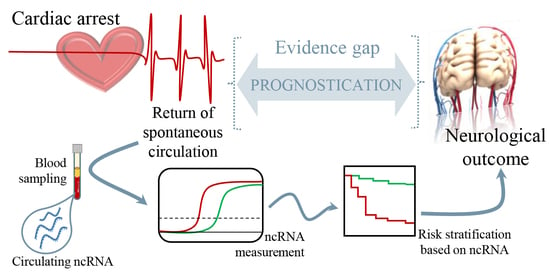
Non-Coding RNAs to Aid in Neurological Prognosis after Cardiac Arrest
Abstract
:1. Background
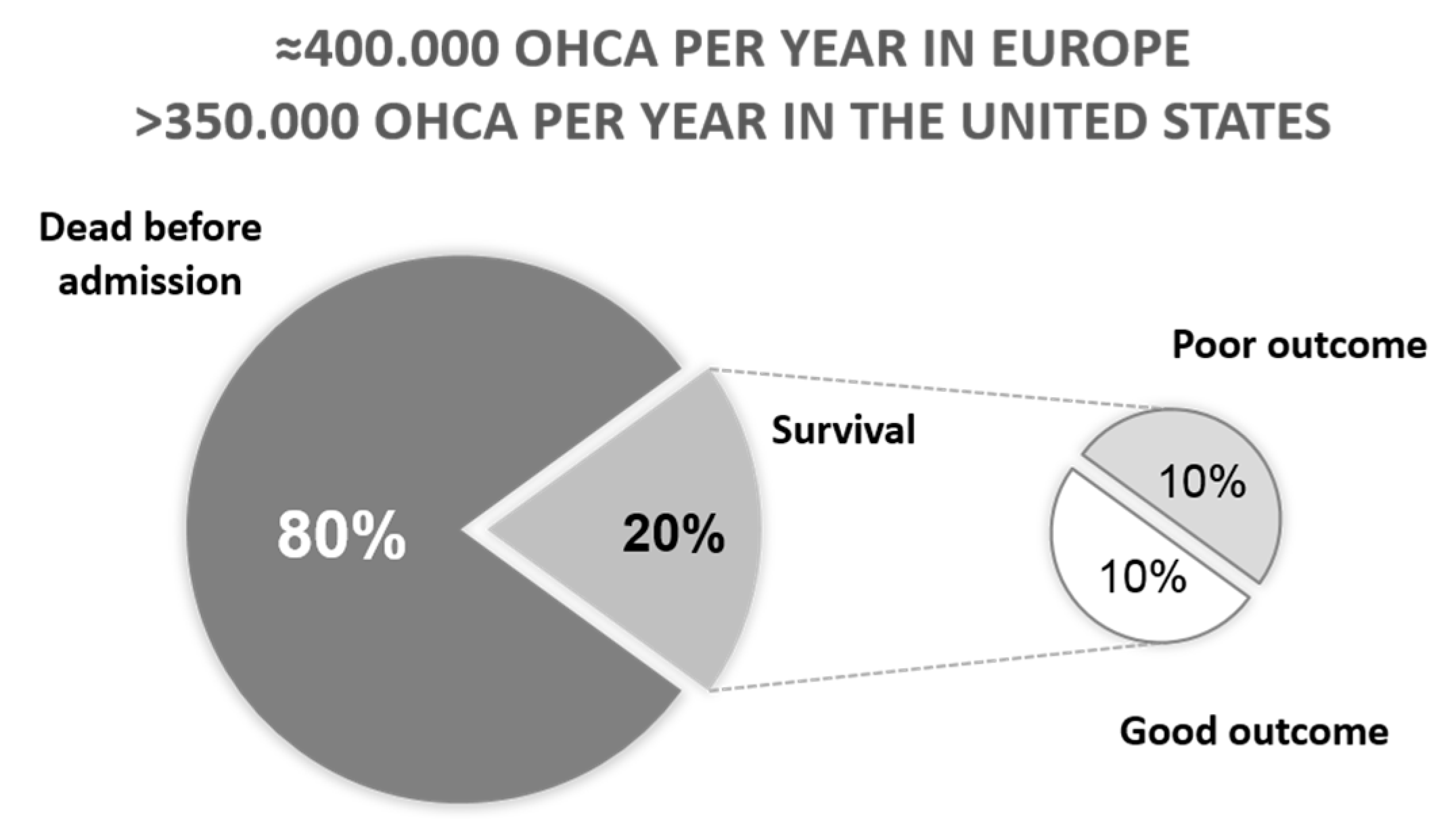
2. Outcome after Cardiac Arrest
3. Prediction of Neurological Outcome after Cardiac Arrest
4. Non-Coding RNAs
4.1. MicroRNAs
4.2. Long Non-Coding RNAs
4.3. Circular RNAs
5. Future Directions and Conclusions
- Further deep RNA sequencing experiments are needed to identify the best candidates for prediction among the currently-identified 2500 human mature miRNAs, 146,000 human annotated lncRNAs and 32,000 predicted circRNAs.
