrossRef] [PubMed]
- Fusieger, A.; Martins, M.C.F.; de Freitas, R.; Nero, L.A.; de Carvalho, A.F. Technological Properties of Lactococcus lactis subsp. lactis bv. diacetylactis Obtained from Dairy and Non-Dairy Niches. Braz. J. Microbiol. 2020, 51, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Perin, L.M.; Miranda, R.O.; Todorov, S.D.; de Melo Franco, B.D.G.; Nero, L.A. Virulence, Antibiotic Resistance and Biogenic Amines of Bacteriocinogenic Lactococci and Enterococci Isolated from Goat Milk. Int. J. Food Microbiol. 2014, 185, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Dan, T.; Chen, Y.; Chen, X.; Sun, C.; Wang, X.; Wang, J.; Zhang, H. Isolation and Characterisation of a Lactobacillus delbrueckii subsp. bulgaricus Mutant with Low H+-ATP Ase Activity. Int. J. Dairy Technol. 2015, 68, 527–532. [Google Scholar] [CrossRef]
- Tripathi, M.K.; Giri, S.K. Probiotic Functional Foods: Survival of Probiotics during Processing and Storage. J. Funct. Foods 2014, 9, 225–241. [Google Scholar] [CrossRef]
- Shori, A.B. The Potential Applications of Probiotics on Dairy and Non-Dairy Foods Focusing on Viability during Storage. Biocatal. Agric. Biotechnol. 2015, 4, 423–431. [Google Scholar] [CrossRef]
- Turgut, T.; Cakmakci, S. Probiotic Strawberry Yogurts: Microbiological, Chemical and Sensory Properties. Probiotics Antimicro. Prot. 2018, 10, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Panghal, A.; Janghu, S.; Virkar, K.; Gat, Y.; Kumar, V.; Chhikara, N. Potential Non-Dairy Probiotic Products—A Healthy Approach. Food Biosci. 2018, 21, 80–89. [Google Scholar] [CrossRef]
- Angmo, K.; Kumari, A.; Savitri; Bhalla, T.C. Probiotic Characterization of Lactic Acid Bacteria Isolated from Fermented Foods and Beverage of Ladakh. LWT-Food Sci. Technol. 2016, 66, 428–435. [Google Scholar] [CrossRef]
- Sokovic Bajic, S.; Djokic, J.; Dinic, M.; Veljovic, K.; Golic, N.; Mihajlovic, S.; Tolinacki, M. GABA-Producing Natural Dairy Isolate From Artisanal Zlatar Cheese Attenuates Gut Inflammation and Strengthens Gut Epithelial Barrier in Vitro. Front. Microbiol. 2019, 10, 527. [Google Scholar] [CrossRef] [PubMed]
- Abushelaibi, A.; Al-Mahadin, S.; El-Tarabily, K.; Shah, N.P.; Ayyash, M. Characterization of Potential Probiotic Lactic Acid Bacteria Isolated from Camel Milk. LWT-Food Sci. Technol. 2017, 79, 316–325. [Google Scholar] [CrossRef]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.-A.G.; Tsakalidou, E.; Nychas, G.-J.E.; Panagou, E.Z.; Tassou, C.C. Selection of Potential Probiotic Lactic Acid Bacteria from Fermented Olives by in Vitro Tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef]
- Sharma, P.; Tomar, S.K.; Goswami, P.; Sangwan, V.; Singh, R. Antibiotic Resistance among Commercially Available Probiotics. Food Res. Int. 2014, 57, 176–195. [Google Scholar] [CrossRef]
- Vasudha, S.; Mishra, H.N. Non Dairy Probiotic Beverages. Int. Food Res. J. 2013, 20, 7. [Google Scholar]
- Kondrotiene, K.; Lauciene, L.; Andruleviciute, V.; Kasetiene, N.; Serniene, L.; Sekmokiene, D.; Malakauskas, M. Safety Assessment and Preliminary In Vitro Evaluation of Probiotic Potential of Lactococcus lactis Strains Naturally Present in Raw and Fermented Milk. Curr. Microbiol. 2020, 77, 3013–3023. [Google Scholar] [CrossRef]
- Akbar, A.; Sadiq, M.B.; Ali, I.; Anwar, M.; Muhammad, N.; Muhammad, J.; Shafee, M.; Ullah, S.; Gul, Z.; Qasim, S.; et al. Lactococcus lactis subsp. lactis Isolated from Fermented Milk Products and Its Antimicrobial Potential. CyTA-J. Food 2019, 17, 214–220. [Google Scholar] [CrossRef]
- Hermanns, G.; Funck, G.D.; Schmidt, J.T.; Pereira, J.Q.; Brandelli, A.; Richards, N.S.P.d.S. Evaluation of Probiotic Characteristics of Lactic Acid Bacteria Isolated from Artisan Cheese. J. Food Saf. 2014, 34, 380–387. [Google Scholar] [CrossRef]
- Das, P.; Khowala, S.; Biswas, S. In Vitro Probiotic Characterization of Lactobacillus casei Isolated from Marine Samples. LWT 2016, 73, 383–390. [Google Scholar] [CrossRef]
- Zielińska, D.; Rzepkowska, A.; Radawska, A.; Zieliński, K. In Vitro Screening of Selected Probiotic Properties of Lactobacillus Strains Isolated from Traditional Fermented Cabbage and Cucumber. Curr. Microbiol. 2015, 70, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Jawan, R.; Abbasiliasi, S.; Mustafa, S.; Kapri, M.R.; Halim, M.; Ariff, A.B. In Vitro Evaluation of Potential Probiotic Strain Lactococcus lactis Gh1 and Its Bacteriocin-Like Inhibitory Substances for Potential Use in the Food Industry. Probiotics Antimicro. Prot. 2021, 13, 422–440. [Google Scholar] [CrossRef] [PubMed]
- Vinderola, C.G.; Reinheimer, J.A. Lactic Acid Starter and Probiotic Bacteria: A Comparative “in Vitro” Study of Probiotic Characteristics and Biological Barrier Resistance. Food Res. Int. 2003, 36, 895–904. [Google Scholar] [CrossRef]
- Jung, M.Y.; Lee, C.; Seo, M.-J.; Roh, S.W.; Lee, S.H. Characterization of a Potential Probiotic Bacterium Lactococcus raffinolactis WiKim0068 Isolated from Fermented Vegetable Using Genomic and in Vitro Analyses. BMC Microbiol. 2020, 20, 136. [Google Scholar] [CrossRef]
- Sałański, P.; Kowalczyk, M.; Bardowski, J.K.; Szczepankowska, A.K. Health-Promoting Nature of Lactococcus lactis IBB109 and Lactococcus lactis IBB417 Strains Exhibiting Proliferation Inhibition and Stimulation of Interleukin-18 Expression in Colorectal Cancer Cells. Front. Microbiol. 2022, 13, 822912. [Google Scholar] [CrossRef]
- Tenea, G.N.; Suárez, J. Probiotic Potential and Technological Properties of Bacteriocinogenic Lactococcus lactis subsp. lactis UTNGt28 from a Native Amazonian Fruit as a Yogurt Starter Culture. Microorganisms 2020, 14, 733. [Google Scholar] [CrossRef]
- Mileriene, J.; Serniene, L.; Kasparaviciene, B.; Lauciene, L.; Kasetiene, N.; Zakariene, G.; Kersiene, M.; Leskauskaite, D.; Viskelis, J.; Kourkoutas, Y.; et al. Exploring the Potential of Sustainable Acid Whey Cheese Supplemented with Apple Pomace and GABA-Producing Indigenous Lactococcus lactis Strain. Microorganisms 2023, 11, 436. [Google Scholar] [CrossRef]
- Hurtado-Romero, A.; Del Toro-Barbosa, M.; Gradilla-Hernández, M.S.; Garcia-Amezquita, L.E.; García-Cayuela, T. Probiotic Properties, Prebiotic Fermentability, and GABA-Producing Capacity of Microorganisms Isolated from Mexican Milk Kefir Grains: A Clustering Evaluation for Functional Dairy Food Applications. Foods 2021, 10, 2275. [Google Scholar] [CrossRef]
- Jaskulski, I.B.; Uecker, J.; Bordini, F.; Moura, F.; Gonçalves, T.; Chaves, N.G.; Camargo, F.; Grecco, F.B.; Fiorentini, Â.M.; da Silva, W.P.; et al. In Vivo Action of Lactococcus lactis subsp. lactis Isolate (R7) with Probiotic Potential in the Stabilization of Cancer Cells in the Colorectal Epithelium. Process Biochem. 2020, 91, 165–171. [Google Scholar] [CrossRef]
- Ramalho, J.B.; Soares, M.B.; Spiazzi, C.C.; Bicca, D.F.; Soares, V.M.; Pereira, J.G.; da Silva, W.P.; Sehn, C.P.; Cibin, F.W.S. In Vitro Probiotic and Antioxidant Potential of Lactococcus lactis subsp. cremoris LL95 and Its Effect in Mice Behaviour. Nutrients 2019, 11, 901. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Singh, N.; Singh, S.; Khare, S.K.; Nain, P.K.S.; Nain, L. Potent γ-Amino Butyric Acid Producing Psychobiotic Lactococcus lactis LP-68 from Non-Rhizospheric Soil of Syzygium Cumini (Black Plum). Arch. Microbiol. 2021, 204, 82. [Google Scholar] [CrossRef] [PubMed]
- da Silva, L.A.; Lopes Neto, J.H.P.; Cardarelli, H.R. Safety and Probiotic Functionality of Isolated Goat Milk Lactic Acid Bacteria. Ann. Microbiol. 2019, 69, 1497–1505. [Google Scholar] [CrossRef]
- Kazancıgil, E.; Demirci, T.; Öztürk-Negiş, H.İ.; Akın, N. Isolation, Technological Characterization and in Vitro Probiotic Evaluation of Lactococcus Strains from Traditional Turkish Skin Bag Tulum Cheeses. Ann. Microbiol. 2019, 69, 1275–1287. [Google Scholar] [CrossRef]
- Colautti, A.; Arnoldi, M.; Comi, G.; Iacumin, L. Antibiotic Resistance and Virulence Factors in Lactobacilli: Something to Carefully Consider. Food Microbiol. 2022, 103, 103934. [Google Scholar] [CrossRef]
- EFSA, E.F.S. Introduction of a Qualified Presumption of Safety (QPS) Approach for Assessment of Selected Microorganisms Referred to EFSA—Opinion of the Scientific Committee. EFSA J. 2007, 5, 587. [Google Scholar] [CrossRef]
- EFSA, E.F.S. EFSA Statement on the Requirements for Whole Genome Sequence Analysis of Microorganisms Intentionally Used in the Food Chain. EFSA J. 2021, 19, e06506. [Google Scholar]
- Herreros, M.A.; Sandoval, H.; González, L.; Castro, J.M.; Fresno, J.M.; Tornadijo, M.E. Antimicrobial Activity and Antibiotic Resistance of Lactic Acid Bacteria Isolated from Armada Cheese (a Spanish Goats’ Milk Cheese). Food Microbiol. 2005, 22, 455–459. [Google Scholar] [CrossRef]
- Colautti, A.; Orecchia, E.; Comi, G.; Iacumin, L. Lactobacilli, a Weapon to Counteract Pathogens through the Inhibition of Their Virulence Factors. J. Bacteriol. 2022, 204, e0027222. [Google Scholar] [CrossRef]
- Pieniz, S.; de Moura, T.M.; Cassenego, A.P.V.; Andreazza, R.; Frazzon, A.P.G.; Camargo, F.A.d.O.; Brandelli, A. Evaluation of Resistance Genes and Virulence Factors in a Food Isolated Enterococcus durans with Potential Probiotic Effect. Food Control 2015, 51, 49–54. [Google Scholar] [CrossRef]
- Sylvere, N.; Mustopa, A.Z.; Budiarti, S.; Meilina, L.; Hertati, A.; Handayani, I. Whole-Genome Sequence Analysis and Probiotic Characteristics of Lactococcus lactis subsp. lactis Strain Lac3 Isolated from Traditional Fermented Buffalo Milk (Dadih). J. Genet. Eng. Biotechnol. 2023, 21, 49. [Google Scholar] [CrossRef] [PubMed]
- Brinkwirth, S.; Ayobami, O.; Eckmanns, T.; Markwart, R. Hospital-Acquired Infections Caused by Enterococci: A Systematic Review and Meta-Analysis, WHO European Region, 1 January 2010 to 4 February 2020. Eurosurveillance 2021, 26, 2001628. [Google Scholar] [CrossRef]
- Șchiopu, P.; Toc, D.A.; Colosi, I.A.; Costache, C.; Ruospo, G.; Berar, G.; Gălbău Ștefan, G.; Ghilea, A.C.; Botan, A.; Pană, A.-G.; et al. An Overview of the Factors Involved in Biofilm Production by the Enterococcus Genus. Int. J. Mol. Sci. 2023, 24, 11577. [Google Scholar] [CrossRef] [PubMed]
- Chajęcka-Wierzchowska, W.; Zadernowska, A.; Łaniewska-Trokenheim, Ł. Virulence Factors of Enterococcus spp. Presented in Food. LWT 2017, 75, 670–676. [Google Scholar] [CrossRef]
- Czarnowska-Kujawska, M.; Paszczyk, B. Changes in the Folate Content and Fatty Acid Profile in Fermented Milk Produced with Different Starter Cultures during Storage. Molecules 2021, 26, 6063. [Google Scholar] [CrossRef] [PubMed]
- Eubank, T.A.; Gonzales-Luna, A.J.; Hurdle, J.G.; Garey, K.W. Genetic Mechanisms of Vancomycin Resistance in Clostridioides Difficile: A Systematic Review. Antibiotics 2022, 11, 258. [Google Scholar] [CrossRef] [PubMed]
- Cavera, V.L.; Arthur, T.D.; Kashtanov, D.; Chikindas, M.L. Bacteriocins and Their Position in the next Wave of Conventional Antibiotics. Int. J. Antimicrob. Agents 2015, 46, 494–501. [Google Scholar] [CrossRef]
- Zycka-Krzesinska, J.; Boguslawska, J.; Aleksandrzak-Piekarczyk, T.; Jopek, J.; Bardowski, J.K. Identification and Characterization of Tetracycline Resistance in Lactococcus lactis Isolated from Polish Raw Milk and Fermented Artisanal Products. Int. J. Food Microbiol. 2015, 211, 134–141. [Google Scholar] [CrossRef]
- Miquel, S.; Beaumont, M.; Martín, R.; Langella, P.; Braesco, V.; Thomas, M. A Proposed Framework for an Appropriate Evaluation Scheme for Microorganisms as Novel Foods with a Health Claim in Europe. Microb. Cell Fact. 2015, 14, 48. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Guidance on the Assessment of Bacterial Susceptibility to Antimicrobials of Human and Veterinary Importance. EFSA J. 2012, 10, 2740.
