Long Non-Coding RNAs as Molecular Signatures for Canine B-Cell Lymphoma Characterization
Abstract
1. Background
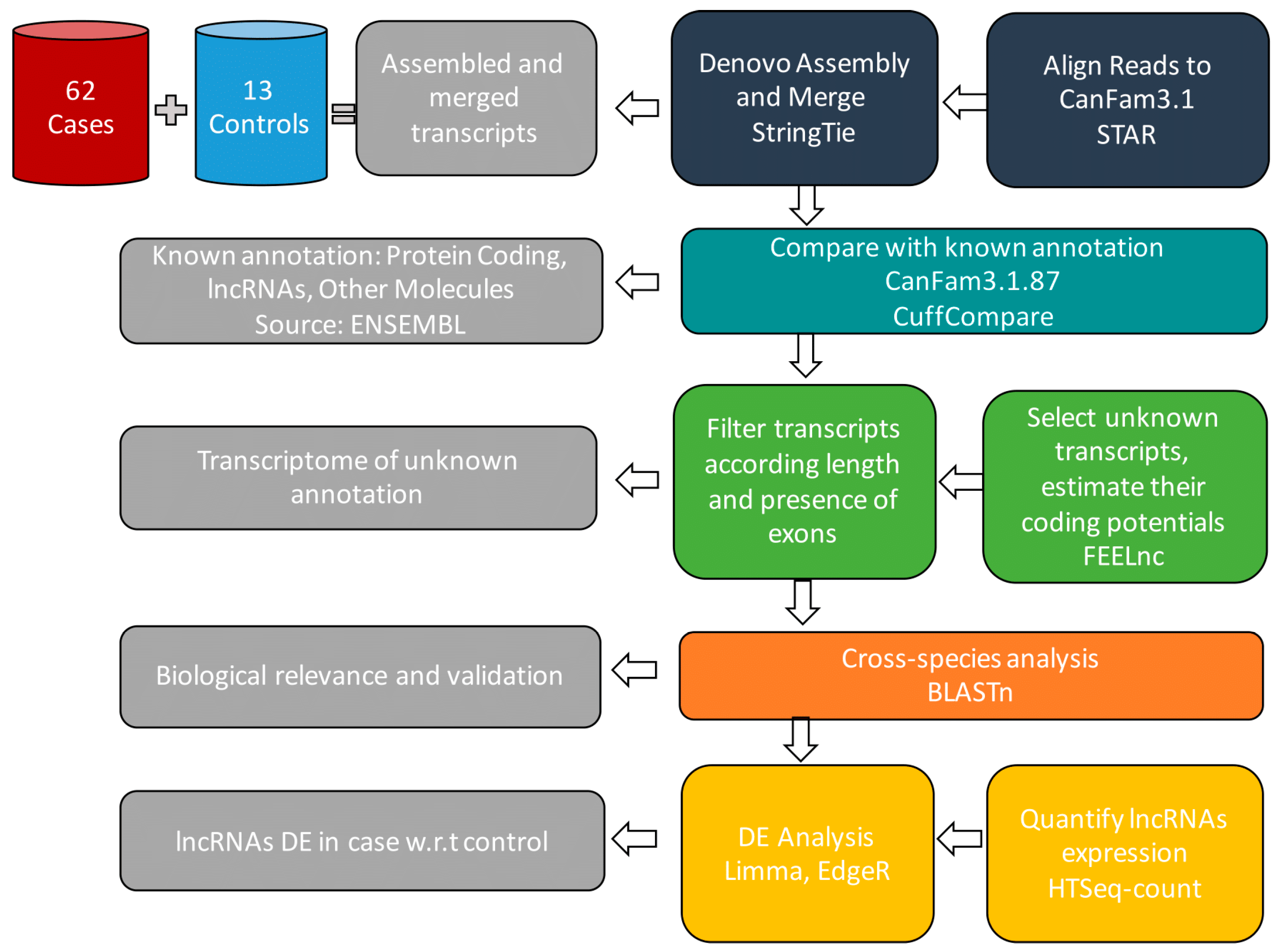
2. Methods
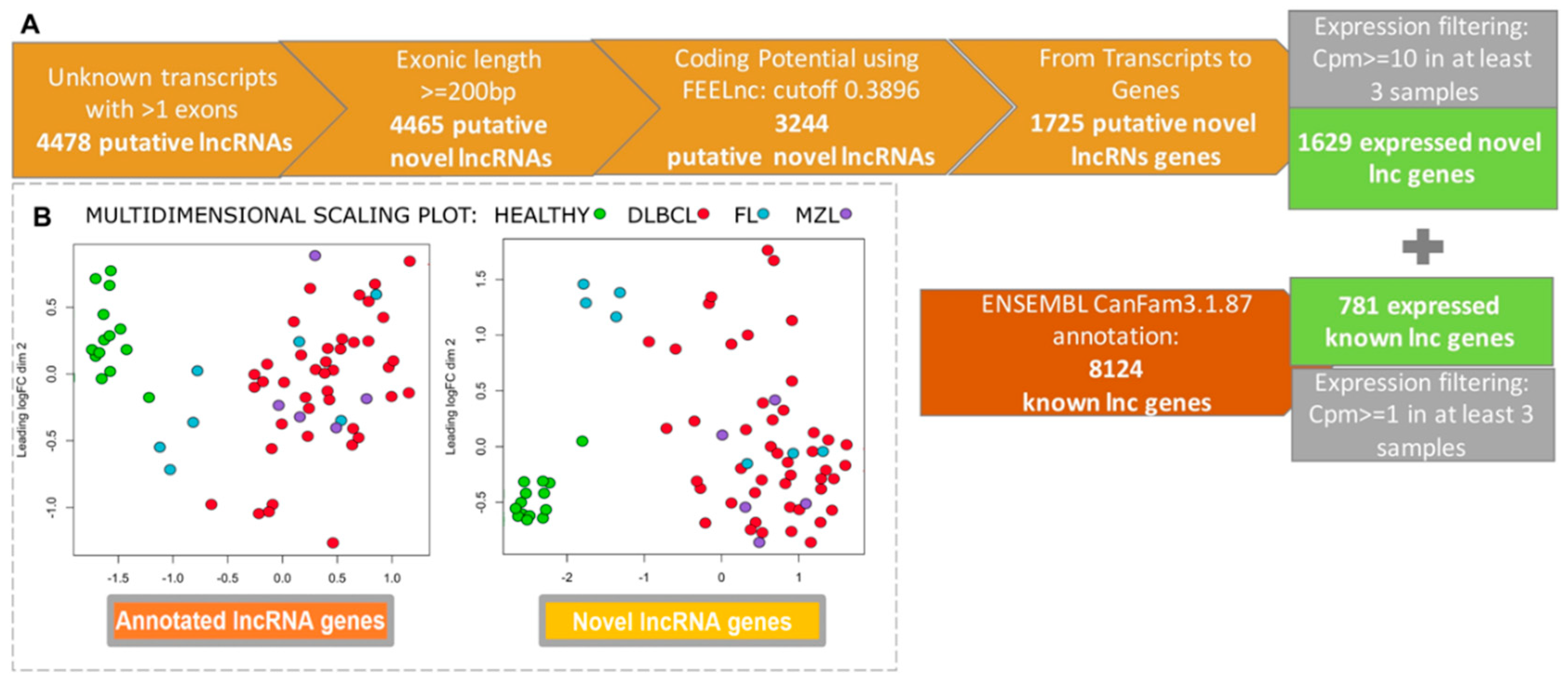
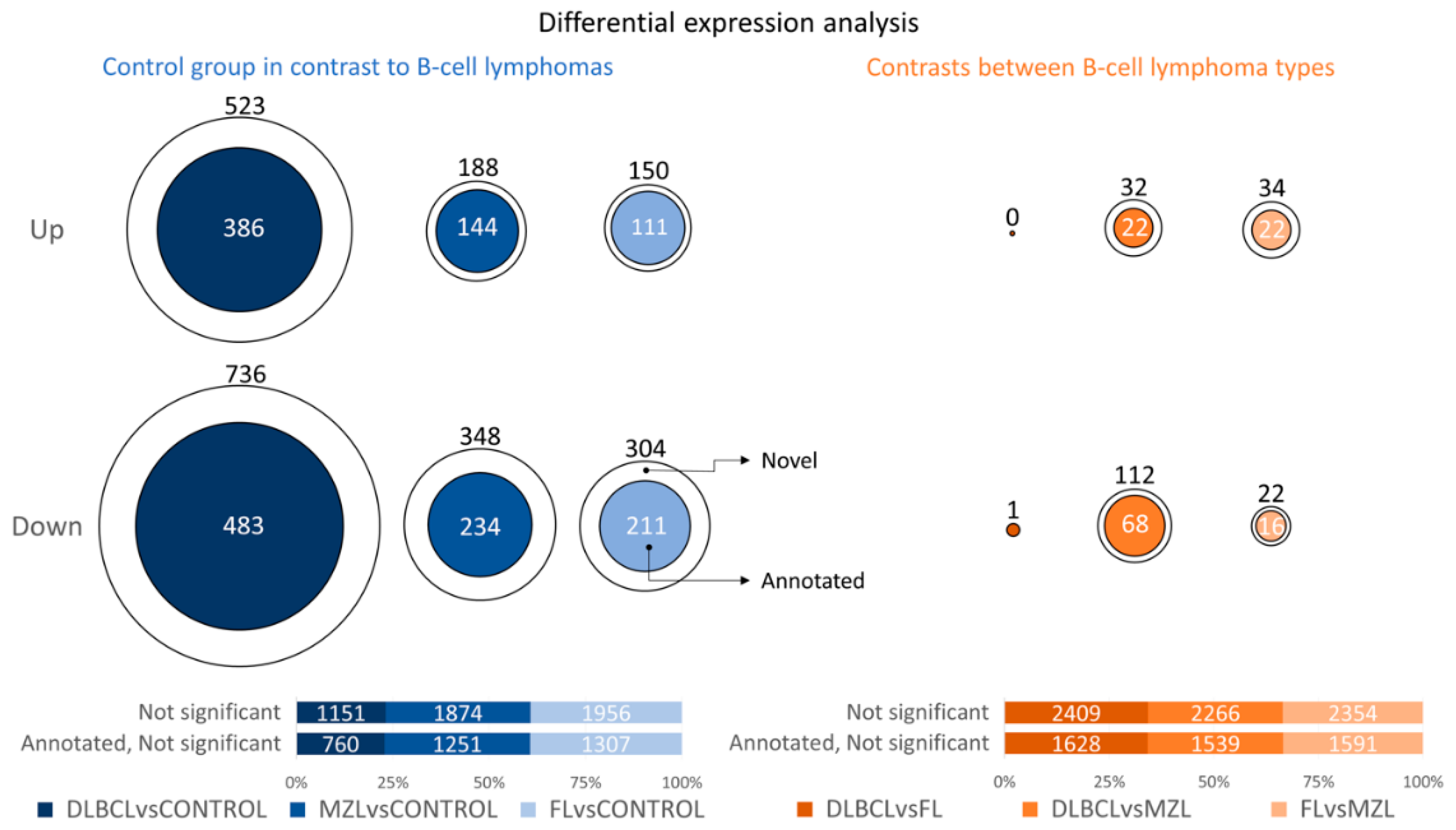
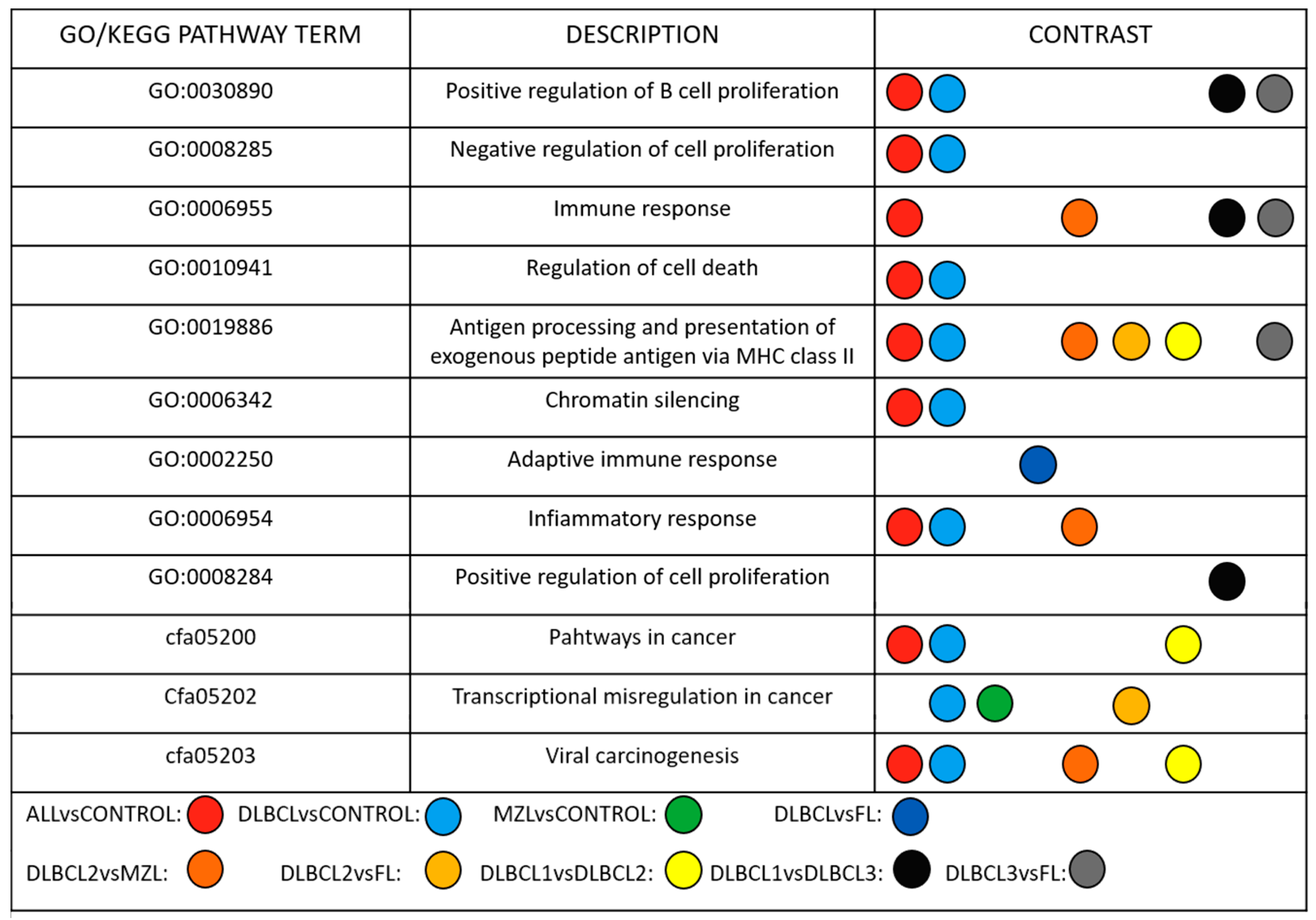
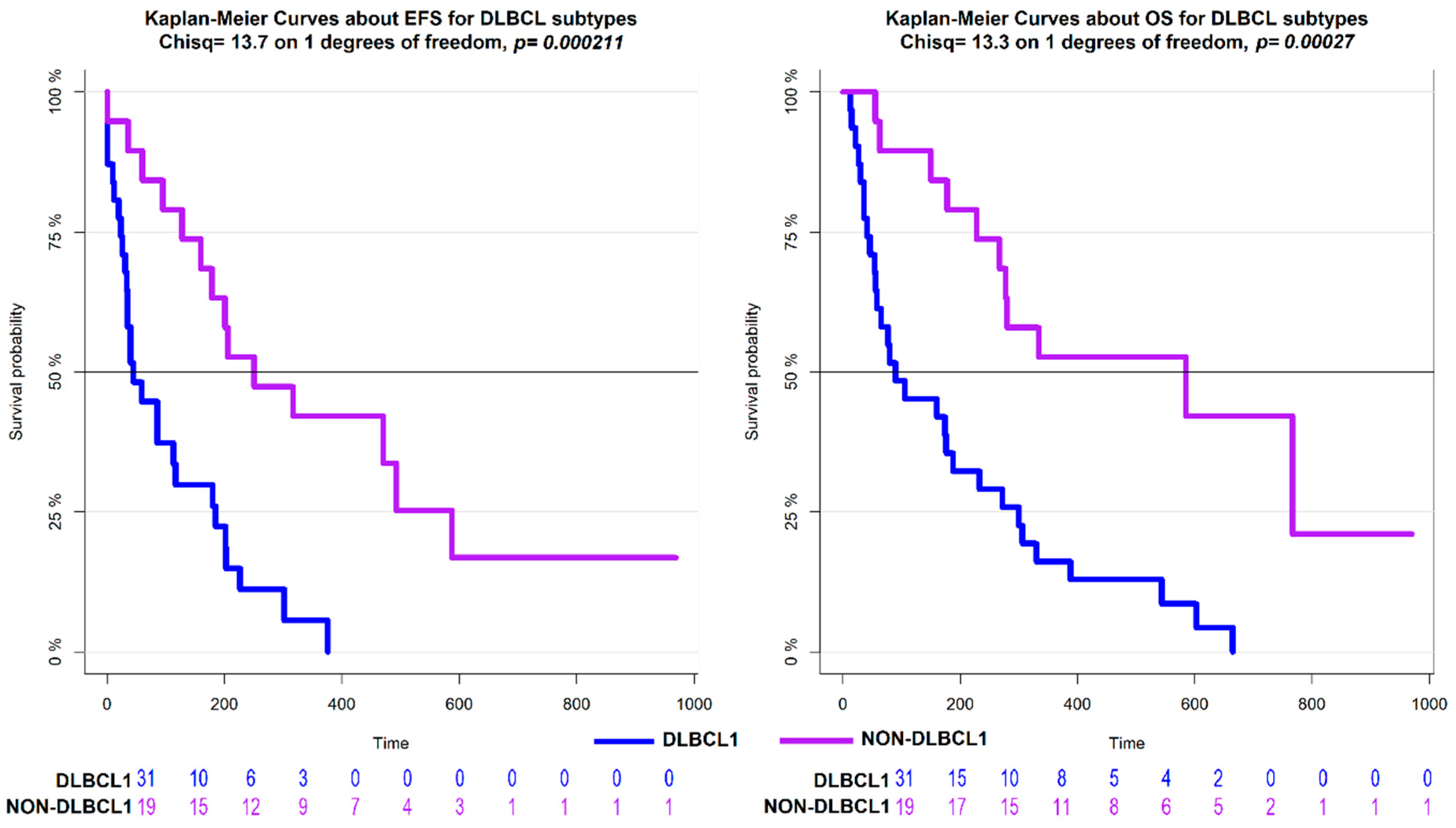
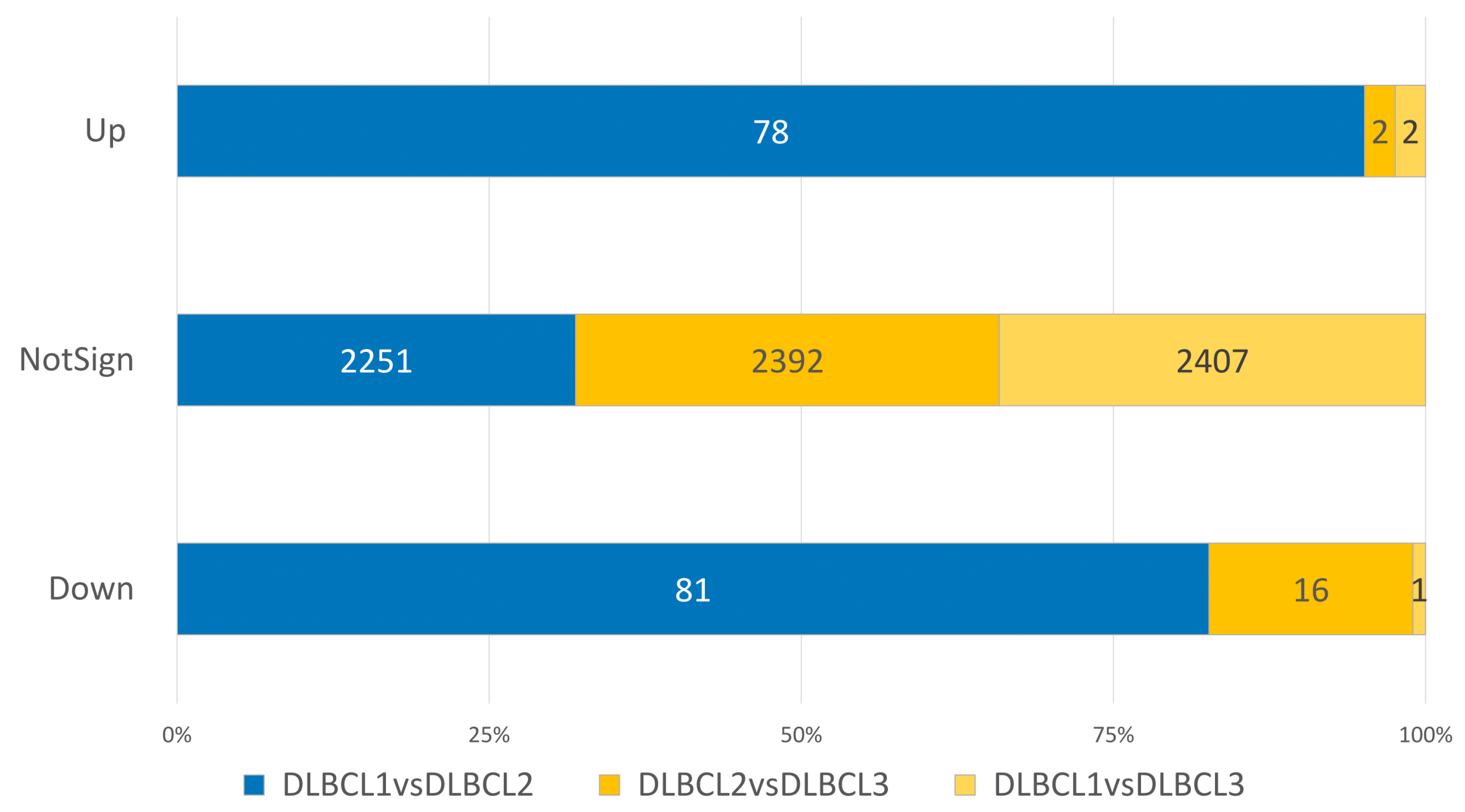
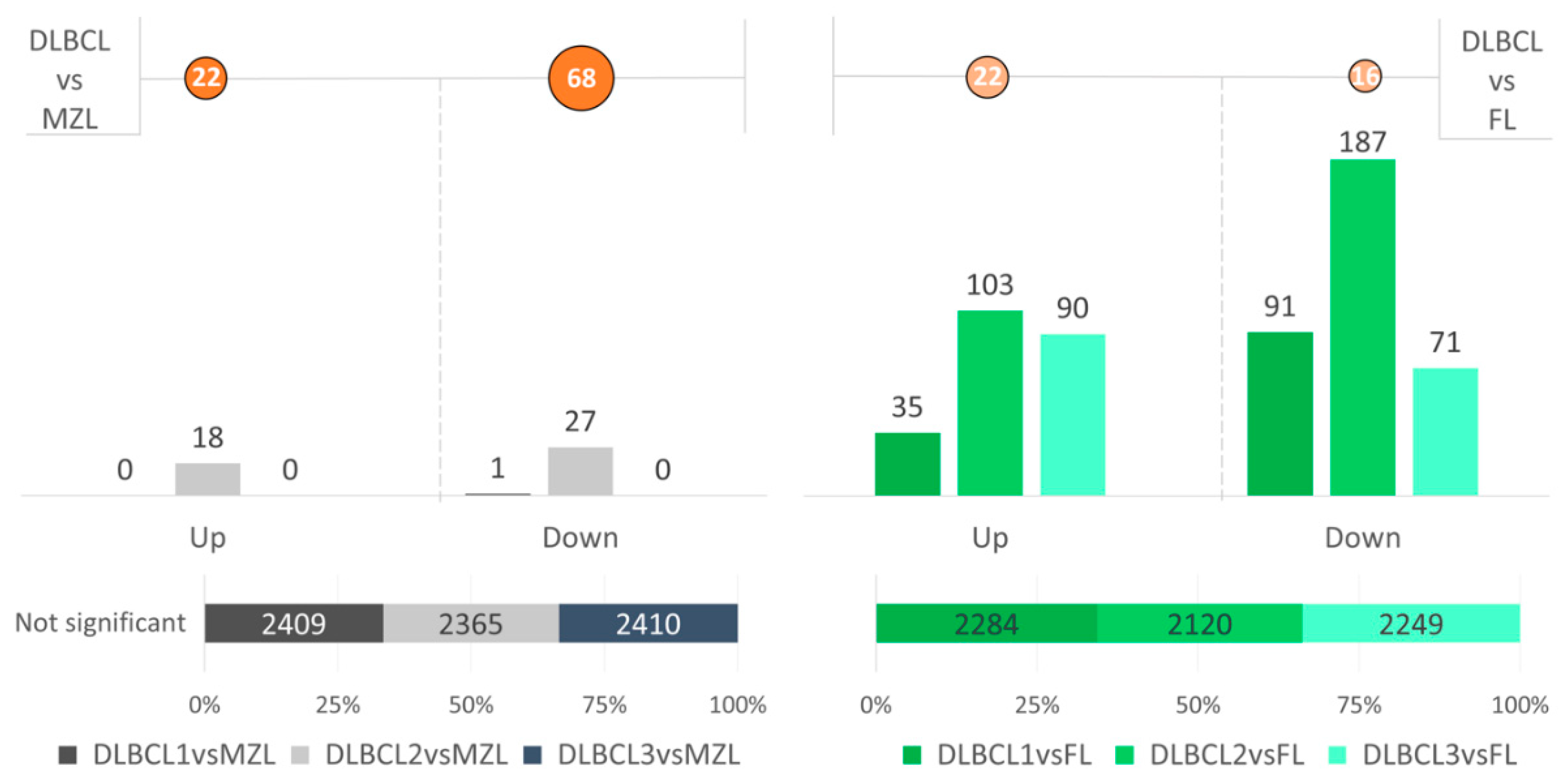
3. Results
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of Data and Materials
Abbreviations
| BCL | B-cell lymphomas |
| DLBCL | Diffuse large B cell lymphoma |
| ncRNA | non-coding RNA |
| MZL | marginal zone lymphoma |
| FL | follicular lymphoma |
| ABC-DLBCL | activated B cell-like DLBCL |
| EFS | event-free survival |
| AIC | Akaike’s Information Criterion |