- Independent validation of candidates in large patient cohorts will be the key to discovering robust and clinically-applicable biomarkers.
- Address gender specificities.
- Test the incremental predictive value of panels of non-coding RNAs using suitable correction strategies to avoid model overfitting.
- Assess the evolution of circulating levels of non-coding RNAs within the few hours/days after CA.
- Assess the influence of co-variates, such as age and target temperature, on circulating levels of non-coding RNAs.
- Define optimized protocols for blood sample collection, handling, storage and processing for RNA biomarkers assessment.
- Design molecular diagnostic assays for RNA assessment at the bedside that will allow clinically-applicable decision support systems combining biomarker assessment, neurophysiology, clinical examination, statistical analysis and risk stratification models.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abreviations
| CA | Cardiac arrest |
| circRNAs | Circular RNAs |
| HUVEC | Human umbilical vein endothelial cells |
| LAD | Left anterior descending |
| lncRNA | Long non-coding RNA |
| MCAO | Middle cerebral artery occlusion |
| miRNA | MicroRNA |
| NFκB | Nuclear Factor kappa B |
| NSE | Neuron Specific Enolase |
| OGD/R | Oxygen and glucose deprivation/reoxygenation |
| OHCA | Out-of-hospital cardiac arrest |
| RNA | Ribonucleic acid |
| RNase | Ribonuclease |
| ROSC | Return Of Spontaneus Circulation |
| TBI | Traumatic brain injury |
| TNF | Tumor Necrosis Factor |
| TTM | Target Temperature Management |
References
- Townsend, N.; Wilson, L.; Bhatnagar, P.; Wickramasinghe, K.; Rayner, M.; Nichols, M. Cardiovascular disease in Europe: Epidemiological update 2016. Eur. Heart J. 2016, 37, 3232–3245. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, E.J.; Blaha, M.J.; Chiuve, S.E.; Cushman, M.; Das, S.R.; Deo, R.; de Ferranti, S.D.; Floyd, J.; Fornage, M.; Gillespie, C.; et al. Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association. Circulation 2017, 135, e146–e603. [Google Scholar] [CrossRef] [PubMed]
- Sandroni, C.; Nolan, J.; Cavallaro, F.; Antonelli, M. In-hospital cardiac arrest: Incidence, prognosis and possible measures to improve survival. Intens. Care Med. 2007, 33, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Wissenberg, M.; Lippert, F.K.; Folke, F.; Weeke, P.; Hansen, C.M.; Christensen, E.F.; Jans, H.; Hansen, P.A.; Lang-Jensen, T.; Olesen, J.B.; et al. Association of national initiatives to improve cardiac arrest management with rates of bystander intervention and patient survival after out-of-hospital cardiac arrest. JAMA 2013, 310, 1377–1384. [Google Scholar] [CrossRef] [PubMed]
- Hasselqvist-Ax, I.; Riva, G.; Herlitz, J.; Rosenqvist, M.; Hollenberg, J.; Nordberg, P.; Ringh, M.; Jonsson, M.; Axelsson, C.; Lindqvist, J.; et al. Early cardiopulmonary resuscitation in out-of-hospital cardiac arrest. N. Engl. J. Med. 2015, 372, 2307–2315. [Google Scholar] [CrossRef] [PubMed]
- Perkins, G.D.; Handley, A.J.; Koster, R.W.; Castren, M.; Smyth, M.A.; Olasveengen, T.; Monsieurs, K.G.; Raffay, V.; Grasner, J.T.; Wenzel, V.; et al. European Resuscitation Council Guidelines for Resuscitation 2015: Section 2. Adult basic life support and automated external defibrillation. Resuscitation 2015, 95, 81–99. [Google Scholar] [CrossRef] [PubMed]
- Fordyce, C.B.; Hansen, C.M.; Kragholm, K.; Dupre, M.E.; Jollis, J.G.; Roettig, M.L.; Becker, L.B.; Hansen, S.M.; Hinohara, T.T.; Corbett, C.C.; et al. Association of Public Health Initiatives with Outcomes for Out-of-Hospital Cardiac Arrest at Home and in Public Locations. JAMA Cardiol. 2017, 2, 1226–1235. [Google Scholar] [CrossRef] [PubMed]
- Larribau, R.; Deham, H.; Niquille, M.; Sarasin, F.P. Improvement of out-of-hospital cardiac arrest survival rate after implementation of the 2010 resuscitation guidelines. PLoS ONE 2018, 13, e0204169. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, N.; Wetterslev, J.; Cronberg, T.; Erlinge, D.; Gasche, Y.; Hassager, C.; Horn, J.; Hovdenes, J.; Kjaergaard, J.; Kuiper, M.; et al. Targeted temperature management at 33 °C versus 36 °C after cardiac arrest. N. Engl. J. Med. 2013, 369, 2197–2206. [Google Scholar] [CrossRef] [PubMed]