- Kim, T.; Mondal, S.C.; Jeong, C.; Kim, S.; Ban, O.; Jung, Y.H.; Yang, J.; Kim, S. Safety Evaluation of Lactococcus lactis IDCC 2301 Isolated from Homemade Cheese. Food Sci. Nutr. 2021, 10, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Shin, E.; Paek, J.J.; Lee, Y. Antimicrobial Resistance of Seventy Lactic Acid Bacteria Isolated from Commercial Probiotics in Korea. J. Microbiol. Biotechnol. 2023, 33, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Araújo, C.; Muñoz-Atienza, E.; Ramírez, M.; Poeta, P.; Igrejas, G.; Hernández, P.E.; Herranz, C.; Cintas, L.M. Safety Assessment, Genetic Relatedness and Bacteriocin Activity of Potential Probiotic Lactococcus lactis Strains from Rainbow Trout (Oncorhynchus Mykiss, Walbaum) and Rearing Environment. Eur. Food Res. Technol. 2015, 241, 647–662. [Google Scholar] [CrossRef]
- Ammor, M.S.; Flórez, A.B.; van Hoek, A.H.A.M.; de Los Reyes-Gavilán, C.G.; Aarts, H.J.M.; Margolles, A.; Mayo, B. Molecular Characterization of Intrinsic and Acquired Antibiotic Resistance in Lactic Acid Bacteria and Bifidobacteria. J. Mol. Microbiol. Biotechnol. 2008, 14, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Casarotti, S.N.; Carneiro, B.M.; Todorov, S.D.; Nero, L.A.; Rahal, P.; Penna, A.L.B. In Vitro Assessment of Safety and Probiotic Potential Characteristics of Lactobacillus Strains Isolated from Water Buffalo Mozzarella Cheese. Ann. Microbiol. 2017, 67, 289–301. [Google Scholar] [CrossRef]
- Doeun, D.; Davaatseren, M.; Chung, M.-S. Biogenic Amines in Foods. Food Sci. Biotechnol. 2017, 26, 1463–1474. [Google Scholar] [CrossRef]
- Barbieri, F.; Montanari, C.; Gardini, F.; Tabanelli, G. Biogenic Amine Production by Lactic Acid Bacteria: A Review. Foods 2019, 8, 17. [Google Scholar] [CrossRef]
- Kononiuk, A.D.; Karwowska, M. Influence of Freeze-Dried Acid Whey Addition on Biogenic Amines Formation in a Beef and Deer Dry Fermented Sausages without Added Nitrite. Asian-Australas J. Anim. Sci. 2020, 33, 332–338. [Google Scholar] [CrossRef]
- Moniente, M.; García-Gonzalo, D.; Llamas-Arriba, M.G.; Garate, J.; Ontañón, I.; Jaureguibeitia, A.; Virto, R.; Pagán, R.; Botello-Morte, L. The Significance of Cheese Sampling in the Determination of Histamine Concentration: Distribution Pattern of Histamine in Ripened Cheeses. LWT 2022, 171, 114099. [Google Scholar] [CrossRef]
- Costantini, A.; Vaudano, E.; Pulcini, L.; Carafa, T.; Garcia-Moruno, E. An Overview on Biogenic Amines in Wine. Beverages 2019, 5, 19. [Google Scholar] [CrossRef]
- Gao, X.; Li, C.; He, R.; Zhang, Y.; Wang, B.; Zhang, Z.-H.; Ho, C.-T. Research Advances on Biogenic Amines in Traditional Fermented Foods: Emphasis on Formation Mechanism, Detection and Control Methods. Food Chem. 2023, 405, 134911. [Google Scholar] [CrossRef]
- Flasarová, R.; Pachlová, V.; Buňková, L.; Menšíková, A.; Georgová, N.; Dráb, V.; Buňka, F. Biogenic Amine Production by Lactococcus lactis subsp. cremoris Strains in the Model System of Dutch-Type Cheese. Food Chem. 2016, 194, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Ladero, V.; Cañedo, E.; Pérez, M.; Martín, M.C.; Fernández, M.; Alvarez, M.A. Multiplex qPCR for the Detection and Quantification of Putrescine-Producing Lactic Acid Bacteria in Dairy Products. Food Control 2012, 27, 307–313. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism Characteristics of Lactic Acid Bacteria and the Expanding Applications in Food Industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef]
- Ladero, V.; Rattray, F.P.; Mayo, B.; Martín, M.C.; Fernández, M.; Alvarez, M.A. Sequencing and Transcriptional Analysis of the Biosynthesis Gene Cluster of Putrescine-Producing Lactococcus lactis. Appl. Environ. Microbiol. 2011, 77, 6409–6418. [Google Scholar] [CrossRef]
- del Rio, B.; Linares, D.M.; Ladero, V.; Redruello, B.; Fernández, M.; Martin, M.C.; Alvarez, M.A. Putrescine Production via the Agmatine Deiminase Pathway Increases the Growth of Lactococcus lactis and Causes the Alkalinization of the Culture Medium. Appl. Microbiol. Biotechnol. 2015, 99, 897–905. [Google Scholar] [CrossRef]
- del Rio, B.; Redruello, B.; Linares, D.M.; Ladero, V.; Ruas-Madiedo, P.; Fernandez, M.; Martin, M.C.; Alvarez, M.A. The Biogenic Amines Putrescine and Cadaverine Show in Vitro Cytotoxicity at Concentrations That Can Be Found in Foods. Sci. Rep. 2019, 9, 120. [Google Scholar] [CrossRef]
- Garbowska, M.; Pluta, A.; Berthold-Pluta, A. Impact of Nisin-Producing Strains of Lactococcus lactis on the Contents of Bioactive Dipeptides, Free Amino Acids, and Biogenic Amines in Dutch-Type Cheese Models. Materials 2020, 13, 1835. [Google Scholar] [CrossRef]
- Spizzirri, U.G.; Restuccia, D.; Curcio, M.; Parisi, O.I.; Iemma, F.; Picci, N. Determination of Biogenic Amines in Different Cheese Samples by LC with Evaporative Light Scattering Detector. J. Food Compos. Anal. 2013, 29, 43–51. [Google Scholar] [CrossRef]
- EFSA Scientific Opinion on Risk Based Control of Biogenic Amine Formation in Fermented foods-European Food Safety Authority Panel on Biological Hazards (BIOHAZ). EFSA J. 2011, 9, 2393–2486. [CrossRef]
- Moret, S.; Bortolomeazzi, R.; Feruglio, M.; Lercker, G. Determination of Biogenic Amines in Italian Cheeses [Amine Biogeniche Nei Formaggi Italiani]. Sci. E Tec. Latt.-Casearia 1992, 43, 187–198. [Google Scholar]
- Zdolec, N.; Bogdanović, T.; Severin, K.; Dobranić, V.; Kazazić, S.; Grbavac, J.; Pleadin, J.; Petričević, S.; Kiš, M. Biogenic Amine Content in Retailed Cheese Varieties Produced with Commercial Bacterial or Mold Cultures. Processes 2022, 10, 10. [Google Scholar] [CrossRef]
- Kandasamy, S.; Yoo, J.; Yun, J.; Kang, H.B.; Seol, K.-H.; Ham, J.-S. Quantitative Analysis of Biogenic Amines in Different Cheese Varieties Obtained from the Korean Domestic and Retail Markets. Metabolites 2021, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Berthoud, H.; Wechsler, D.; Irmler, S. Production of Putrescine and Cadaverine by Paucilactobacillus Wasatchensis. Front. Microbiol. 2022, 13, 842403. [Google Scholar] [CrossRef] [PubMed]
- Buňka, F.; Zálešáková, L.; Flasarová, R.; Pachlová, V.; Budinský, P.; Buňková, L. Biogenic Amines Content in Selected Commercial Fermented Products of Animal Origin. J. Microbiol. Biotechnol. Food Sci. 2012, 2, 209–218. [Google Scholar]
- Novella-Rodríguez, S.; Veciana-Nogués, M.T.; Roig-Sagués, A.X.; Trujillo-Mesa, A.J.; Vidal-Carou, M.C. Evaluation of Biogenic Amines and Microbial Counts throughout the Ripening of Goat Cheeses from Pasteurized and Raw Milk. J. Dairy Res. 2004, 71, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Marijan, A.; Džaja, P.; Bogdanović, T.; Škoko, I.; Cvetnić, Ž.; Dobranić, V.; Zdolec, N.; Šatrović, E.; Severin, K. Influence of Ripening Time on the Amount of Certain Biogenic Amines in Rind and Core of Cow Milk Livno Cheese. Mljekarstvo Časopis Za Unaprjeđenje Proizv. I Prerade Mlijeka 2014, 64, 159–169. [Google Scholar]
- Bogdanović, T.; Petričević, S.; Brkljača, M.; Listeš, I.; Pleadin, J. Biogenic Amines in Selected Foods of Animal Origin Obtained from the Croatian Retail Market. Food Addit. Contam. Part A 2020, 37, 815–830. [Google Scholar] [CrossRef]
- Shalaby, M.A.; Kassem, M.A.; Morsy, O.; Mohamed, N.M. Anti-Tyramine Potential of Lactobacillus rhamnosus (LGG®) in Cheese Samples Collected from Alexandria, Egypt. Food Biotechnol. 2020, 34, 243–261. [Google Scholar] [CrossRef]
- Schirone, M.; Esposito, L.; D’Onofrio, F.; Visciano, P.; Martuscelli, M.; Mastrocola, D.; Paparella, A. Biogenic Amines in Meat and Meat Products: A Review of the Science and Future Perspectives. Foods 2022, 11, 788. [Google Scholar] [CrossRef]
- Decadt, H.; Weckx, S.; De Vuyst, L. The Rotation of Primary Starter Culture Mixtures Results in Batch-to-Batch Variations during Gouda Cheese Production. Front. Microbiol. 2023, 14, 1128394. [Google Scholar] [CrossRef] [PubMed]
- Ladero, V.; Martín, M.C.; Redruello, B.; Mayo, B.; Flórez, A.B.; Fernández, M.; Alvarez, M.A. Genetic and Functional Analysis of Biogenic Amine Production Capacity among Starter and Non-Starter Lactic Acid Bacteria Isolated from Artisanal Cheeses. Eur. Food Res. Technol. 2015, 241, 377–383. [Google Scholar] [CrossRef]
- Perin, L.M.; Dal Bello, B.; Belviso, S.; Zeppa, G.; de Carvalho, A.F.; Cocolin, L.; Nero, L.A. Microbiota of Minas Cheese as Influenced by the Nisin Producer Lactococcus lactis subsp. lactis GLc05. Int. J. Food Microbiol. 2015, 214, 159–167. [Google Scholar] [CrossRef]
- Alvarez, M.A.; Moreno-Arribas, M.V. The Problem of Biogenic Amines in Fermented Foods and the Use of Potential Biogenic Amine-Degrading Microorganisms as a Solution. Trends Food Sci. Technol. 2014, 39, 146–155. [Google Scholar] [CrossRef]
- Omer, A.K.; Mohammed, R.R.; Ameen, P.S.M.; Abas, Z.A.; Ekici, K. Presence of Biogenic Amines in Food and Their Public Health Implications: A Review. J. Food Prot. 2021, 84, 1539–1548. [Google Scholar] [CrossRef]
- Dala-Paula, B.M.; Custódio, F.B.; Gloria, M.B. Health Concerns Associated with Biogenic Amines in Food and Interaction with Amine Oxidase Drugs. Curr. Opin. Food Sci. 2023, 54, 101090. [Google Scholar] [CrossRef]
- Fernández, M.; Linares, D.M.; Del Río, B.; Ladero, V.; Alvarez, M.A. HPLC Quantification of Biogenic Amines in Cheeses: Correlation with PCR-Detection of Tyramine-Producing Microorganisms. J. Dairy Res. 2007, 74, 276–282. [Google Scholar] [CrossRef]
- Barone, C.; Barebera, M.; Barone, M.; Parisi, S.; Zaccheo, A.; Barone, C.; Barbera, M.; Barone, M.; Parisi, S.; Zaccheo, A. Biogenic Amines in Cheeses: Types and Typical Amounts. In Chemical Evolution of Nitrogen-based Compounds in Mozzarella Cheeses; Springer: Cham, Switzerland, 2018; pp. 1–18. [Google Scholar] [CrossRef]
- Perez, R.H.; Ancuelo, A.E.; Perez, R.H.; Ancuelo, A.E. Diverse Bioactive Molecules from the Genus Lactobacillus. In Lactobacillus—A Multifunctional Genus; IntechOpen: London, UK, 2022; ISBN 978-1-80355-445-7. [Google Scholar]
- Nehal, F.; Sahnoun, M.; Smaoui, S.; Jaouadi, B.; Bejar, S.; Mohammed, S. Characterization, High Production and Antimicrobial Activity of Exopolysaccharides from Lactococcus lactis F-Mou. Microb. Pathog. 2019, 132, 10–19. [Google Scholar] [CrossRef]