| CPE | concordance probability estimates |
| GSEA | Gene Set Enrichment Analysis |
| MSigDB | Molecular Signatures Database |
References
- Armitage, J.O.; Gascoyne, R.D.; Lunning, M.A.; Cavalli, F. Non-Hodgkin lymphoma. Lancet 2017, 390, 298–310. [Google Scholar] [CrossRef]
- Aresu, L. Canine Lymphoma, More Than a Morphological Diagnosis: What We Have Learned about Diffuse Large B-Cell Lymphoma. Front. Veter Sci. 2016, 3, 348. [Google Scholar] [CrossRef] [PubMed]
- Richards, K.L.; Suter, S.E. Man’s best friend: what can pet dogs teach us about non-Hodgkin’s lymphoma? Immunol. Rev. 2015, 263, 173–191. [Google Scholar] [CrossRef] [PubMed]
- Frantz, A.M.; Sarver, A.L.; Ito, D.; Phang, T.L.; Karimpour-Fard, A.; Scott, M.C.; Valli, V.E.O.; Lindblad-Toh, K.; Burgess, K.E.; Husbands, B.D.; et al. Molecular Profiling Reveals Prognostically Significant Subtypes of Canine Lymphoma. Veter Pathol. 2012, 50, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Richards, K.L.; Motsinger-Reif, A.A.; Chen, H.W.; Fedoriw, Y.; Fan, C.; Nielsen, D.M.; Small, G.W.; Thomas, R.; Smith, C.; Dave, S.S.; et al. Gene profiling of canine B-cell lymphoma reveals germinal center and post-germinal center subtypes with different survival times, modeling human DLBCL. Cancer Res. 2013, 73, 5029–5039. [Google Scholar] [CrossRef] [PubMed]
- Aresu, L.; Ferraresso, S.; Marconato, L.; Cascione, L.; Napoli, S.; Gaudio, E.; Kwee, I.; Tarantelli, C.; Testa, A.; Maniaci, C.; et al. New molecular and therapeutic insights into canine diffuse large B cell lymphoma elucidates the role of the dog as a model for human disease. Haematologica 2019. [Google Scholar] [CrossRef]