- Moulaert, V.R.; Verbunt, J.A.; van Heugten, C.M.; Wade, D.T. Cognitive impairments in survivors of out-of-hospital cardiac arrest: A systematic review. Resuscitation 2009, 80, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Dragancea, I.; Horn, J.; Kuiper, M.; Friberg, H.; Ullen, S.; Wetterslev, J.; Cranshaw, J.; Hassager, C.; Nielsen, N.; Cronberg, T.; et al. Neurological prognostication after cardiac arrest and targeted temperature management 33 °C versus 36 °C: Results from a randomised controlled clinical trial. Resuscitation 2015, 93, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Winther-Jensen, M.; Pellis, T.; Kuiper, M.; Koopmans, M.; Hassager, C.; Nielsen, N.; Wetterslev, J.; Cronberg, T.; Erlinge, D.; Friberg, H.; et al. Mortality and neurological outcome in the elderly after target temperature management for out-of-hospital cardiac arrest. Resuscitation 2015, 91, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, V.; Dankiewicz, J.; Nielsen, N.; Kern, K.B.; Mooney, M.R.; Riker, R.R.; Rubertsson, S.; Seder, D.B.; Stammet, P.; Sunde, K.; et al. Association of gender to outcome after out-of-hospital cardiac arrest—A report from the International Cardiac Arrest Registry. Crit. Care 2015, 19, 182. [Google Scholar] [CrossRef] [PubMed]
- Gupta, T.; Kolte, D.; Khera, S.; Aronow, W.S.; Palaniswamy, C.; Mujib, M.; Jain, D.; Sule, S.; Ahmed, A.; Iwai, S.; et al. Relation of smoking status to outcomes after cardiopulmonary resuscitation for in-hospital cardiac arrest. Am. J. Cardiol. 2014, 114, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Gupta, T.; Kolte, D.; Mohananey, D.; Khera, S.; Goel, K.; Mondal, P.; Aronow, W.S.; Jain, D.; Cooper, H.A.; Iwai, S.; et al. Relation of Obesity to Survival After In-Hospital Cardiac Arrest. Am. J. Cardiol. 2016, 118, 662–667. [Google Scholar] [CrossRef] [PubMed]
- Matinrazm, S.; Ladejobi, A.; Pasupula, D.K.; Javed, A.; Durrani, A.; Ahmad, S.; Munir, M.B.; Adelstein, E.; Jain, S.K.; Saba, S. Effect of body mass index on survival after sudden cardiac arrest. Clin. Cardiol. 2018, 41, 46–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stammet, P.; Collignon, O.; Hassager, C.; Wise, M.P.; Hovdenes, J.; Aneman, A.; Horn, J.; Devaux, Y.; Erlinge, D.; Kjaergaard, J.; et al. Neuron-Specific Enolase as a Predictor of Death or Poor Neurological Outcome After Out-of-Hospital Cardiac Arrest and Targeted Temperature Management at 33 °C and 36 °C. J. Am. Coll. Cardiol. 2015, 65, 2104–2114. [Google Scholar] [CrossRef] [PubMed]
- Devaux, Y.; Stammet, P.; Friberg, H.; Hassager, C.; Kuiper, M.A.; Wise, M.P.; Nielsen, N.; Biomarker subcommittee of TTM trial (Target Temperature Management After Cardiac Arrest, NCT01020916). MicroRNAs: New biomarkers and therapeutic targets after cardiac arrest? Crit. Care 2015, 19, 54. [Google Scholar] [CrossRef] [PubMed]
- Deakin, C.D.; Nolan, J.P.; Soar, J.; Sunde, K.; Koster, R.W.; Smith, G.B.; Perkins, G.D. European Resuscitation Council Guidelines for Resuscitation 2010 Section 4. Adult advanced life support. Resuscitation 2010, 81, 1305–1352. [Google Scholar] [CrossRef] [PubMed]
- Nolan, J.P.; Soar, J.; Cariou, A.; Cronberg, T.; Moulaert, V.R.; Deakin, C.D.; Bottiger, B.W.; Friberg, H.; Sunde, K.; Sandroni, C.; et al. European Resuscitation Council and European Society of Intensive Care Medicine 2015 guidelines for post-resuscitation care. Intens. Care Med. 2015, 41, 2039–2056. [Google Scholar] [CrossRef] [PubMed]
- Zandbergen, E.G.; Hijdra, A.; Koelman, J.H.; Hart, A.A.; Vos, P.E.; Verbeek, M.M.; de Haan, R.J.; Group, P.S. Prediction of poor outcome within the first 3 days of postanoxic coma. Neurology 2006, 66, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Nolan, J.P.; Soar, J.; Cariou, A.; Cronberg, T.; Moulaert, V.R.; Deakin, C.D.; Bottiger, B.W.; Friberg, H.; Sunde, K.; Sandroni, C. European Resuscitation Council and European Society of Intensive Care Medicine Guidelines for Post-resuscitation Care 2015: Section 5 of the European Resuscitation Council Guidelines for Resuscitation 2015. Resuscitation 2015, 95, 202–222. [Google Scholar] [CrossRef] [PubMed]