- Tiwari, O.N.; Sasmal, S.; Kataria, A.K.; Devi, I. Application of Microbial Extracellular Carbohydrate Polymeric Substances in Food and Allied Industries. 3 Biotech 2020, 10, 221. [Google Scholar] [CrossRef]
- Costa, N.E.; Hannon, J.A.; Guinee, T.P.; Auty, M.A.E.; McSweeney, P.L.H.; Beresford, T.P. Effect of Exopolysaccharide Produced by Isogenic Strains of Lactococcus lactis on Half-Fat Cheddar Cheese. J. Dairy Sci. 2010, 93, 3469–3486. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, Y.; Ye, L.; Wang, C. The Anti-Cancer Effects and Mechanisms of Lactic Acid Bacteria Exopolysaccharides in Vitro: A Review. Carbohydr. Polym. 2021, 253, 117308. [Google Scholar] [CrossRef] [PubMed]
- Muscariello, L.; De Siena, B.; Marasco, R. Lactobacillus Cell Surface Proteins Involved in Interaction with Mucus and Extracellular Matrix Components. Curr. Microbiol. 2020, 77, 3831–3841. [Google Scholar] [CrossRef] [PubMed]
- Nugroho, A.D.W.; Kleerebezem, M.; Bachmann, H. A Novel Method for Long-Term Analysis of Lactic Acid and Ammonium Production in Non-Growing Lactococcus lactis Reveals Pre-Culture and Strain Dependence. Front. Bioeng. Biotechnol. 2020, 8, 580090. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.N.; Awang Adeni, D.S.; Suhaili, N.; Bujang, K. Optimisation of Pre-Harvest Sago Frond Sap for the Production of l-Lactic Acid Using Lactococcus lactis IO-1. Biocatal. Agric. Biotechnol. 2022, 43, 102435. [Google Scholar] [CrossRef]
- Azhar, N.S.; Zin, N.H.M.; Hamid, T.H.T.A. Lactococcus Lactis Strain A5 Producing Nisin-like Bacteriocin Active against Gram Positive and Negative Bacteria. Trop. Life Sci. Res. 2017, 28, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Acedo, J.Z.; Chiorean, S.; Vederas, J.C.; van Belkum, M.J. The Expanding Structural Variety among Bacteriocins from Gram-Positive Bacteria. FEMS Microbiol. Rev. 2018, 42, 805–828. [Google Scholar] [CrossRef] [PubMed]
- Takala, T.M.; Mokhtari, S.; Ahonen, S.L.; Wan, X.; Saris, P.E.J. Wild-Type Lactococcus lactis Producing Bacteriocin-like Prophage Lysins. Front. Microbiol. 2023, 14, 1219723. [Google Scholar] [CrossRef]
- Chandran, C.; Tham, H.Y.; Rahim, R.A.; Lim, S.H.E.; Yusoff, K.; Song, A.A.-L. Lactococcus lactis Secreting Phage Lysins as a Potential Antimicrobial against Multi-Drug Resistant Staphylococcus aureus. PeerJ 2022, 10, e12648. [Google Scholar] [CrossRef]
- Nebbia, S.; Lamberti, C.; Lo Bianco, G.; Cirrincione, S.; Laroute, V.; Cocaign-Bousquet, M.; Cavallarin, L.; Giuffrida, M.G.; Pessione, E. Antimicrobial Potential of Food Lactic Acid Bacteria: Bioactive Peptide Decrypting from Caseins and Bacteriocin Production. Microorganisms 2020, 9, 65. [Google Scholar] [CrossRef]
- Kurbanova, M.; Voroshilin, R.; Kozlova, O.; Atuchin, V. Effect of Lactobacteria on Bioactive Peptides and Their Sequence Identification in Mature Cheese. Microorganisms 2022, 10, 2068. [Google Scholar] [CrossRef] [PubMed]
- Kocak, A.; Sanli, T.; Anli, E.A.; Hayaloglu, A.A. Role of Using Adjunct Cultures in Release of Bioactive Peptides in White-Brined Goat-Milk Cheese. LWT 2020, 123, 109127. [Google Scholar] [CrossRef]
- Cirrincione, S.; Luganini, A.; Lamberti, C.; Manfredi, M.; Cavallarin, L.; Giuffrida, M.G.; Pessione, E. Donkey Milk Fermentation by Lactococcus lactis subsp. cremoris and Lactobacillus rhamnosus Affects the Antiviral and Antibacterial Milk Properties. Molecules 2021, 26, 5100. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Liu, R.h. Increase of Conjugated Linoleic Acid Content in Milk by Fermentation with Lactic Acid Bacteria. J. Food Sci. 2002, 67, 1731–1737. [Google Scholar] [CrossRef]
- Puniya, A.K.; Chaitanya, S.; Tyagi, A.K.; De, S.; Singh, K. Conjugated Linoleic Acid Producing Potential of Lactobacilli Isolated from the Rumen of Cattle. J. Ind. Microbiol. Biotechnol. 2008, 35, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Renes, E.; Linares, D.M.; González, L.; Fresno, J.M.; Tornadijo, M.E.; Stanton, C. Production of Conjugated Linoleic Acid and Gamma-Aminobutyric Acid by Autochthonous Lactic Acid Bacteria and Detection of the Genes Involved. J. Funct. Foods 2017, 34, 340–346. [Google Scholar] [CrossRef]
- Laroute, V.; Mazzoli, R.; Loubière, P.; Pessione, E.; Cocaign-Bousquet, M. Environmental Conditions Affecting GABA Production in Lactococcus lactis NCDO 2118. Microorganisms 2021, 9, 122. [Google Scholar] [CrossRef]
- Khalili, M.; Rad, A.H.; Khosroushahi, A.Y.; Khosravi, H.; Jafarzadeh, S. Application of Probiotics in Folate Bio-Fortification of Yoghurt. Probiotics Antimicrob. Proteins 2020, 12, 756–763. [Google Scholar] [CrossRef]
- Liu, Y.; van Bennekom, E.O.; Zhang, Y.; Abee, T.; Smid, E.J. Long-Chain Vitamin K2 Production in Lactococcus lactis Is Influenced by Temperature, Carbon Source, Aeration and Mode of Energy Metabolism. Microb. Cell Factories 2019, 18, 129. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, S.; Hao, G.; Yu, H.; Tian, H.; Zhao, G. Role of Lactic Acid Bacteria on the Yogurt Flavour: A Review. Int. J. Food Prop. 2017, 20, S316–S330. [Google Scholar] [CrossRef]
- Bøe, C.A.; Holo, H. Engineering Lactococcus lactis for Increased Vitamin K2 Production. Front. Bioeng. Biotechnol. 2020, 8, 191. [Google Scholar] [CrossRef] [PubMed]
- Mir Khan, U.; Selamoglu, Z. Use of Enzymes in Dairy Industry: A Review of Current Progress. Arch. Razi Inst. 2020, 75, 131–136. [Google Scholar] [CrossRef]
- Hernandez-Valdes, J.A.; Solopova, A.; Kuipers, O.P. Development of Lactococcus lactis Biosensors for Detection of Diacetyl. Front. Microbiol. 2020, 11, 1032. [Google Scholar] [CrossRef] [PubMed]
- Román Naranjo, D.; Callanan, M.; Thierry, A.; McAuliffe, O. Superior Esterolytic Activity in Environmental Lactococcus lactis Strains Is Linked to the Presence of the SGNH Hydrolase Family of Esterases. JDS Commun. 2020, 1, 25–28. [Google Scholar] [CrossRef]
- Park, Y.W.; Nam, M.S. Bioactive Peptides in Milk and Dairy Products: A Review. Korean J. Food Sci. Anim. Resour. 2015, 35, 831. [Google Scholar] [CrossRef]
- Ham, S.; Bhatia, S.K.; Gurav, R.; Choi, Y.-K.; Jeon, J.-M.; Yoon, J.-J.; Choi, K.-Y.; Ahn, J.; Kim, H.T.; Yang, Y.-H. Gamma Aminobutyric Acid (GABA) Production in Escherichia coli with Pyridoxal Kinase (pdxY) Based Regeneration System. Enzym. Microb. Technol. 2022, 155, 109994. [Google Scholar] [CrossRef] [PubMed]
- Santos-Espinosa, A.; Beltrán-Barrientos, L.M.; Reyes-Díaz, R.; Mazorra-Manzano, M.Á.; Hernández-Mendoza, A.; González-Aguilar, G.A.; Sáyago-Ayerdi, S.G.; Vallejo-Cordoba, B.; González-Córdova, A.F. Gamma-Aminobutyric Acid (GABA) Production in Milk Fermented by Specific Wild Lactic Acid Bacteria Strains Isolated from Artisanal Mexican Cheeses. Ann. Microbiol. 2020, 70, 12. [Google Scholar] [CrossRef]
- Pouliot-Mathieu, K.; Gardner-Fortier, C.; Lemieux, S.; St-Gelais, D.; Champagne, C.P.; Vuillemard, J.-C. Effect of Cheese Containing Gamma-Aminobutyric Acid-Producing Lactic Acid Bacteria on Blood Pressure in Men. PharmaNutrition 2013, 1, 141–148. [Google Scholar] [CrossRef]
- Hudec, J.; Kobida, Ľ.; Čanigová, M.; Lacko-Bartošová, M.; Ložek, O.; Chlebo, P.; Mrázová, J.; Ducsay, L.; Bystrická, J. Production of γ-Aminobutyric Acid by Microorganisms from Different Food Sources. J. Sci. Food Agric. 2015, 95, 1190–1198. [Google Scholar] [CrossRef]
- Sarasa, S.B.; Mahendran, R.; Muthusamy, G.; Thankappan, B.; Selta, D.R.F.; Angayarkanni, J. A Brief Review on the Non-Protein Amino Acid, Gamma-Amino Butyric Acid (GABA): Its Production and Role in Microbes. Curr. Microbiol. 2020, 77, 534–544. [Google Scholar] [CrossRef]
- Dhakal, R.; Bajpai, V.K.; Baek, K.-H. Production of Gaba (γ-Aminobutyric Acid) by Microorganisms: A Review. Braz. J. Microbiol. 2012, 43, 1230–1241. [Google Scholar] [CrossRef] [PubMed]
- Redruello, B.; Saidi, Y.; Sampedro, L.; Ladero, V.; del Rio, B.; Alvarez, M.A. GABA-Producing Lactococcus lactis Strains Isolated from Camel’s Milk as Starters for the Production of GABA-Enriched Cheese. Foods 2021, 10, 633. [Google Scholar] [CrossRef] [PubMed]
- Diana, M.; Quílez, J.; Rafecas, M. Gamma-Aminobutyric Acid as a Bioactive Compound in Foods: A Review. J. Funct. Foods 2014, 10, 407–420. [Google Scholar] [CrossRef]
- Inoue, K.; Shirai, T.; Ochiai, H.; Kasao, M.; Hayakawa, K.; Kimura, M.; Sansawa, H. Blood-Pressure-Lowering Effect of a Novel Fermented Milk Containing γ-Aminobutyric Acid (GABA) in Mild Hypertensives. Eur. J. Clin. Nutr. 2003, 57, 490–495. [Google Scholar] [CrossRef]
- Ruas-Madiedo, P.; de los Reyes-Gavilán, C.G. Invited Review: Methods for the Screening, Isolation, and Characterization of Exopolysaccharides Produced by Lactic Acid Bacteria. J. Dairy Sci. 2005, 88, 843–856. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, Z. A Functional and Genetic Overview of Exopolysaccharides Produced by Lactobacillus plantarum. J. Funct. Foods 2018, 47, 229–240. [Google Scholar] [CrossRef]
- Jeong, H.; Kim, S.; Hwang, U.-S.; Choi, H.; Park, Y.-S. Immunostimulatory Activity of Lactococcus lactis subsp. lactis CAB701 Isolated from Jeju Cabbage. Microorganisms 2023, 11, 1718. [Google Scholar] [CrossRef]
- Lynch, K.M.; Coffey, A.; Arendt, E.K. Exopolysaccharide Producing Lactic Acid Bacteria: Their Techno-Functional Role and Potential Application in Gluten-Free Bread Products. Food Res. Int. 2018, 110, 52–61. [Google Scholar] [CrossRef]
- Knoshaug, E.P.; Ahlgren, J.A.; Trempy, J.E. Exopolysaccharide Expression in Lactococcus lactis subsp. cremoris Ropy352: Evidence for Novel Gene Organization. Appl. Environ. Microbiol. 2007, 73, 897–905. [Google Scholar] [CrossRef]
- Surber, G.; Schäper, C.; Wefers, D.; Rohm, H.; Jaros, D. Exopolysaccharides from Lactococcus lactis Affect Manufacture, Texture and Sensory Properties of Concentrated Acid Milk Gel Suspensions (Fresh Cheese). Int. Dairy J. 2021, 112, 104854. [Google Scholar] [CrossRef]
- Zhou, Y.; Cui, Y.; Qu, X. Exopolysaccharides of Lactic Acid Bacteria: Structure, Bioactivity and Associations: A Review. Carbohydr. Polym. 2019, 207, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Kristo, E.; Miao, Z.; Corredig, M. The Role of Exopolysaccharide Produced by Lactococcus lactis subsp. cremoris in Structure Formation and Recovery of Acid Milk Gels. Int. Dairy J. 2011, 21, 656–662. [Google Scholar] [CrossRef]
- Poulsen, V.K.; Derkx, P.; Oregaard, G. High-Throughput Screening for Texturing Lactococcus Strains. FEMS Microbiol. Lett. 2019, 366, fnz001. [Google Scholar] [CrossRef] [PubMed]