- Giannuzzi, D.; Giudice, L.; Marconato, L. Integrated analysis of RNA-seq, MBD-seq and copy number variations in canine marginal zone and follicular lymphoma. In Proceedings of the Veterinary Cancer Society Annual Conference, Houston, TX, USA, 17–19 October 2019. [Google Scholar]
- Ferraresso, S.; Aricò, A.; Sanavia, T.; Da Ros, S.; Milan, M.; Cascione, L.; Comazzi, S.; Martini, V.; Giantin, M.; Di Camillo, B.; et al. DNA methylation profiling reveals common signatures of tumorigenesis and defines epigenetic prognostic subtypes of canine Diffuse Large B-cell Lymphoma. Sci. Rep. 2017, 7, 11591. [Google Scholar] [CrossRef]
- Elvers, I.; Turner-Maier, J.; Swofford, R.; Koltookian, M.; Johnson, J.; Stewart, C.; Zhang, C.-Z.; Schumacher, S.E.; Beroukhim, R.; Rosenberg, M.; et al. Exome sequencing of lymphomas from three dog breeds reveals somatic mutation patterns reflecting genetic background. Genome Res. 2015, 25, 1634–1645. [Google Scholar] [CrossRef]
- Ponting, C.P.; Belgard, T.G. Transcribed dark matter: meaning or myth? Hum. Mol. Genet. 2010, 19, R162–R168. [Google Scholar] [CrossRef]
- Sanchez Calle, A.; Kawamura, Y.; Yamamoto, Y.; Takeshita, F.; Ochiya, T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018, 109, 2093–2100. [Google Scholar] [CrossRef]
- Le Béguec, C.; Wucher, V.; Lagoutte, L.; Cadieu, E.; Botherel, N.; Hédan, B.; De Brito, C.; Guillory, A.S.; Andre, C.; Derrien, T.; et al. Characterisation and functional predictions of canine long non-coding RNAs. Sci. Rep. 2018, 8, 13444. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Jiang, Y.; Du, W.; Fairchild, L.; Melnick, A.; Elemento, O. Transcriptome sequencing reveals thousands of novel long non-coding RNAs in B cell lymphoma. Genome Med. 2015, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Wucher, V.; Legeai, F.; Hédan, B.; Rizk, G.; Lagoutte, L.; Leeb, T.; Jagannathan, V.; Cadieu, E.; David, A.; Lohi, H.; et al. FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017, 45, e57. [Google Scholar] [CrossRef] [PubMed]
- Giannuzzi, D.; Marconato, L.; Cascione, L.; Comazzi, S.; Elgendy, R.; Pegolo, S.; Cecchinato, A.; Bertoni, F.; Aresu, L.; Ferraresso, S. Mutational landscape of canine B-cell lymphoma profiled at single nucleotide resolution by RNA-seq. PLoS ONE 2019, 14, e0215154. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. Stringtie enables improved reconstruction of a transcriptome from rna-seq reads. Nat. Biotechnol. 2015, 33, 290. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by rna-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2014, 31, 166–169. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Gordon, K.S. edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [PubMed]
- Cassisi, C.; Ferro, A.; Giugno, R.; Pigola, G.; Pulvirenti, A. Enhancing density-based clustering: Parameter reduction and outlier detection. Inf. Syst. 2013, 38, 317–330. [Google Scholar] [CrossRef]
- Hahsler, M.; Piekenbrock, M. dbscan: Density Based Clustering of Applications with Noise (DBSCAN) and Related Algorithms. R Package Version 1.1-1, 2017. Available online: https://rdrr.io/cran/dbscan/ (accessed on 19 September 2019).