- Storm, C.; Nee, J.; Jorres, A.; Leithner, C.; Hasper, D.; Ploner, C.J. Serial measurement of neuron specific enolase improves prognostication in cardiac arrest patients treated with hypothermia: A prospective study. Scand. J. Trauma Resusc. Emerg. Med. 2012, 20, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoerkhuber, W.; Kittler, H.; Sterz, F.; Behringer, W.; Holzer, M.; Frossard, M.; Spitzauer, S.; Laggner, A.N. Time course of serum neuron-specific enolase. A predictor of neurological outcome in patients resuscitated from cardiac arrest. Stroke 1999, 30, 1598–1603. [Google Scholar] [CrossRef] [PubMed]
- Rech, T.H.; Vieira, S.R.; Nagel, F.; Brauner, J.S.; Scalco, R. Serum neuron-specific enolase as early predictor of outcome after in-hospital cardiac arrest: A cohort study. Crit. Care 2006, 10, R133. [Google Scholar] [CrossRef] [PubMed]
- Rundgren, M.; Karlsson, T.; Nielsen, N.; Cronberg, T.; Johnsson, P.; Friberg, H. Neuron specific enolase and S-100B as predictors of outcome after cardiac arrest and induced hypothermia. Resuscitation 2009, 80, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Burghuber, O.C.; Worofka, B.; Schernthaner, G.; Vetter, N.; Neumann, M.; Dudczak, R.; Kuzmits, R. Serum neuron-specific enolase is a useful tumor marker for small cell lung cancer. Cancer 1990, 65, 1386–1390. [Google Scholar] [CrossRef]
- DeGiorgio, C.M.; Gott, P.S.; Rabinowicz, A.L.; Heck, C.N.; Smith, T.D.; Correale, J.D. Neuron-specific enolase, a marker of acute neuronal injury, is increased in complex partial status epilepticus. Epilepsia 1996, 37, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Pelinka, L.E.; Jafarmadar, M.; Redl, H.; Bahrami, S. Neuron-specific-enolase is increased in plasma after hemorrhagic shock and after bilateral femur fracture without traumatic brain injury in the rat. Shock 2004, 22, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Park, K.; Ryu, S.; Kang, T.; Kim, H.; Cho, S.; Oh, S. Use of S-100B, NSE, CRP and ESR to predict neurological outcomes in patients with return of spontaneous circulation and treated with hypothermia. Emerg. Med. J. 2016, 33, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Mussack, T.; Biberthaler, P.; Kanz, K.G.; Wiedemann, E.; Gippner-Steppert, C.; Mutschler, W.; Jochum, M. Serum S-100B and interleukin-8 as predictive markers for comparative neurologic outcome analysis of patients after cardiac arrest and severe traumatic brain injury. Crit. Care Med. 2002, 30, 2669–2674. [Google Scholar] [CrossRef] [PubMed]
- Randall, J.; Mortberg, E.; Provuncher, G.K.; Fournier, D.R.; Duffy, D.C.; Rubertsson, S.; Blennow, K.; Zetterberg, H.; Wilson, D.H. Tau proteins in serum predict neurological outcome after hypoxic brain injury from cardiac arrest: Results of a pilot study. Resuscitation 2013, 84, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Mortberg, E.; Zetterberg, H.; Nordmark, J.; Blennow, K.; Catry, C.; Decraemer, H.; Vanmechelen, E.; Rubertsson, S. Plasma tau protein in comatose patients after cardiac arrest treated with therapeutic hypothermia. Acta Anaesthesiol. Scand. 2011, 55, 1132–1138. [Google Scholar] [CrossRef] [PubMed]
- Rana, O.R.; Schroder, J.W.; Baukloh, J.K.; Saygili, E.; Mischke, K.; Schiefer, J.; Weis, J.; Marx, N.; Rassaf, T.; Kelm, M.; et al. Neurofilament light chain as an early and sensitive predictor of long-term neurological outcome in patients after cardiac arrest. Int. J. Cardiol. 2013, 168, 1322–1327. [Google Scholar] [CrossRef] [PubMed]
- Fries, M.; Kunz, D.; Gressner, A.M.; Rossaint, R.; Kuhlen, R. Procalcitonin serum levels after out-of-hospital cardiac arrest. Resuscitation 2003, 59, 105–109. [Google Scholar] [CrossRef]