- Allam, M.G.; Darwish, A.M.; Ayad, E.H.; Shokery, E.S.; Darwish, S.M. Lactococcus Species for Conventional Karish Cheese Conservation. LWT-Food Sci. Technol. 2017, 79, 625–631. [Google Scholar] [CrossRef]
- Mende, S.; Rohm, H.; Jaros, D. Influence of Exopolysaccharides on the Structure, Texture, Stability and Sensory Properties of Yoghurt and Related Products. Int. Dairy J. 2016, 52, 57–71. [Google Scholar] [CrossRef]
- Mailam, M.; Abd El-Razek, H.; A Shaaban, H. Effect of Exopolysaccharides-Producing Starter Culture on the Flavor Profile and Characteristics of Low Fat Ras Cheese. Pak. J. Biol Sci. 2020, 23, 691–700. [Google Scholar] [CrossRef]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C.G. Lactic Acid Bacteria in Raw-Milk Cheeses: From Starter Cultures to Probiotic Functions. Foods 2022, 11, 2276. [Google Scholar] [CrossRef]
- Werning, M.L.; Hernández-Alcántara, A.M.; Ruiz, M.J.; Soto, L.P.; Dueñas, M.T.; López, P.; Frizzo, L.S. Biological Functions of Exopolysaccharides from Lactic Acid Bacteria and Their Potential Benefits for Humans and Farmed Animals. Foods 2022, 11, 1284. [Google Scholar] [CrossRef]
- De Vuyst, L.; Degeest, B. Heteropolysaccharides from Lactic Acid Bacteria. FEMS Microbiol. Rev. 1999, 23, 153–177. [Google Scholar] [CrossRef]
- Jurášková, D.; Ribeiro, S.C.; Silva, C.C.G. Exopolysaccharides Produced by Lactic Acid Bacteria: From Biosynthesis to Health-Promoting Properties. Foods 2022, 11, 156. [Google Scholar] [CrossRef]
- Broadbent, J.R.; Barnes, M.; Brennand, C.; Strickland, M.; Houck, K.; Johnson, M.E.; Steele, J.L. Contribution of Lactococcus lactis Cell Envelope Proteinase Specificity to Peptide Accumulation and Bitterness in Reduced-Fat Cheddar Cheese. Appl. Environ. Microbiol. 2002, 68, 1778–1785. [Google Scholar] [CrossRef] [PubMed]
- Mozzi, F.; Vaningelgem, F.; Hébert, E.M.; Van der Meulen, R.; Foulquié Moreno, M.R.; Font de Valdez, G.; De Vuyst, L. Diversity of Heteropolysaccharide-Producing Lactic Acid Bacterium Strains and Their Biopolymers. Appl. Environ. Microbiol. 2006, 72, 4431–4435. [Google Scholar] [CrossRef] [PubMed]
- Gruter, M.; Leeflang, B.R.; Kuiper, J.; Kamerling, J.P.; Vliegenthart, J.F. Structure of the Exopolysaccharide Produced by Lactococcus lactis subspecies cremoris H414 Grown in a Defined Medium or Skimmed Milk. Carbohydr. Res. 1992, 231, 273–291. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van Kranenburg, R.; Vos, H.R.; van Swam, I.I.; Kleerebezem, M.; de Vos, W.M. Functional Analysis of Glycosyltransferase Genes from Lactococcus lactis and Other Gram-Positive Cocci: Complementation, Expression, and Diversity. J. Bacteriol. 1999, 181, 6347–6353. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Leroy, F. Functional Role of Yeasts, Lactic Acid Bacteria and Acetic Acid Bacteria in Cocoa Fermentation Processes. FEMS Microbiol. Rev. 2020, 44, 432–453. [Google Scholar] [CrossRef] [PubMed]
- Prete, R.; Alam, M.K.; Perpetuini, G.; Perla, C.; Pittia, P.; Corsetti, A. Lactic Acid Bacteria Exopolysaccharides Producers: A Sustainable Tool for Functional Foods. Foods 2021, 10, 1653. [Google Scholar] [CrossRef]
- Torino, M.; Font de Valdez, G.; Mozzi, F. Biopolymers from Lactic Acid Bacteria. Novel Applications in Foods and Beverages. Front. Microbiol. 2015, 6, 834. [Google Scholar] [CrossRef]
- Li, M.; Li, W.; Li, D.; Tian, J.; Xiao, L.; Kwok, L.-Y.; Li, W.; Sun, Z. Structure Characterization, Antioxidant Capacity, Rheological Characteristics and Expression of Biosynthetic Genes of Exopolysaccharides Produced by Lactococcus lactis subsp. lactis IMAU11823. Food Chem. 2022, 384, 132566. [Google Scholar] [CrossRef]
- Lo, T.C.T.; Kang, M.W.; Wang, B.C.; Chang, C.A. Glycosyl Linkage Characteristics and Classifications of Exo-Polysaccharides of Some Regionally Different Strains of Lentinula Edodes by Amplified Fragment Length Polymorphism Assay and Cluster Analysis. Anal. Chim. Acta 2007, 592, 146–153. [Google Scholar] [CrossRef]
- Guo, Y.; Pan, D.; Sun, Y.; Xin, L.; Li, H.; Zeng, X. Antioxidant Activity of Phosphorylated Exopolysaccharide Produced by Lactococcus lactis subsp. lactis. Carbohydr. Polym. 2013, 97, 849–854. [Google Scholar] [CrossRef]
- Nguyen, A.T.-B.; Picart-Palmade, L.; Nigen, M.; Jimenez, L.; Ait-Abderahim, H.; Marchesseau, S. Effect of Mono or Co-Culture of EPS-Producing Streptococcus thermophilus Strains on the Formation of Acid Milk Gel and the Appearance of Texture Defects. Int. Dairy J. 2019, 98, 17–24. [Google Scholar] [CrossRef]
- Feng, J.; Cai, Z.; Chen, Y.; Zhu, H.; Chang, X.; Wang, X.; Liu, Z.; Zhang, J.; Nie, G. Effects of an Exopolysaccharide from Lactococcus lactis Z-2 on Innate Immune Response, Antioxidant Activity, and Disease Resistance against Aeromonas Hydrophila in Cyprinus Carpio L. Fish Shellfish Immunol. 2020, 98, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Dowdell, P.; Chankhamhaengdecha, S.; Panbangred, W.; Janvilisri, T.; Aroonnual, A. Probiotic Activity of Enterococcus Faecium and Lactococcus lactis Isolated from Thai Fermented Sausages and Their Protective Effect Against Clostridium Difficile. Probiotics Antimicro. Prot. 2020, 12, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, Y.; Suzuki, S.; Amako, M.; Kitamura, S.; Toda, T. Effect of Orally Administered Exopolysaccharides Produced by Lactococcus lactis subsp. cremoris FC on a Mouse Model of Dermatitis Induced by Repeated Exposure to 2,4,6-Trinitro-1-Chlorobenzene. J. Funct. Foods 2017, 35, 43–50. [Google Scholar] [CrossRef]
- Cho, Y.; Han, H.T.; Kim, T.; Sohn, M.; Park, Y.-S. Immunostimulatory Activity of Lactococcus lactis LM1185 Isolated from Hydrangea Macrophylla. Food Sci. Biotechnol. 2023, 32, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Holst, O.A. Molinaro in Microbial Glycobiology; Academic Press: San Diego, CA, USA, 2010. [Google Scholar]
- Şanlier, N.; Gökcen, B.B.; Sezgin, A.C. Health Benefits of Fermented Foods. Crit. Rev. Food Sci. Nutr. 2019, 59, 506–527. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Vestergaard, M.; Jensen, T.G.; Shen, J.; Dufva, M.; Solem, C.; Jensen, P.R. Finding the Needle in the Haystack—The Use of Microfluidic Droplet Technology to Identify Vitamin-Secreting Lactic Acid Bacteria. mBio 2017, 8, 10–1128. [Google Scholar] [CrossRef]
- Pohanka, M. D-Lactic Acid as a Metabolite: Toxicology, Diagnosis, and Detection. Biomed. Res. Int. 2020, 2020, 3419034. [Google Scholar] [CrossRef]
- Remund, B.; Yilmaz, B.; Sokollik, C. D-Lactate: Implications for Gastrointestinal Diseases. Children 2023, 10, 945. [Google Scholar] [CrossRef]
- Weiss, G.A.; Hennet, T. Mechanisms and Consequences of Intestinal Dysbiosis. Cell. Mol. Life Sci. 2017, 74, 2959–2977. [Google Scholar] [CrossRef]
- Mishima, Y.; Sartor, R.B. Manipulating Resident Microbiota to Enhance Regulatory Immune Function to Treat Inflammatory Bowel Diseases. J. Gastroenterol. 2020, 55, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Kalliomäki, M.; Carmen Collado, M.; Salminen, S.; Isolauri, E. Early Differences in Fecal Microbiota Composition in Children May Predict Overweight2. Am. J. Clin. Nutr. 2008, 87, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Maślak, E.; Złoch, M.; Arendowski, A.; Sugajski, M.; Janczura, I.; Rudnicka, J.; Walczak-Skierska, J.; Buszewska-Forajta, M.; Rafińska, K.; Pomastowski, P.; et al. Isolation and Identification of Lactococcus lactis and Weissella Cibaria Strains from Fermented Beetroot and an Investigation of Their Properties as Potential Starter Cultures and Probiotics. Foods 2022, 11, 2257. [Google Scholar] [CrossRef] [PubMed]
- Hatti-Kaul, R.; Chen, L.; Dishisha, T.; Enshasy, H.E. Lactic Acid Bacteria: From Starter Cultures to Producers of Chemicals. FEMS Microbiol. Lett. 2018, 365, fny213. [Google Scholar] [CrossRef] [PubMed]
- Gandini, C.; Tarraran, L.; Kalemasi, D.; Pessione, E.; Mazzoli, R. Recombinant Lactococcus lactis for Efficient Conversion of Cellodextrins into L-Lactic Acid. Biotechnol. Bioeng. 2017, 114, 2807–2817. [Google Scholar] [CrossRef]
- Bintsis, T. Lactic Acid Bacteria as Starter Cultures: An Update in Their Metabolism and Genetics. AIMS Microbiol. 2018, 4, 665. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.; Vázquez, L.; Flórez, A.B.; Mayo, B. Phenotype Testing, Genome Analysis, and Metabolic Interactions of Three Lactic Acid Bacteria Strains Existing as a Consortium in a Naturally Fermented Milk. Front. Microbiol. 2022, 13, 1000683. [Google Scholar] [CrossRef]
- Iskandar, C.F.; Cailliez-Grimal, C.; Borges, F.; Revol-Junelles, A.-M. Review of Lactose and Galactose Metabolism in Lactic Acid Bacteria Dedicated to Expert Genomic Annotation. Trends Food Sci. Technol. 2019, 88, 121–132. [Google Scholar] [CrossRef]
- Fortina, M.G.; Ricci, G.; Borgo, F. A Study of Lactose Metabolism in Lactococcus garvieae Reveals a Genetic Marker for Distinguishing between Dairy and Fish Biotypes. J. Food Prot. 2009, 72, 1248–1254. [Google Scholar] [CrossRef]
- Bukvicki, D.; Siroli, L.; D’Alessandro, M.; Cosentino, S.; Fliss, I.; Said, L.B.; Hassan, H.; Lanciotti, R.; Patrignani, F. Unravelling the Potential of Lactococcus lactis Strains to Be Used in Cheesemaking Production as Biocontrol Agents. Foods 2020, 9, 1815. [Google Scholar] [CrossRef]
- Poudel, R.; Thunell, R.K.; Oberg, C.J.; Overbeck, S.; Lefevre, M.; Oberg, T.S.; McMahon, D.J. Comparison of Growth and Survival of Single Strains of Lactococcus lactis and Lactococcus cremoris during Cheddar Cheese Manufacture. J. Dairy Sci. 2022, 105, 2069–2081. [Google Scholar] [CrossRef] [PubMed]
- Taïbi, A.; Dabour, N.; Lamoureux, M.; Roy, D.; LaPointe, G. Comparative Transcriptome Analysis of Lactococcus lactis subsp. cremoris Strains under Conditions Simulating Cheddar Cheese Manufacture. Int. J. Food Microbiol. 2011, 146, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Vedamuthu, E.R.; Sandine, W.E.; Elliker, P.R. Flavor and Texture in Cheddar Cheese. I. Role of Mixed Strain Lactic Starter Cultures. J. Dairy Sci. 1966, 49, 144–150. [Google Scholar] [CrossRef]
- Li, A.; Zheng, J.; Han, X.; Yang, S.; Cheng, S.; Zhao, J.; Zhou, W.; Lu, Y. Advances in Low-Lactose/Lactose-Free Dairy Products and Their Production. Foods 2023, 12, 2553. [Google Scholar] [CrossRef]