- Kaplan, E.L.; Meier, P. Nonparametric Estimation from Incomplete Observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Altman, D.G.; Bland, J.M. Diagnostic tests 2: Predictive values. BMJ 1994, 309, 102. [Google Scholar] [CrossRef] [PubMed]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Gönen, M.H.G. Concordance probability and discriminatory power in proportional hazards regression. Biometrika 2005, 92, 965–970. [Google Scholar] [CrossRef]
- Harrell, F.E. Regression Modeling Strategies with Applications to Linear Models, Logistic Regression, and Survival Analysis; Springer: New York, NY, USA, 2001. [Google Scholar]
- Wong, E.S.; Schmitt, B.M.; Kazachenka, A.; Thybert, D.; Redmond, A.; Connor, F.; Rayner, T.F.; Feig, C.; Ferguson-Smith, A.C.; Marioni, J.C.; et al. Interplay of cis and trans mechanisms driving transcription factor binding and gene expression evolution. Nat. Commun. 2017, 8, 1092. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; A Lempicki, R. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Boil. 2007, 8, R183. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Tarantelli, C.; Gaudio, E.; Arribas, A.J.; Kwee, I.; Hillmann, P.; Rinaldi, A.; Cascione, L.; Spriano, F.; Bernasconi, E.; Guidetti, F.; et al. PQR309 Is a Novel Dual PI3K/mTOR Inhibitor with Preclinical Antitumor Activity in Lymphomas as a Single Agent and in Combination Therapy. Clin. Cancer Res. 2018, 24, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Bonnici, V.; Busato, F.; Aldegheri, S.; Akhmedov, M.; Cascione, L.; Carmena, A.A.; Bertoni, F.; Bombieri, F.; Kwee, I.; Giugno, R. cuRnet: An R package for graph traversing on GPU. BMC Bioinform. 2018, 19, 221. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef] [PubMed]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular Subtypes of Diffuse Large B-cell Lymphoma are Associated with Distinct Pathogenic Mechanisms and Outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cascione, L.; Giudice, L.; Ferraresso, S.; Marconato, L.; Giannuzzi, D.; Napoli, S.; Bertoni, F.; Giugno, R.; Aresu, L. Long Non-Coding RNAs as Molecular Signatures for Canine B-Cell Lymphoma Characterization. Non-Coding RNA 2019, 5, 47. https://doi.org/10.3390/ncrna5030047
Cascione L, Giudice L, Ferraresso S, Marconato L, Giannuzzi D, Napoli S, Bertoni F, Giugno R, Aresu L. Long Non-Coding RNAs as Molecular Signatures for Canine B-Cell Lymphoma Characterization. Non-Coding RNA. 2019; 5(3):47. https://doi.org/10.3390/ncrna5030047
Chicago/Turabian StyleCascione, Luciano, Luca Giudice, Serena Ferraresso, Laura Marconato, Diana Giannuzzi, Sara Napoli, Francesco Bertoni, Rosalba Giugno, and Luca Aresu. 2019. "Long Non-Coding RNAs as Molecular Signatures for Canine B-Cell Lymphoma Characterization" Non-Coding RNA 5, no. 3: 47. https://doi.org/10.3390/ncrna5030047
APA StyleCascione, L., Giudice, L., Ferraresso, S., Marconato, L., Giannuzzi, D., Napoli, S., Bertoni, F., Giugno, R., & Aresu, L. (2019). Long Non-Coding RNAs as Molecular Signatures for Canine B-Cell Lymphoma Characterization. Non-Coding RNA, 5(3), 47. https://doi.org/10.3390/ncrna5030047