- Stammet, P.; Devaux, Y.; Azuaje, F.; Werer, C.; Lorang, C.; Gilson, G.; Max, M. Assessment of procalcitonin to predict outcome in hypothermia-treated patients after cardiac arrest. Crit. Care Res. Pract. 2011, 2011, 631062. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, H.; Kaneko, T.; Kasaoka, S.; Oshima, C.; Miyauchi, T.; Fujita, M.; Oda, Y.; Tsuruta, R.; Maekawa, T. Comparison of the predictability of neurological outcome by serum procalcitonin and glial fibrillary acidic protein in postcardiac-arrest patients. Neurocrit. Care 2010, 12, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Assicot, M.; Gendrel, D.; Carsin, H.; Raymond, J.; Guilbaud, J.; Bohuon, C. High serum procalcitonin concentrations in patients with sepsis and infection. Lancet 1993, 341, 515–518. [Google Scholar] [CrossRef]
- Wiberg, S.; Hassager, C.; Stammet, P.; Winther-Jensen, M.; Thomsen, J.H.; Erlinge, D.; Wanscher, M.; Nielsen, N.; Pellis, T.; Aneman, A.; et al. Single versus Serial Measurements of Neuron-Specific Enolase and Prediction of Poor Neurological Outcome in Persistently Unconscious Patients after Out-Of-Hospital Cardiac Arrest—A TTM-Trial Substudy. PLoS ONE 2017, 12, e0168894. [Google Scholar] [CrossRef] [PubMed]
- Sandroni, C.; Cariou, A.; Cavallaro, F.; Cronberg, T.; Friberg, H.; Hoedemaekers, C.; Horn, J.; Nolan, J.P.; Rossetti, A.O.; Soar, J. Prognostication in comatose survivors of cardiac arrest: An advisory statement from the European Resuscitation Council and the European Society of Intensive Care Medicine. Resuscitation 2014, 85, 1779–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung-Esaki, H.M.; Mui, G.; Mlynash, M.; Eyngorn, I.; Catabay, K.; Hirsch, K.G. The neuron specific enolase (NSE) ratio offers benefits over absolute value thresholds in post-cardiac arrest coma prognosis. J. Clin. Neurosci. 2018, 57, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Stammet, P.; Wagner, D.R.; Gilson, G.; Devaux, Y. Modeling serum level of s100beta and bispectral index to predict outcome after cardiac arrest. J. Am. Coll. Cardiol. 2013, 62, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Stammet, P.; Dankiewicz, J.; Nielsen, N.; Fays, F.; Collignon, O.; Hassager, C.; Wanscher, M.; Unden, J.; Wetterslev, J.; Pellis, T.; et al. Protein S100 as outcome predictor after out-of-hospital cardiac arrest and targeted temperature management at 33 °C and 36 °C. Crit. Care 2017, 21, 153. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, H.; Johnsson, P.; Hoglund, P.; Alling, C.; Blomquist, S. Elimination of S100B and renal function after cardiac surgery. J. Cardiothorac. Vasc. Anesth. 2000, 14, 698–701. [Google Scholar] [CrossRef] [PubMed]
- Bloomfield, S.M.; McKinney, J.; Smith, L.; Brisman, J. Reliability of S100B in predicting severity of central nervous system injury. Neurocrit. Care 2007, 6, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Bro-Jeppesen, J.; Kjaergaard, J.; Stammet, P.; Wise, M.P.; Hovdenes, J.; Aneman, A.; Horn, J.; Devaux, Y.; Erlinge, D.; Gasche, Y.; et al. Predictive value of interleukin-6 in post-cardiac arrest patients treated with targeted temperature management at 33 °C or 36 °C. Resuscitation 2016, 98, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dankiewicz, J.; Nielsen, N.; Linder, A.; Kuiper, M.; Wise, M.P.; Cronberg, T.; Erlinge, D.; Gasche, Y.; Harmon, M.B.; Hassager, C.; et al. Infectious complications after out-of-hospital cardiac arrest-A comparison between two target temperatures. Resuscitation 2017, 113, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Frydland, M.; Kjaergaard, J.; Erlinge, D.; Stammet, P.; Nielsen, N.; Wanscher, M.; Pellis, T.; Friberg, H.; Hovdenes, J.; Horn, J.; et al. Usefulness of Serum B-Type Natriuretic Peptide Levels in Comatose Patients Resuscitated from Out-of-Hospital Cardiac Arrest to Predict Outcome. Am. J. Cardiol. 2016, 118, 998–1005. [Google Scholar] [CrossRef] [PubMed]