- Serna Cock, L.; Rodriguez De Stouvenel, A. Lactic Acid Production by a Strain of Lactococcus lactis subs lactis Isolated from Sugar Cane Plants. Electron. J. Biotechnol. 2006, 9, 40–45. [Google Scholar] [CrossRef]
- Petrov, K.; Urshev, Z.; Petrova, P. L(+)-Lactic Acid Production from Starch by a Novel Amylolytic Lactococcus lactis subsp. lactis B84. Food Microbiol. 2008, 25, 550–557. [Google Scholar] [CrossRef]
- Sánchez, C.; Neves, A.R.; Cavalheiro, J.; dos Santos, M.M.; García-Quintáns, N.; López, P.; Santos, H. Contribution of Citrate Metabolism to the Growth of Lactococcus lactis CRL264 at Low pH. Appl. Environ. Microbiol. 2008, 74, 1136–1144. [Google Scholar] [CrossRef]
- Magni, C.; de Mendoza, D.; Konings, W.N.; Lolkema, J.S. Mechanism of Citrate Metabolism in Lactococcus lactis: Resistance against Lactate Toxicity at Low pH. J. Bacteriol. 1999, 181, 1451–1457. [Google Scholar] [CrossRef]
- Laëtitia, G.; Pascal, D.; Yann, D. The Citrate Metabolism in Homo- and Heterofermentative LAB: A Selective Means of Becoming Dominant over Other Microorganisms in Complex Ecosystems. Food Nutr. Sci. 2014, 5, 953–969. [Google Scholar] [CrossRef]
- Monnet, C.; Aymes, F.; Corrieu, G. Diacetyl and α-Acetolactate Overproduction by Lactococcus lactis subsp. lactis biovar diacetylactis Mutants That Are Deficient in α-Acetolactate Decarboxylase and Have a Low Lactate Dehydrogenase Activity. Appl. Environ. Microbiol. 2000, 66, 5518–5520. [Google Scholar] [CrossRef]
- Shin, J.M.; Gwak, J.W.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Biomedical Applications of Nisin. J. Appl. Microbiol. 2016, 120, 1449–1465. [Google Scholar] [CrossRef] [PubMed]
- Kajwadkar, R.; Shin, J.M.; Lin, G.-H.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. High-Purity Nisin Alone or in Combination with Sodium Hypochlorite Is Effective against Planktonic and Biofilm Populations of Enterococcus faecalis. J. Endod. 2017, 43, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Setiarto, R.H.B.; Anshory, L.; Wardana, A.A. Biosynthesis of Nisin, Antimicrobial Mechanism and Its Applications as a Food Preservation: A Review. IOP Conf. Ser. Earth Environ. Sci. 2023, 1169, 012105. [Google Scholar] [CrossRef]
- Hassan, H.; St-Gelais, D.; Gomaa, A.; Fliss, I. Impact of Nisin and Nisin-Producing Lactococcus lactis ssp. lactis on Clostridium tyrobutyricum and Bacterial Ecosystem of Cheese Matrices. Foods 2021, 10, 898. [Google Scholar] [CrossRef]
- Senan, S.; El-aal, H.A.; Dave, R.; Hassan, A. Production and Stability of Nisin in Whey Protein Concentrate. LWT-Food Sci. Technol. 2016, 71, 125–129. [Google Scholar] [CrossRef]
- EFSA Panel on Food Additives and Nutrient Sources added to Food (ANS); Younes, M.; Aggett, P.; Aguilar, F.; Crebelli, R.; Dusemund, B.; Filipič, M.; Frutos, M.J.; Galtier, P.; Gundert-Remy, U.; et al. Safety of Nisin (E 234) as a Food Additive in the Light of New Toxicological Data and the Proposed Extension of Use. EFSA J. 2017, 15, e05063. [Google Scholar] [CrossRef]
- Novickij, V.; Stanevičienė, R.; Staigvila, G.; Gruškienė, R.; Sereikaitė, J.; Girkontaitė, I.; Novickij, J.; Servienė, E. Effects of Pulsed Electric Fields and Mild Thermal Treatment on Antimicrobial Efficacy of Nisin-Loaded Pectin Nanoparticles for Food Preservation. LWT 2020, 120, 108915. [Google Scholar] [CrossRef]
- Gharsallaoui, A.; Oulahal, N.; Joly, C.; Degraeve, P. Nisin as a Food Preservative: Part 1: Physicochemical Properties, Antimicrobial Activity, and Main Uses. Crit. Rev. Food Sci. Nutr. 2016, 56, 1262–1274. [Google Scholar] [CrossRef]
- Belfiore, C.; Castellano, P.; Vignolo, G. Reduction of Escherichia Coli Population Following Treatment with Bacteriocins from Lactic Acid Bacteria and Chelators. Food Microbiol. 2007, 24, 223–229. [Google Scholar] [CrossRef]
- Delves-Broughton, J. 16-Use of the Natural Food Preservatives, Nisin and Natamycin, to Reduce Detrimental Thermal Impact on Product Quality. In In-Pack Processed Foods; Richardson, P., Ed.; Woodhead Publishing Series in Food Science, Technology and Nutrition; Woodhead Publishing: Thorston, UK, 2008; pp. 319–337. ISBN 978-1-84569-246-9. [Google Scholar]
- Junior, B.R.D.C.L.; Tribst, A.A.L. Use of Nisin and Bioprotective Lactic Cultures to Extend the Shelf Life of Sheep and Goat Cheese Whey. Food Biosci. 2022, 50, 102096. [Google Scholar] [CrossRef]
- Popa, E.E.; Miteluț, A.C.; Râpă, M.; Popescu, P.A.; Drăghici, M.C.; Geicu-Cristea, M.; Popa, M.E. Antimicrobial Active Packaging Containing Nisin for Preservation of Products of Animal Origin: An Overview. Foods 2022, 11, 3820. [Google Scholar] [CrossRef] [PubMed]
- Ibarra-Sánchez, L.A.; El-Haddad, N.; Mahmoud, D.; Miller, M.J.; Karam, L. Invited Review: Advances in Nisin Use for Preservation of Dairy Products. J. Dairy Sci. 2020, 103, 2041–2052. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, N.; Otani, T.; Inai, T. Nisin, a Food Preservative Produced by Lactococcus lactis, Affects the Localization Pattern of Intermediate Filament Protein in HaCaT Cells. Anat. Sci. Int. 2019, 94, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Smaoui, S.; Echegaray, N.; Kumar, M.; Chaari, M.; D’Amore, T.; Shariati, M.A.; Rebezov, M.; Lorenzo, J.M. Beyond Conventional Meat Preservation: Saddling the Control of Bacteriocin and Lactic Acid Bacteria for Clean Label and Functional Meat Products. Appl. Biochem. Biotechnol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Santiesteban-López, N.A.; Gómez-Salazar, J.A.; Santos, E.M.; Campagnol, P.C.B.; Teixeira, A.; Lorenzo, J.M.; Sosa-Morales, M.E.; Domínguez, R. Natural Antimicrobials: A Clean Label Strategy to Improve the Shelf Life and Safety of Reformulated Meat Products. Foods 2022, 11, 2613. [Google Scholar] [CrossRef] [PubMed]
- Abbasiliasi, S.; Shun Tan, J.; Ibrahim, T.A.T.; Bashokouh, F.; Ramanan Ramakrishnan, N.; Mustafa, S.; Ariff, A.B. Fermentation Factors Influencing the Production of Bacteriocins by Lactic Acid Bacteria: A Review. RSC Adv. 2017, 7, 29395–29420. [Google Scholar] [CrossRef]
- Modugno, C.; Loupiac, C.; Bernard, A.; Jossier, A.; Neiers, F.; Perrier-Cornet, J.-M.; Simonin, H. Effect of High Pressure on the Antimicrobial Activity and Secondary Structure of the Bacteriocin Nisin. Innov. Food Sci. Emerg. Technol. 2018, 47, 9–15. [Google Scholar] [CrossRef]
- Jung, D.-S.; Bodyfelt, F.W.; Daeschel, M.A. Influence of Fat and Emulsifiers on the Efficacy of Nisin in Inhibiting Listeria monocytogenes in Fluid Milk. J. Dairy Sci. 1992, 75, 387–393. [Google Scholar] [CrossRef]
- Chen, R.; Skeens, J.W.; Wiedmann, M.; Guariglia-Oropeza, V. The Efficacy of Nisin against Listeria monocytogenes on Cold-Smoked Salmon at Natural Contamination Levels Is Concentration-Dependent and Varies by Serotype. Front. Microbiol. 2022, 13, 930400. [Google Scholar] [CrossRef]
- Soltani, S.; Hammami, R.; Cotter, P.D.; Rebuffat, S.; Said, L.B.; Gaudreau, H.; Bédard, F.; Biron, E.; Drider, D.; Fliss, I. Bacteriocins as a New Generation of Antimicrobials: Toxicity Aspects and Regulations. FEMS Microbiol. Rev. 2021, 45, fuaa039. [Google Scholar] [CrossRef]
- Zapico, P.; de Paz, M.; Medina, M.; Nuñez, M. The Effect of Homogenization of Whole Milk, Skim Milk and Milk Fat on Nisin Activity against Listeria Innocua. Int. J. Food Microbiol. 1999, 46, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ramos, A.; Madi-Moussa, D.; Coucheney, F.; Drider, D. Current Knowledge of the Mode of Action and Immunity Mechanisms of LAB-Bacteriocins. Microorganisms 2021, 9, 2107. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Hao, L.; Li, J.; Yi, J.; Kang, Q.; Liu, X.; Lu, L.; Lu, J. An Innovative Method to Enhance Protease Tolerance of Nisin in Endogenous Proteases. J. Dairy Sci. 2020, 103, 3038–3044. [Google Scholar] [CrossRef] [PubMed]
- Aasen, I.M.; Markussen, S.; Møretrø, T.; Katla, T.; Axelsson, L.; Naterstad, K. Interactions of the Bacteriocins Sakacin P and Nisin with Food Constituents. Int. J. Food Microbiol. 2003, 87, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zang, M.; Wang, S.; Zhao, B.; Bai, J.; Xu, C.; Shi, Y.; Qiao, X. Nisin: From a Structural and Meat Preservation Perspective. Food Microbiol. 2023, 111, 104207. [Google Scholar] [CrossRef]
- Ucar, Y.; Ozogul, Y.; Durmuş, M.; Ozogul, F. The Effects of Nisin on the Growth of Foodborne Pathogens and Biogenic Amine Formation: In Vivo and in Vitro Studies. Food Biosci. 2021, 43, 101266. [Google Scholar] [CrossRef]
- Felicio, B.A.; Pinto, M.S.; Oliveira, F.S.; Lempk, M.W.; Pires, A.C.S.; Lelis, C.A. Effects of Nisin on Staphylococcus aureus Count and Physicochemical Properties of Minas Frescal Cheese. J. Dairy Sci. 2015, 98, 4364–4369. [Google Scholar] [CrossRef]
- Oppegård, C.; Rogne, P.; Kristiansen, P.E.; Nissen-Meyer, J. Structure Analysis of the Two-Peptide Bacteriocin Lactococcin G by Introducing d-Amino Acid Residues. Microbiology 2010, 156, 1883–1889. [Google Scholar] [CrossRef]
- Moll, G.; Hildeng-Hauge, H.; Nissen-Meyer, J.; Nes, I.F.; Konings, W.N.; Driessen, A.J. Mechanistic Properties of the Two-Component Bacteriocin Lactococcin G. J. Bacteriol. 1998, 180, 96–99. [Google Scholar] [CrossRef]
- Moll, G.; Ubbink-Kok, T.; Hildeng-Hauge, H.; Nissen-Meyer, J.; Nes, I.F.; Konings, W.N.; Driessen, A.J. Lactococcin G Is a Potassium Ion-Conducting, Two-Component Bacteriocin. J. Bacteriol. 1996, 178, 600–605. [Google Scholar] [CrossRef]
- Zendo, T.; Koga, S.; Shigeri, Y.; Nakayama, J.; Sonomoto, K. Lactococcin Q, a Novel Two-Peptide Bacteriocin Produced by Lactococcus lactis QU 4. Appl. Environ. Microbiol. 2006, 72, 3383–3389. [Google Scholar] [CrossRef] [PubMed]
- Rendón-Rosales, M.Á.; Torres-Llanez, M.J.; Mazorra-Manzano, M.A.; González-Córdova, A.F.; Hernández-Mendoza, A.; Vallejo-Cordoba, B. In Vitro and in Silico Evaluation of Multifunctional Properties of Bioactive Synthetic Peptides Identified in Milk Fermented with Lactococcus lactis NRRL B-50571 and NRRL B-50572. LWT 2022, 154, 112581. [Google Scholar] [CrossRef]
- Guha, S.; Sharma, H.; Deshwal, G.K.; Rao, P.S. A Comprehensive Review on Bioactive Peptides Derived from Milk and Milk Products of Minor Dairy Species. Food Prod. Process. Nutr. 2021, 3, 2. [Google Scholar] [CrossRef]