- Gilje, P.; Koul, S.; Thomsen, J.H.; Devaux, Y.; Friberg, H.; Kuiper, M.; Horn, J.; Nielsen, N.; Pellis, T.; Stammet, P.; et al. High-sensitivity troponin-T as a prognostic marker after out-of-hospital cardiac arrest—A targeted temperature management (TTM) trial substudy. Resuscitation 2016, 107, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Frydland, M.; Kjaergaard, J.; Erlinge, D.; Wanscher, M.; Nielsen, N.; Pellis, T.; Aneman, A.; Friberg, H.; Hovdenes, J.; Horn, J.; et al. Target temperature management of 33 °C and 36 °C in patients with out-of-hospital cardiac arrest with initial non-shockable rhythm—A TTM sub-study. Resuscitation 2015, 89, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Hasslacher, J.; Lehner, G.F.; Harler, U.; Beer, R.; Ulmer, H.; Kirchmair, R.; Fischer-Colbrie, R.; Bellmann, R.; Dunzendorfer, S.; Joannidis, M. Secretoneurin as a marker for hypoxic brain injury after cardiopulmonary resuscitation. Intens. Care Med. 2014, 40, 1518–1527. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Goretti, E.; Wagner, D.R.; Devaux, Y. miRNAs as biomarkers of myocardial infarction: A step forward towards personalized medicine? Trends Mol. Med. 2014, 20, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Stammet, P.; Goretti, E.; Vausort, M.; Zhang, L.; Wagner, D.R.; Devaux, Y. Circulating microRNAs after cardiac arrest. Crit. Care Med. 2012, 40, 3209–3214. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Li, Y.; Chen, X.; Yang, S.; Zhang, J. Microarray based analysis of microRNA expression in rat cerebral cortex after traumatic brain injury. Brain Res. 2009, 1284, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Andersson, P.; Gidlof, O.; Braun, O.O.; Gotberg, M.; van der Pals, J.; Olde, B.; Erlinge, D. Plasma levels of liver-specific miR-122 is massively increased in a porcine cardiogenic shock model and attenuated by hypothermia. Shock 2012, 37, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Wander, P.L.; Enquobahrie, D.A.; Pritchard, C.C.; McKnight, B.; Rice, K.; Christiansen, M.; Lemaitre, R.N.; Rea, T.; Siscovick, D.; Sotoodehnia, N. Circulating microRNAs and sudden cardiac arrest outcomes. Resuscitation 2016, 106, 96–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braza-Boils, A.; Mari-Alexandre, J.; Molina, P.; Arnau, M.A.; Barcelo-Molina, M.; Domingo, D.; Girbes, J.; Giner, J.; Martinez-Dolz, L.; Zorio, E. Deregulated hepatic microRNAs underlie the association between non-alcoholic fatty liver disease and coronary artery disease. Liver Int. 2016, 36, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Gilje, P.; Gidlof, O.; Rundgren, M.; Cronberg, T.; Al-Mashat, M.; Olde, B.; Friberg, H.; Erlinge, D. The brain-enriched microRNA miR-124 in plasma predicts neurological outcome after cardiac arrest. Crit. Care 2014, 18, R40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laterza, O.F.; Lim, L.; Garrett-Engele, P.W.; Vlasakova, K.; Muniappa, N.; Tanaka, W.K.; Johnson, J.M.; Sina, J.F.; Fare, T.L.; Sistare, F.D.; et al. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin. Chem. 2009, 55, 1977–1983. [Google Scholar] [CrossRef] [PubMed]
- Patz, S.; Trattnig, C.; Grunbacher, G.; Ebner, B.; Gully, C.; Novak, A.; Rinner, B.; Leitinger, G.; Absenger, M.; Tomescu, O.A.; et al. More than cell dust: Microparticles isolated from cerebrospinal fluid of brain injured patients are messengers carrying mRNAs, miRNAs, and proteins. J. Neurotrauma 2013, 30, 1232–1242. [Google Scholar] [CrossRef] [PubMed]
- Weng, H.; Shen, C.; Hirokawa, G.; Ji, X.; Takahashi, R.; Shimada, K.; Kishimoto, C.; Iwai, N. Plasma miR-124 as a biomarker for cerebral infarction. Biomed. Res. 2011, 32, 135–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rink, C.; Khanna, S. MicroRNA in ischemic stroke etiology and pathology. Physiol. Genom. 2011, 43, 521–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devaux, Y.; Dankiewicz, J.; Salgado-Somoza, A.; Stammet, P.; Collignon, O.; Gilje, P.; Gidlof, O.; Zhang, L.; Vausort, M.; Hassager, C.; et al. Association of Circulating MicroRNA-124-3p Levels With Outcomes After Out-of-Hospital Cardiac Arrest: A Substudy of a Randomized Clinical Trial. JAMA Cardiol. 2016, 1, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Devaux, Y.; Salgado-Somoza, A.; Dankiewicz, J.; Boileau, A.; Stammet, P.; Schritz, A.; Zhang, L.; Vausort, M.; Gilje, P.; Erlinge, D.; et al. Incremental Value of Circulating MiR-122-5p to Predict Outcome after Out of Hospital Cardiac Arrest. Theranostics 2017, 7, 2555–2564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Cao, F.; Ran, Q.; Wang, F. Long non-coding RNA Gm4419 promotes trauma-induced astrocyte apoptosis by targeting tumor necrosis factor alpha. Biochem. Biophys. Res. Commun. 2017, 491, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Volders, P.J.; Verheggen, K.; Menschaert, G.; Vandepoele, K.; Martens, L.; Vandesompele, J.; Mestdagh, P. An update on LNCipedia: A database for annotated human lncRNA sequences. Nucleic Acids Res. 2015, 43, D174–D180. [Google Scholar] [CrossRef] [PubMed]
- Devaux, Y.; Zangrando, J.; Schroen, B.; Creemers, E.E.; Pedrazzini, T.; Chang, C.P.; Dorn, G.W., 2nd; Thum, T.; Heymans, S.; Cardiolinc, N. Long noncoding RNAs in cardiac development and ageing. Nat. Rev. Cardiol. 2015, 12, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, C.P.C.; Spencer, H.; Ford, K.L.; Michel, L.Y.M.; Baker, A.H.; Emanueli, C.; Balligand, J.L.; Devaux, Y.; Cardiolinc, N. The Function and Therapeutic Potential of Long Non-coding RNAs in Cardiovascular Development and Disease. Mol. Therapy Nucleic Acids 2017, 8, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Salgado Somoza, A.; Devaux, Y.; Martelli, F. Long Noncoding RNAs and Cardiac Disease. Antioxid. Redox Signal. 2017. [Google Scholar] [CrossRef] [PubMed]
- Vausort, M.; Wagner, D.R.; Devaux, Y. Long noncoding RNAs in patients with acute myocardial infarction. Circul. Res. 2014, 115, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, J. LncRNAs: Macromolecules with big roles in neurobiology and neurological diseases. Metab. Brain Dis. 2017, 32, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Q.; Zhang, K.S.; Hu, B.; Niu, X.; Zhou, S.M.; Li, S.G.; Luo, Y.P.; Wang, Y.; Deng, Z.F. Downregulation of the Long Non-Coding RNA Meg3 Promotes Angiogenesis After Ischemic Brain Injury by Activating Notch Signaling. Mol. Neurobiol. 2017, 54, 8179–8190. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.L.; Kim, T.; Vemuganti, R. Long Noncoding RNA FosDT Promotes Ischemic Brain Injury by Interacting with REST-Associated Chromatin-Modifying Proteins. J. Neurosci. 2015, 35, 16443–16449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Deng, F.; Xing, Z.; Wu, Z.; Cen, B.; Xu, S.; Zhao, Z.; Nepomuceno, R.; Bhuiyan, M.I.; Sun, D.; et al. Long non-coding RNA C2dat1 regulates CaMKIIdelta expression to promote neuronal survival through the NFκB signaling pathway following cerebral ischemia. Cell Death Dis. 2016, 7, e2173. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Yu, Y.; Fu, X. LncRNA Gm4419 contributes to OGD/R injury of cerebral microglial cells via IκB phosphorylation and NFκB activation. Biochem. Biophys. Res. Commun. 2017, 487, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Liao, X.; Li, X.; Wei, H.; Liang, Q.; Zhang, Z.; Yin, M.; Zeng, X.; Liang, Z.; Hu, C. Expression profiles of long noncoding RNAs and mRNAs in post-cardiac arrest rat brains. Mol. Med. Rep. 2018, 17, 6413–6424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, J.; Jiang, L.; Huang, Z.; Zhang, H.; Cheng, C.; Liu, H.; He, J.; Wu, J.; Darwazeh, R.; Wu, Y.; et al. The long non-coding RNA Neat1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain Behav. Immun. 2017, 65, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Jiang, L.; Cheng, C.; Huang, Z.; Zhang, H.; Liu, H.; He, J.; Cao, F.; Peng, J.; Jiang, Y.; et al. Altered expression of long non-coding RNA and mRNA in mouse cortex after traumatic brain injury. Brain Res. 2016, 1646, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.F.; Zhao, C.C.; Weng, W.J.; Lei, J.; Lin, Y.; Mao, Q.; Gao, G.Y.; Feng, J.F.; Jiang, J.Y. Alteration in Long Non-Coding RNA Expression after Traumatic Brain Injury in Rats. J. Neurotrauma 2017, 34, 2100–2108. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Liu, H.; Wang, R.; Xu, S.; Liu, Y.; Wang, K.; Hou, X.; Shen, C.; Wu, J.; Chen, X.; et al. Expression signatures of long non-coding RNAs in early brain injury following experimental subarachnoid hemorrhage. Mol. Med. Rep. 2015, 12, 967–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, J.; Wu, Y.; Tian, X.; Pang, J.; Kuai, L.; Cao, F.; Qin, X.; Zhong, J.; Li, X.; Li, Y.; et al. High-Throughput Sequencing and Co-Expression Network Analysis of lncRNAs and mRNAs in Early Brain Injury Following Experimental Subarachnoid Haemorrhage. Sci. Rep. 2017, 7, 46577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Memczak, S.; Papavasileiou, P.; Peters, O.; Rajewsky, N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS ONE 2015, 10, e0141214. [Google Scholar] [CrossRef] [PubMed]