- Raveschot, C.; Cudennec, B.; Coutte, F.; Flahaut, C.; Fremont, M.; Drider, D.; Dhulster, P. Production of Bioactive Peptides by Lactobacillus Species: From Gene to Application. Front. Microbiol. 2018, 9, 2354. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, D.; Masoodi, F.A.; Wani, S.M.; Mir, S.A. Bioactive Peptides from Fermented Foods and Their Relevance in COVID-19 Mitigation. Food Prod. Process. Nutr. 2023, 5, 53. [Google Scholar] [CrossRef]
- Huang, C.; Kok, J. Editing of the Proteolytic System of Lactococcus lactis Increases Its Bioactive Potential. Appl. Environ. Microbiol. 2020, 86, e01319-20. [Google Scholar] [CrossRef] [PubMed]
- Forler, B.; Horstmann, G.; Schäfer, J.; Michel, C.; Weiss, A.; Stressler, T.; Fischer, L.; Hinrichs, J.; Schmidt, H. Effects of Protein, Calcium, and pH on Gene Transcription, Cell-Envelope Peptidase Activity of Lactococcus lactis Strains, and the Formation of Bitter Peptides. Foods 2021, 10, 1588. [Google Scholar] [CrossRef]
- Guédon, E.; Renault, P.; Ehrlich, S.D.; Delorme, C. Transcriptional Pattern of Genes Coding for the Proteolytic System of Lactococcus lactis and Evidence for Coordinated Regulation of Key Enzymes by Peptide Supply. J. Bacteriol. 2001, 183, 3614–3622. [Google Scholar] [CrossRef]
- den Hengst, C.D.; van Hijum, S.A.F.T.; Geurts, J.M.W.; Nauta, A.; Kok, J.; Kuipers, O.P. The Lactococcus lactis CodY Regulon: Identification of a conserved Cis-Regulatory Element*[Boxs]. J. Biol. Chem. 2005, 280, 34332–34342. [Google Scholar] [CrossRef]
- Bounouala, F.Z.; Roudj, S.; Karam, N.-E.; Recio, I.; Miralles, B. Casein Hydrolysates by Lactobacillus Brevis and Lactococcus Lactis Proteases: Peptide Profile Discriminates Strain-Dependent Enzyme Specificity. J. Agric. Food Chem. 2017, 65, 9324–9332. [Google Scholar] [CrossRef]
- Han, R.; Maycock, J.; Murray, B.S.; Boesch, C. Identification of Angiotensin Converting Enzyme and Dipeptidyl Peptidase-IV Inhibitory Peptides Derived from Oilseed Proteins Using Two Integrated Bioinformatic Approaches. Food Res. Int. 2019, 115, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Feng, L.; Wang, Z.; Qiao, M.; Shahidi, F.; Lu, W.; Du, M. Sequence Analysis and Molecular Docking of Antithrombotic Peptides from Casein Hydrolysate by Trypsin Digestion. J. Funct. Foods 2017, 32, 313–323. [Google Scholar] [CrossRef]
- Figueroa-Hernández, C.; Cruz-Guerrero, A.; Rodríguez-Serrano, G.; Gómez-Ruiz, L.; García-Garibay, M.; Jiménez-Guzmán, J. Calcium and Iron Binding Peptides Production by Lactococcus lactis subsp. cremoris NCFB 712. Rev. Mex. Ing. Química 2012, 11, 259–267. [Google Scholar]
- Dimitrov, Z. Characterization of Bioactive Peptides with Calcium-Binding Activity Released by Specially Designed Cheese Starter. Biotechnol. Biotechnol. Equip. 2009, 23, 927–930. [Google Scholar] [CrossRef]
- Raw Milk Kefir: Microbiota, Bioactive Peptides, and Immune Modulation. Food Funct. 2023, 14, 1648–1661. [CrossRef] [PubMed]
- Gobbetti, M.; Ferranti, P.; Smacchi, E.; Goffredi, F.; Addeo, F. Production of Angiotensin-I-Converting-Enzyme-Inhibitory Peptides in Fermented Milks Started by Lactobacillus delbrueckii subsp. bulgaricus SS1 and Lactococcus lactis subsp. cremoris FT4. Appl. Environ. Microbiol. 2000, 66, 3898–3904. [Google Scholar] [CrossRef] [PubMed]
- Gagnaire, V.; Jardin, J.; Jan, G.; Lortal, S. Invited Review: Proteomics of Milk and Bacteria Used in Fermented Dairy Products: From Qualitative to Quantitative Advances. J. Dairy Sci. 2009, 92, 811–825. [Google Scholar] [CrossRef]
- Sedaghati, M.; Ezzatpanah, H.; Boojar, M.M.A.; Ebrahimi, M.T.; Kobarfard, F. Isolation and Identification of Some Antibacterial Peptides in the Plasmin-Digest of β-Casein. LWT-Food Sci. Technol. 2016, 68, 217–225. [Google Scholar] [CrossRef]
- Choi, J.; Sabikhi, L.; Hassan, A.; Anand, S. Bioactive Peptides in Dairy Products. Int. J. Dairy Technol. 2012, 65, 1–12. [Google Scholar] [CrossRef]
- Rafiq, S.; Gulzar, N.; Sameen, A.; Huma, N.; Hayat, I.; Ijaz, R. Functional Role of Bioactive Peptides with Special Reference to Cheeses. Int. J. Dairy Technol. 2021, 74, 1–16. [Google Scholar] [CrossRef]
- Rafiq, S.; Huma, N.; Pasha, I.; Shahid, M.; Xiao, H. Angiotensin-converting Enzyme-inhibitory and Antithrombotic Activities of Soluble Peptide Extracts from Buffalo and Cow Milk Cheddar Cheeses. Int. J. Dairy Technol. 2017, 70, 380–388. [Google Scholar] [CrossRef]
- Kim, H.D.; Lee, H.J.; Shin, Z.I.; Nam, H.S.; Woo, H.J. Anticancer Effects of Hydrophobic Peptides Derived from a Cheese Slurry. Food Sci. Biotechnol. 1995, 4, 268–272. [Google Scholar]
- Santos, M.I.; Lima, A.; Mota, J.; Rebelo, P.; Ferreira, R.B.; Pedroso, L.; Ferreira, M.A.; Sousa, I. Extended Cheese Whey Fermentation Produces a Novel Casein-Derived Antibacterial Polypeptide That Also Inhibits Gelatinases MMP-2 and MMP-9. Int. J. Mol. Sci. 2021, 22, 11130. [Google Scholar] [CrossRef] [PubMed]
- González-González, F.; Delgado, S.; Ruiz, L.; Margolles, A.; Ruas-Madiedo, P. Functional Bacterial Cultures for Dairy Applications: Towards Improving Safety, Quality, Nutritional and Health Benefit Aspects. J. Appl. Microbiol. 2022, 133, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Acevedo-Rocha, C.G.; Gronenberg, L.S.; Mack, M.; Commichau, F.M.; Genee, H.J. Microbial Cell Factories for the Sustainable Manufacturing of B Vitamins. Curr. Opin. Biotechnol. 2019, 56, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.; Wang, S.; Guan, J.; Shi, J.; Evivie, S.E.; Yan, F.; Li, N.; Chen, J.; Li, B.; Huo, G. Milk Fermented with Lactococcus lactis KLDS4.0325 Alleviates Folate Status in Deficient Mice. Food Funct. 2020, 11, 4571–4581. [Google Scholar] [CrossRef] [PubMed]
- Sybesma, W.; Starrenburg, M.; Kleerebezem, M.; Mierau, I.; de Vos, W.M.; Hugenholtz, J. Increased Production of Folate by Metabolic Engineering of Lactococcus lactis. Appl. Environ. Microbiol. 2003, 69, 3069–3076. [Google Scholar] [CrossRef] [PubMed]
- Gangadharan, D.; Nampoothiri, K.M. Folate Production Using Lactococcus Lactis Ssp Cremoris with Implications for Fortification of Skim Milk and Fruit Juices. LWT-Food Sci. Technol. 2011, 44, 1859–1864. [Google Scholar] [CrossRef]
- Divya, J.B.; Nampoothiri, K.M. Folate Fortification of Skim Milk by a Probiotic Lactococcus lactis CM28 and Evaluation of Its Stability in Fermented Milk on Cold Storage. J. Food Sci. Technol. 2015, 52, 3513–3519. [Google Scholar] [CrossRef][Green Version]
- Burgess, C.M.; Slotboom, D.J.; Geertsma, E.R.; Duurkens, R.H.; Poolman, B.; van Sinderen, D. The Riboflavin Transporter RibU in Lactococcus lactis: Molecular Characterization of Gene Expression and the Transport Mechanism. J. Bacteriol. 2006, 188, 2752–2760. [Google Scholar] [CrossRef]
- Shulpekova, Y.; Nechaev, V.; Kardasheva, S.; Sedova, A.; Kurbatova, A.; Bueverova, E.; Kopylov, A.; Malsagova, K.; Dlamini, J.C.; Ivashkin, V. The Concept of Folic Acid in Health and Disease. Molecules 2021, 26, 3731. [Google Scholar] [CrossRef]
- Albu, C.-C.; Bencze, M.-A.; Dragomirescu, A.-O.; Suciu, I.; Tănase, M.; Albu, Ş.-D.; Russu, E.-A.; Ionescu, E. Folic Acid and Its Role in Oral Health: A Narrative Review. Processes 2023, 11, 1994. [Google Scholar] [CrossRef]
- Pacheco Da Silva, F.F.; Biscola, V.; LeBlanc, J.G.; Gombossy de Melo Franco, B.D. Effect of Indigenous Lactic Acid Bacteria Isolated from Goat Milk and Cheeses on Folate and Riboflavin Content of Fermented Goat Milk. LWT-Food Sci. Technol. 2016, 71, 155–161. [Google Scholar] [CrossRef]
- Mahabadi, N.; Bhusal, A.; Banks, S.W. Riboflavin Deficiency. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Hariri, E.; Kassis, N.; Iskandar, J.-P.; Schurgers, L.J.; Saad, A.; Abdelfattah, O.; Bansal, A.; Isogai, T.; Harb, S.C.; Kapadia, S. Vitamin K2-a Neglected Player in Cardiovascular Health: A Narrative Review. Open Heart 2021, 8, e001715. [Google Scholar] [CrossRef]
- Saubade, F.; Hemery, Y.M.; Guyot, J.-P.; Humblot, C. Lactic Acid Fermentation as a Tool for Increasing the Folate Content of Foods. Crit. Rev. Food Sci. Nutr. 2017, 57, 3894–3910. [Google Scholar] [CrossRef]
- Mahara, F.A.; Nuraida, L.; Lioe, H. Fermentation of Milk Using Folate-Producing Lactic Acid Bacteria to Increase Natural Folate Content: A Review. J. Apple Biotechnol. Rep. 2019, 6, 129–136. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Laiño, J.E.; del Valle, M.J.; Vannini, V.; van Sinderen, D.; Taranto, M.P.; de Valdez, G.F.; de Giori, G.S.; Sesma, F. B-Group Vitamin Production by Lactic Acid Bacteria—Current Knowledge and Potential Applications. J. Appl. Microbiol. 2011, 111, 1297–1309. [Google Scholar] [CrossRef]
- Averianova, L.A.; Balabanova, L.A.; Son, O.M.; Podvolotskaya, A.B.; Tekutyeva, L.A. Production of Vitamin B2 (Riboflavin) by Microorganisms: An Overview. Front. Bioeng. Biotechnol. 2020, 8, 1172. [Google Scholar] [CrossRef]
- Gu, Q.; Li, P.; Gu, Q.; Li, P. Biosynthesis of Vitamins by Probiotic Bacteria. In Probiotics and Prebiotics in Human Nutrition and Health; IntechOpen: London, UK, 2016; ISBN 978-953-51-2476-4. [Google Scholar]
- Nayik, G.A.; Jagdale, Y.D.; Gaikwad, S.A.; Devkatte, A.N.; Dar, A.H.; Ansari, M.J. Nutritional Profile, Processing and Potential Products: A Comparative Review of Goat Milk. Dairy 2022, 3, 44. [Google Scholar] [CrossRef]
- Manoury, E.; Jourdon, K.; Boyaval, P.; Fourcassié, P. Quantitative Measurement of Vitamin K2 (Menaquinones) in Various Fermented Dairy Products Using a Reliable High-Performance Liquid Chromatography Method. J. Dairy Sci. 2013, 96, 1335–1346. [Google Scholar] [CrossRef]
- Vermeer, C.; Raes, J.; Van’t Hoofd, C.; Knapen, M.H.; Xanthoulea, S. Menaquinone Content of Cheese. Nutrients 2018, 10, 446. [Google Scholar] [CrossRef]
- Walther, B.; Guggisberg, D.; Schmidt, R.S.; Portmann, R.; Risse, M.-C.; Badertscher, R.; Chollet, M. Quantitative Analysis of Menaquinones (Vitamin K2) in Various Types of Cheese from Switzerland. Int. Dairy J. 2021, 112, 104853. [Google Scholar] [CrossRef]
- Walther, B.; Karl, J.P.; Booth, S.L.; Boyaval, P. Menaquinones, Bacteria, and the Food Supply: The Relevance of Dairy and Fermented Food Products to Vitamin K Requirements. Adv. Nutr. 2013, 4, 463–473. [Google Scholar] [CrossRef]
- Homayouni Rad, A.; Yari Khosroushahi, A.; Khalili, M.; Jafarzadeh, S. Folate Bio-Fortification of Yoghurt and Fermented Milk: A Review. Dairy Sci. Technol. 2016, 96, 427–441. [Google Scholar] [CrossRef]
- Sybesma, W.; Burgess, C.; Starrenburg, M.; van Sinderen, D.; Hugenholtz, J. Multivitamin Production in Lactococcus lactis Using Metabolic Engineering. Metab. Eng. 2004, 6, 109–115. [Google Scholar] [CrossRef]
- Thakur, K.; Tomar, S.K.; De, S. Lactic Acid Bacteria as a Cell Factory for Riboflavin Production. Microb. Biotechnol. 2016, 9, 441–451. [Google Scholar] [CrossRef]
- Green, J.M.; Matthews, R.G. Folate Biosynthesis, Reduction, and Polyglutamylation and the Interconversion of Folate Derivatives. EcoSal Plus 2007, 2, 10-1128. [Google Scholar] [CrossRef]
- Klaus, S.M.; Wegkamp, A.; Sybesma, W.; Hugenholtz, J.; Gregory, J.F.; Hanson, A.D. A Nudix Enzyme Removes Pyrophosphate from Dihydroneopterin Triphosphate in the Folate Synthesis Pathway of Bacteria and Plants. J. Biol. Chem. 2005, 280, 5274–5280. [Google Scholar] [CrossRef]
- Strandler, H.S.; Patring, J.; Jägerstad, M.; Jastrebova, J. Challenges in the Determination of Unsubstituted Food Folates: Impact of Stabilities and Conversions on Analytical Results. J. Agric. Food Chem. 2015, 63, 2367–2377. [Google Scholar] [CrossRef]
- Patel, A.; Shah, N.; Prajapati, J.B. Biosynthesis of Vitamins and Enzymes in Fermented Foods by Lactic Acid Bacteria and Related Genera—A Promising Approach. Croat. J. Food Sci. Technol. 2013, 5, 85–91. [Google Scholar]
- Brooijmans, R.; Smit, B.; Santos, F.; van Riel, J.; de Vos, W.M.; Hugenholtz, J. Heme and Menaquinone Induced Electron Transport in Lactic Acid Bacteria. Microb. Cell Factories 2009, 8, 28. [Google Scholar] [CrossRef]
- Vieira, C.P.; Cabral, C.C.; da Costa Lima, B.R.C.; Paschoalin, V.M.F.; Leandro, K.C.; Conte-Junior, C.A. Lactococcus lactis ssp. cremoris MRS47, a Potential Probiotic Strain Isolated from Kefir Grains, Increases Cis-9, Trans-11-CLA and PUFA Contents in Fermented Milk. J. Funct. Foods 2017, 31, 172–178. [Google Scholar] [CrossRef]
- den Hartigh, L.J. Conjugated Linoleic Acid Effects on Cancer, Obesity, and Atherosclerosis: A Review of Pre-Clinical and Human Trials with Current Perspectives. Nutrients 2019, 11, 370. [Google Scholar] [CrossRef]
- Dachev, M.; Bryndová, J.; Jakubek, M.; Moučka, Z.; Urban, M. The Effects of Conjugated Linoleic Acids on Cancer. Processes 2021, 9, 454. [Google Scholar] [CrossRef]
- Nasrollahzadeh, A.; Tavani, S.M.; Arjeh, E.; Jafari, S.M. Production of Conjugated Linoleic Acid by Lactic Acid Bacteria; Important Factors and Optimum Conditions. Food Chem. X 2023, 20, 100942. [Google Scholar] [CrossRef]
- Gong, M.; Hu, Y.; Wei, W.; Jin, Q.; Wang, X. Production of Conjugated Fatty Acids: A Review of Recent Advances. Biotechnol. Adv. 2019, 37, 107454. [Google Scholar] [CrossRef]
- Abd El-Salam, M.H.; el-Shafei, K.; Sharaf, O.M.; Effat, B.A.; Asem, F.M.; el-Aasar, M. Screening of Some Potentially Probiotic Lactic Acid Bacteria for Their Ability to Synthesis Conjugated Linoleic Acid. Int. J. Dairy Technol. 2010, 63, 62–69. [Google Scholar] [CrossRef]
- Renes, E.; Ladero, V.; Tornadijo, M.E.; Fresno, J.M. Production of Sheep Milk Cheese with High γ-Aminobutyric Acid and Ornithine Concentration and with Reduced Biogenic Amines Level Using Autochthonous Lactic Acid Bacteria Strains. Food Microbiol. 2019, 78, 1–10. [Google Scholar] [CrossRef]
- Konkit, M.; Kim, W. Activities of Amylase, Proteinase, and Lipase Enzymes from Lactococcus chungangensis and Its Application in Dairy Products. J. Dairy Sci. 2016, 99, 4999–5007. [Google Scholar] [CrossRef]
- Farkye, N.Y. Cheese Technology. Int. J. Dairy Technol. 2004, 57, 91–98. [Google Scholar] [CrossRef]
- Demarigny, Y. LACTOCOCCUS|Lactococcus lactis subspecies lactis and cremoris. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: Oxford, UK, 2014; pp. 442–446. ISBN 978-0-12-384733-1. [Google Scholar]
- Rani, S.; Jagtap, S. Acceleration of Swiss Cheese Ripening by Microbial Lipase without Affecting Its Quality Characteristics. J. Food Sci. Technol. 2019, 56, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Yvon, M.; Thirouin, S.; Rijnen, L.; Fromentier, D.; Gripon, J.C. An Aminotransferase from Lactococcus lactis Initiates Conversion of Amino Acids to Cheese Flavor Compounds. Appl. Environ. Microbiol. 1997, 63, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, C.; Irmler, S.; Bisig, W.; Guggisberg, D.; Roetschi, A.; Portmann, R.; Wechsler, D.; Fröhlich-Wyder, M.-T. The Effect of Starters with a Functional Arginine Deiminase Pathway on Cheese Ripening and Quality. Int. Dairy J. 2018, 85, 191–200. [Google Scholar] [CrossRef]
- van Mastrigt, O.; Mager, E.E.; Jamin, C.; Abee, T.; Smid, E.J. Citrate, Low pH and Amino Acid Limitation Induce Citrate Utilization in Lactococcus lactis biovar diacetylactis. Microb. Biotechnol. 2018, 11, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Medina, R.B.; Katz, M.B.; González, S.; Oliver, G. Determination of Esterolytic and Lipolytic Activities of Lactic Acid Bacteria. Public Health Microbiol. Methods Protoc. 2004, 268, 465–470. [Google Scholar]
- Harper, A.R.; Dobson, R.C.J.; Morris, V.K.; Moggré, G. Fermentation of Plant-based Dairy Alternatives by Lactic Acid Bacteria. Microb. Biotechnol. 2022, 15, 1404–1421. [Google Scholar] [CrossRef]
- Mardis, E.R. The Impact of Next-Generation Sequencing Technology on Genetics. Trends Genet. 2008, 24, 133–141. [Google Scholar] [CrossRef]
- Gautam, S.S.; KC, R.; Leong, K.W.; Aogáin, M.M.; O’Toole, R.F. A Step-by-Step Beginner. J. Biol. Methods 2019, 6, e110. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Bortolaia, V.; Tate, H.; Tyson, G.H.; Aarestrup, F.M.; McDermott, P.F. Using Genomics to Track Global Antimicrobial Resistance. Front. Public Health 2019, 7, 242. [Google Scholar] [CrossRef]
- Lal Gupta, C.; Kumar Tiwari, R.; Cytryn, E. Platforms for Elucidating Antibiotic Resistance in Single Genomes and Complex Metagenomes. Environ. Int. 2020, 138, 105667. [Google Scholar] [CrossRef]
- Seoane, A.; Bou, G. Bioinformatics Approaches to the Study of Antimicrobial Resistance. Rev. Esp. Quim. 2021, 34, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Tóth, A.G.; Csabai, I.; Judge, M.F.; Maróti, G.; Becsei, Á.; Spisák, S.; Solymosi, N. Mobile Antimicrobial Resistance Genes in Probiotics. Antibiotics 2021, 10, 1287. [Google Scholar] [CrossRef] [PubMed]
- Fatahi-Bafghi, M.; Naseri, S.; Alizehi, A. Genome Analysis of Probiotic Bacteria for Antibiotic Resistance Genes. Antonie Van Leeuwenhoek 2022, 115, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. ABRicate. 2023. Available online: https://bio.tools/ABRicate (accessed on 12 November 2023).
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a New Bioinformatic Tool To Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Bonin, N.; Doster, E.; Worley, H.; Pinnell, L.J.; Bravo, J.E.; Ferm, P.; Marini, S.; Prosperi, M.; Noyes, N.; Morley, P.S.; et al. MEGARes and AMR++, v3.0: An Updated Comprehensive Database of Antimicrobial Resistance Determinants and an Improved Software Pipeline for Classification Using High-Throughput Sequencing. Nucleic Acids Res. 2022, 51, D744–D752. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2022, 51, D690–D699. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Tenea, G.N. Metabiotics Signature through Genome Sequencing and In Vitro Inhibitory Assessment of a Novel Lactococcus lactis Strain UTNCys6-1 Isolated from Amazonian Camu-Camu Fruits. Int. J. Mol. Sci. 2023, 24, 6127. [Google Scholar] [CrossRef] [PubMed]
- Wissel, E.F.; Talbot, B.M.; Toyosato, N.A.B.; Petit, R.A.; Hertzberg, V.; Dunlop, A.; Read, T.D. hAMRoaster: A Tool for Comparing Performance of AMR Gene Detection Software. bioRxiv 2023. bioRxiv:2022.01.13.476279. [Google Scholar]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for Taxonomy-Based Analysis of Pathways and Genomes. Nucleic Acids Res. 2022, 51, D587–D592. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Altman, T.; Billington, R.; Dreher, K.; Foerster, H.; Fulcher, C.A.; Holland, T.A.; Keseler, I.M.; Kothari, A.; Kubo, A.; et al. The MetaCyc Database of Metabolic Pathways and Enzymes and the BioCyc Collection of Pathway/Genome Databases. Nucleic Acids Res. 2014, 42, D459–D471. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, X.; Hong, R.; Liu, F. Combined Proteomics and Transcriptomics Analysis of Lactococcus lactis under Different Culture Conditions. J. Dairy Sci. 2021, 104, 2564–2580. [Google Scholar] [CrossRef]
- Camacho, C.; Boratyn, G.M.; Joukov, V.; Vera Alvarez, R.; Madden, T.L. ElasticBLAST: Accelerating Sequence Search via Cloud Computing. BMC Bioinform. 2023, 24, 117. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Sun, Y.; Li, H.; Zheng, L.; Li, J.; Hong, Y.; Liang, P.; Kwok, L.-Y.; Zuo, Y.; Zhang, W.; Zhang, H. iProbiotics: A Machine Learning Platform for Rapid Identification of Probiotic Properties from Whole-Genome Primary Sequences. Brief. Bioinform. 2022, 23, bbab477. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- Hartleb, D.; Fritzemeier, C.J.; Lercher, M.J. Automated High-Quality Reconstruction of Metabolic Networks from High-Throughput Data. bioRxiv 2018. [Google Scholar] [CrossRef]
- Henry, C.S.; DeJongh, M.; Best, A.A.; Frybarger, P.M.; Linsay, B.; Stevens, R.L. High-Throughput Generation, Optimization and Analysis of Genome-Scale Metabolic Models. Nat. Biotechnol. 2010, 28, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, E.; Schulz, C.; Almaas, E. Automated Generation of Genome-Scale Metabolic Draft Reconstructions Based on KEGG. BMC Bioinform. 2018, 19, 467. [Google Scholar] [CrossRef] [PubMed]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Akhter, S.; Miller, J.H. BaPreS: A Software Tool for Predicting Bacteriocins Using an Optimal Set of Features. BMC Bioinform. 2023, 24, 313. [Google Scholar] [CrossRef] [PubMed]
- Hammami, R.; Zouhir, A.; Le Lay, C.; Ben Hamida, J.; Fliss, I. BACTIBASE Second Release: A Database and Tool Platform for Bacteriocin Characterization. BMC Microbiol. 2010, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A More Efficient Report with Usability Improvements. Nucleic Acids Res. 2013, 41, W29–W33. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Eddy, S.R. HMMER User’s Guide. 2023. Available online: http://eddylab.org/software/hmmer/Userguide.pdf (accessed on 12 November 2023).
- Bhagwat, M.; Aravind, L. PSI-BLAST Tutorial. In Comparative Genomics: Volumes 1 and 2; Humana Press: Totowa, NJ, USA, 2007. [Google Scholar]
- Yoon, B.-J. Hidden Markov Models and Their Applications in Biological Sequence Analysis. Curr. Genom. 2009, 10, 402–415. [Google Scholar] [CrossRef]
- Walsh, C.J.; Guinane, C.M.; O’Toole, P.W.; Cotter, P.D. A Profile Hidden Markov Model to Investigate the Distribution and Frequency of LanB-Encoding Lantibiotic Modification Genes in the Human Oral and Gut Microbiome. PeerJ 2017, 5, e3254. [Google Scholar] [CrossRef]
- Golneshin, A.; Gor, M.-C.; Williamson, N.; Vezina, B.; Van, T.T.H.; May, B.K.; Smith, A.T. Discovery and Characterisation of Circular Bacteriocin Plantacyclin B21AG from Lactiplantibacillus plantarum B21. Heliyon 2020, 6, e04715. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, L.L.; Hoffmann, C.; Lacorte, G.A.; de Melo Franco, B.D.G.; Todorov, S.D. Genomic and Functional Characterization of Bacteriocinogenic Lactic Acid Bacteria Isolated from Boza, a Traditional Cereal-Based Beverage. Sci. Rep. 2022, 12, 1460. [Google Scholar] [CrossRef]
- Vezina, B.; Rehm, B.H.A.; Smith, A.T. Bioinformatic Prospecting and Phylogenetic Analysis Reveals 94 Undescribed Circular Bacteriocins and Key Motifs. BMC Microbiol. 2020, 20, 77. [Google Scholar] [CrossRef] [PubMed]
- Ladjouzi, R.; Dussert, E.; Teiar, R.; Belguesmia, Y.; Drider, D. A Review on Enterocin DD14, the Leaderless Two-Peptide Bacteriocin with Multiple Biological Functions and Unusual Transport Pathway. Antibiotics 2023, 12, 1188. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Krawczyk, P.S.; Lipinski, L.; Dziembowski, A. PlasFlow: Predicting Plasmid Sequences in Metagenomic Data Using Genome Signatures. Nucleic Acids Res. 2018, 46, e35. [Google Scholar] [CrossRef]
- Antipov, D.; Hartwick, N.; Shen, M.; Raiko, M.; Lapidus, A.; Pevzner, P.A. plasmidSPAdes: Assembling Plasmids from Whole Genome Sequencing Data. Bioinformatics 2016, 32, 3380–3387. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S. IslandViewer 4: Expanded Prediction of Genomic Islands for Larger-Scale Datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an Update of CRISRFinder, Includes a Portable Version, Enhanced Performance and Integrates Search for Cas Proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez, R.L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. mobileOG-Db: A Manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements. Appl. Environ. Microbiol. 2022, 88, e00991-22. [Google Scholar] [CrossRef]
- McGuffie, M.J.; Barrick, J.E. pLannotate: Engineered Plasmid Annotation. Nucleic Acids Res. 2021, 49, W516–W522. [Google Scholar] [CrossRef]
- Stothard, P.; Grant, J.R.; Van Domselaar, G. Visualizing and Comparing Circular Genomes Using the CGView Family of Tools. Brief. Bioinform. 2017, 20, 1576–1582. [Google Scholar] [CrossRef]
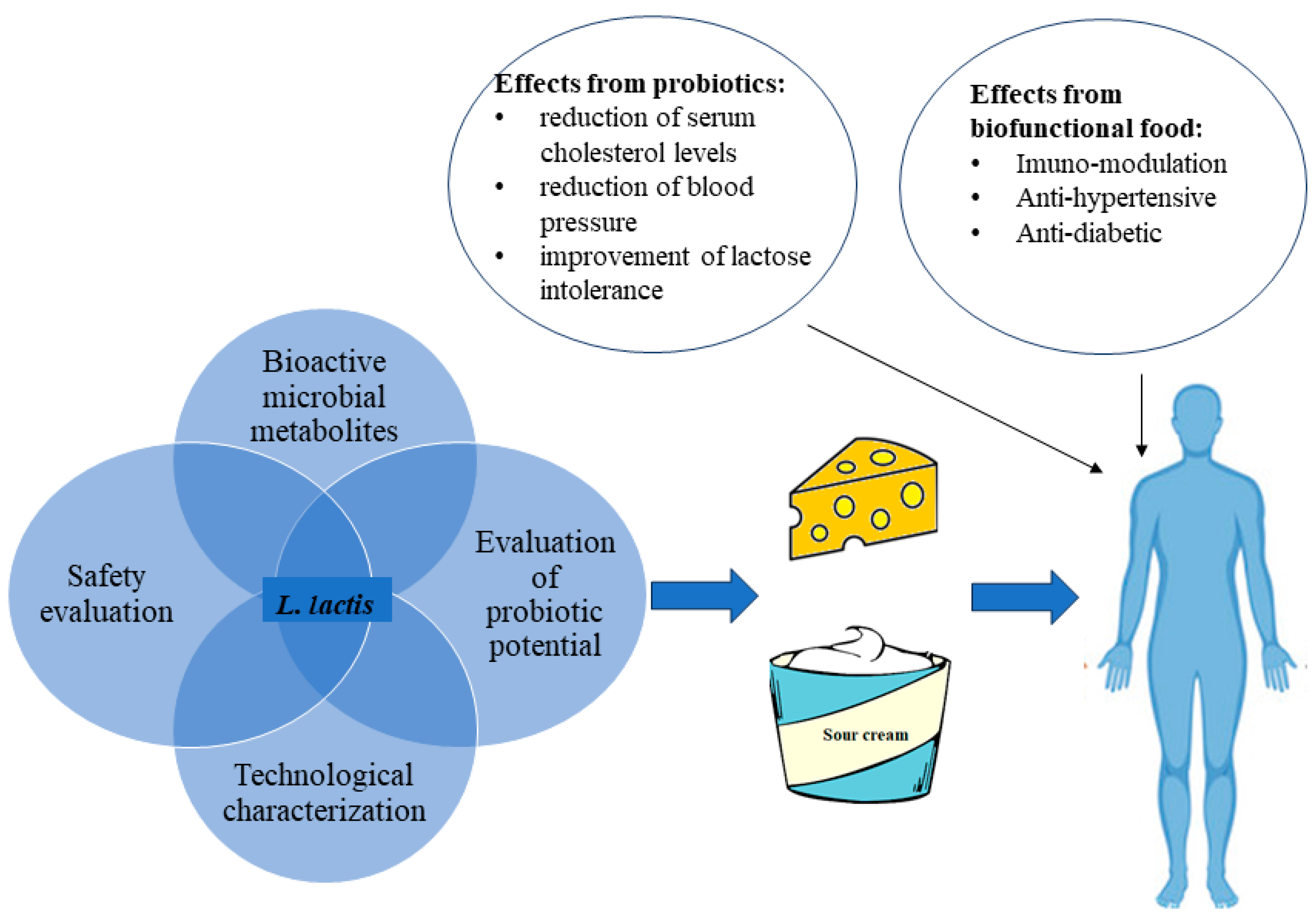

| Source of Isolation | Strain | Application | Properties | Reference |
|---|---|---|---|---|
| Traditionally fermented dairy foods | L. lactis subsp. lactis (IMAU11823) L. lactis subsp. lactis (IMAU11919) | Fermented milk | Good fermentation capacity, good sensory profiles | Li et al. [2] |
| Raw bovine milk | L. lactis subsp. lactis LL16 | Fermented milk | Positive impact on sensory properties and increased shelf life of cheese | Mileriene et al. [45,46] |
| Indigenous Montenegrin dairy products | L. lactis subsp. lactis | Tested as starter cultures for cheese production | Possible starter cultures for the production of traditional Montenegrin cheese as strains displayed rapid acidification ability and proteolysis | Bojanic Rasovic et al. [15] |
| Goat milk Cow milk | L. hircilactis L. laudensis | Strains were tested as acidifying and/or flavoring cultures in small-scale cheesemaking | Interesting technological and functional characteristics (production of aromatic compounds, ability to enhance the flavor in cheese, antioxidant activity) suitable for an application as complementary cultures in the dairy industry, in particular, as flavoring cultures in cheese and butter production | Tidona et al. [35] |
| Raw milk | L. lactis subsp. cremoris M104 L. lactis subsp. cremoris M78 | Graviera mini cheeses | L. monocytogenes count reduction | Samelis et al. [37] |
| Fresh Feta cheese | L. lactis subsp. lactis L. lactis subsp. garvieae | Strains were tested as starter cultures for the production of cheese | Possible starter cultures for the production of cheese (evaluation of acidifying and proteolytic activities, autolysis) | Bozoudi et al. [39] |
| Traditional Spanish, starter-free cheese made of raw milk | L. lactis subsp. lactis L. lactis subsp. cremoris | Strains were tested for bacteriocin production for application in fermented dairy products | Production of bacteriocins, possible application as protective cultures for cheese and other fermented products, possible application as adjunct cultures aimed at improving and accelerating cheese ripening | Alegria et al. [43] |
| Dairy products (milk and cheese) | L. lactis subsp. lactis biovar. diacetylactis | Strains were tested as starter cultures for the application in dairy food products | Interesting technological features for potential application in fermented food production (ability to produce proteases, acidifying activity, growth at different NaCl concentrations) | Fusieger et al. [47] |
| Raw goat milk | L. lactis | Tested for application as starter or biopreservative cultures | Possible starter or biopreservative cultures in fermented milk production | Perin et al. [48] |
| Source of Isolation | Strain | Features | Reference | |
|---|---|---|---|---|
| Kefir grains | L. lactis subsp. lactis | High hydrophobicity, bile salt deconjugation | Yerlikaya [3] | |
| Raw bovine milk | L. lactis subsp. lactis LL16 | GABA production in fermented milk, safety confirmation, probiotic property confirmation | Mileriene et al. [70] | |
| Fermented milk | L. lactis subsp. lactis | Good antimicrobial activity, probiotic potential | Akbar et al. [61] | |
| Mexican artisanal milk kefir grains | L. lactis subsp. lactis | Probiotic potential including antibiotic susceptibility, GABA production | Hurtado-Romero et al. [71] | |
| Camel milk | L. lactis KX881782 L. lactis KX881768 L. lactis KX881782 | Probiotic characteristics including auto-aggregation ability, high cholesterol removal ability, high co-aggregation, strong antimicrobial activity, and EPS production. KX881768 and KX881782 exhibited remarkable cholesterol removal abilities | Abushelaibi et al. [56] | |
| Raw and fermented milk | L. lactis subsp. lactis | Probiotic characteristics including antibiotic resistance, enzymatic activity, hemolytic and gelatinase activities, resistance to bile salts and acid, growth in bile acids and cholesterol, cell surface hydrophobicity | Kondrotiene et al. [60] | |
| Ricotta cheese | L. lactis subsp. lactis R7 | Anti-carcinogenic potential against colorectal cancer, an immune response was observed, and the biochemical parameters showed that L. lactis strain reversed the stress caused by 1,2-dimethylhydrazine and a hypercaloric diet in rats | Jaskulski et al. [72] | |
| Sliced mozzarella cheese | L. lactis subsp. cremoris LL95 | Probiotic properties such as resistance in a simulated gastric tract model and survival at different concentrations of NaCl and bile salts, and antioxidant activity. Depressive- and anxiety-like behavior in mice was proven | Ramalho et al. [73] | |
| Camel milk | L. lactis | Probiotic properties including tolerance and deconjugation of bile salts, antimicrobial activity, surface hydrophobicity, and adhesion potential | Sharma et al. [74] | |
| Artisanal, home-made products or raw milk | L. lactis IBB109 L. lactis IBB417 | Probiotic properties including bile salts and acid tolerance, adhesion properties, functional and safety aspects | Sałański et al. [68] | |
| Goat milk | L. lactis DF04Mi | Safety-related virulence factors (hemolytic activity, gelatinase production, coagulase, and sensitivity to antibiotics), functionality (exopolysaccharide (EPS) production, proteolytic activity, auto-aggregation, gas production, survival in the gastrointestinal tract, and antimicrobial activity against bacteria that impair oral health) | Silva et al. [75] | |
| Tulum cheeses | L. lactis | Probiotic properties including resistance to acid and bile salt; resistance to gastric and pancreatic juices; resistance to antibiotics; auto-aggregation; co-aggregation; diacetyl, hydrogen peroxide, and exopolysaccharide productions | Kazancıgil et al. [76] | |
| Antibiotic | Number of Resistance L. lactis Strains/Number of Tested L. lactis Strains (Percentage of Resistance L. lactis Strains) | Confirmed MIC Range (μg/mL) | Bacteria Isolation Source | Reference | EFSA Cutoff Value (μg mL−1) |
|---|---|---|---|---|---|
| Streptomycin | 4/6 (66.67%) | 128 | Commercial probiotics | Shin et al. [94] | 32 |
| 8/33 (24.24%) | 96−>1024 | Raw and fermented milk | Kondrotiene et al. [60] | ||
| 11/75 (14.67%) | 64 | Rainbow trout and rearing environment | Araujo [95] | ||
| 14/40 (35%) | 64–256 | Montenegrin dairy products | Bojanic Rastovic et al. [15] | ||
| Tetracycline | 6/33 (18.18%) | 12–32 | Raw and fermented milk | Kondrotiene et al. [60] | 4 |
| 3/50 (6%) | 64–128 | Dairy and different environments | Ammor et al. [96] | ||
| 18/75 (24%) | 8–64 | Rainbow trout and rearing environment | Araujo [95] | ||
| Vancomycin | 3/75 (4%) | 64 | 4 | ||
| Gentamicin | 1/75 (1.33%) | 256 | 32 | ||
| Kanamycin | 8/75 (10.67%) | 128 | 64 | ||
| Clindamycin | 6/75 (8%) | 6 | 1 | ||
| Erythromycin | 2/40 (5%) | 2 | Montenegrin dairy products | Bojanic Rastovic et al. [15] | 1 |
| 1 | #FILE | L. lactis_16.fasta | Indication of the Filename or Name of the File Related to the Sequence |
|---|---|---|---|
| 2 | SEQUENCE | NODE_1_length_50065_cov_17.999680 | Specifies details of the sequence |
| 3 | START | 38,077 | Indicates the start coordinate of the sequence |
| 4 | END | 40,071 | Indicates the end coordinate of the sequence |
| 5 | STRAND | - | Denotes the directionality of the sequence, typically represented as + (positive) or − (negative) |
| 6 | GENE | lmrD | Specifies the name of the gene |
| 7 | COVERAGE | 1-1995/1995 | Provides details on the coverage of the sequence in the specific segment, often represented as the proportion of the gene in the sequence |
| 8 | COVERAGE_MAP | =============== | Represents a visual representation of the alignment status; ==—denotes aligned regions, .=—indicates unaligned segments, and /=—signifies regions with gaps |
| 9 | GAPS | 0/0 | Indicates the presence of any gaps or openings in the subject and query sequences, potentially suggesting the existence of a pseudogene |
| 10 | %COVERAGE | 100.00 | Represents the percentage of the gene covered by the sequence |
| 11 | %IDENTITY | 99.80 | Indicates the percentage of exact nucleotide matches found in the sequence |
| 12 | DATABASE | card | Specifies the database from which this particular sequence comes from |
| 13 | ACCESSION | CP033607.1:310893-312888 | Provides details about the genomic source of the sequence |
| 14 | PRODUCT | lmrD is a chromosomally encoded efflux pump that confers resistance to lincosamides in Streptomyces lincolnensis and L. lactis. It can dimerize with lmrC | Describes the product or function of the gene, including its role in conferring resistance to certain antibiotics and any known associations or characteristics of the encoded protein |
| 15 | RESISTANCE | lincosamide | Specifies the putative antibiotic resistance phenotype associated with the gene |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondrotiene, K.; Zavistanaviciute, P.; Aksomaitiene, J.; Novoslavskij, A.; Malakauskas, M. Lactococcus lactis in Dairy Fermentation—Health-Promoting and Probiotic Properties. Fermentation 2024, 10, 16. https://doi.org/10.3390/fermentation10010016
Kondrotiene K, Zavistanaviciute P, Aksomaitiene J, Novoslavskij A, Malakauskas M. Lactococcus lactis in Dairy Fermentation—Health-Promoting and Probiotic Properties. Fermentation. 2024; 10(1):16. https://doi.org/10.3390/fermentation10010016
Chicago/Turabian StyleKondrotiene, Kristina, Paulina Zavistanaviciute, Jurgita Aksomaitiene, Aleksandr Novoslavskij, and Mindaugas Malakauskas. 2024. "Lactococcus lactis in Dairy Fermentation—Health-Promoting and Probiotic Properties" Fermentation 10, no. 1: 16. https://doi.org/10.3390/fermentation10010016
APA StyleKondrotiene, K., Zavistanaviciute, P., Aksomaitiene, J., Novoslavskij, A., & Malakauskas, M. (2024). Lactococcus lactis in Dairy Fermentation—Health-Promoting and Probiotic Properties. Fermentation, 10(1), 16. https://doi.org/10.3390/fermentation10010016