- Devaux, Y.; Creemers, E.E.; Boon, R.A.; Werfel, S.; Thum, T.; Engelhardt, S.; Dimmeler, S.; Squire, I.; Cardiolinc, n. Circular RNAs in heart failure. Eur. J. Heart Fail. 2017, 19, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.Q.; Malhotra, A.; Mayeda, A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006, 34, e63. [Google Scholar] [CrossRef] [PubMed]
- Vausort, M.; Salgado-Somoza, A.; Zhang, L.; Leszek, P.; Scholz, M.; Teren, A.; Burkhardt, R.; Thiery, J.; Wagner, D.R.; Devaux, Y. Myocardial Infarction-Associated Circular RNA Predicting Left Ventricular Dysfunction. J. Am. Coll. Cardiol. 2016, 68, 1247–1248. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, C.; Yang, J.; Geng, X.; Du, H.; Ji, X.; Zhao, H. Screening circular RNA expression patterns following focal cerebral ischemia in mice. Oncotarget 2017, 8, 86535–86547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, S.L.; Pandi, G.; Vemuganti, R. Circular RNA Expression Profiles Alter Significantly in Mouse Brain After Transient Focal Ischemia. Stroke 2017, 48, 2541–2548. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Wang, Y.; Lin, Y.; Zhao, C.C.; Mao, Q.; Feng, J.F.; Cao, J.; Gao, G.Y.; Jiang, J. Circular RNA Expression Profiles Alter Significantly After Traumatic Brain Injury in Rats. J. Neurotrauma 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Zhou, J.; Dong, X.; Bi, C.; Jiang, R.; Dong, J.; Tian, Y.; Yuan, H.; Zhang, J.N. Circular RNA expression alteration in exosomes from the brain extracellular space after traumatic brain injury in mice. J. Neurotrauma 2018. [Google Scholar] [CrossRef] [PubMed]
| ID | Specie | Disease | Experimental Model | Observation | Ref. |
|---|---|---|---|---|---|
| miR-21 | Human | OHCA | --- | Elevated plasma levels in patients with poor neurological outcome | [54] |
| Rat | TBI | Fluid percussion injury | Elevated serum levels in rats with poor outcome | [55] | |
| miR-34a | Human | SCD | Coronary artery and non-alcoholic fatty liver disease | Higher hepatic levels in coronary artery disease-related SCD | [58] |
| miR-122 | Human | OHCA | --- | Elevated serum levels in patients with poor neurological outcome | [54,65] |
| SCA | Ventricular tachycardia-derived cardiac arrest | Elevated in plasma from patients compared to controls. Elevated in successfully resuscitated or discharged alive only versus patients died in the field. | [57] | ||
| SCD | Coronary artery and non-alcoholic fatty liver disease | Lower hepatic levels in coronary artery disease-related SCD | [58] | ||
| Pig | Cardiogenic shock | LAD artery occlusion | Elevated plasma levels after injury. Attenuation by hypothermia. | [56] | |
| miR-124 | Human | OHCA | --- | Elevated serum and plasma levels in patients with poor neurological outcome | [59,64] |
| Rat | Ischemic brain damage | MCAO | Plasma biomarker of ischemic brain damage | [60,62] | |
| miR-466l-3p | Mouse | Mechanical injury | Primary astrocytes | Inhibited Tnfα expression after stretch injury and interacted with lncRNA Gm4419 | [66] |
| Other | Human | SCA | Ventricular tachycardia-derived cardiac arrest | Expression levels of plasmatic miRs were higher (n = 17) or lower (n = 3) in CA patients compared to controls. Mir-122 and miR-205 were elevated in patients successfully resuscitated versus death in the field. Lower levels of cardiac enriched microRNAs were observed in patients discharged alive versus the ones who died in the field. | [57] |
| ID | Specie | Disease | Experimental Model | Observation | Ref. |
|---|---|---|---|---|---|
| C2dat1 | Mouse | Ischemic brain damage | MCAO | Upregulated after transient focal ischemia | [78] |
| FosDT | Rat | Ischemic brain damage | MCAO in spontaneous hypertensive rats | Increased after ischemic brain injury. Potentially regulated brain damage by association with key elements of the Rest complex, upstream of NFκB | [77] |
| Gm4419 | Rat | Ischemia | OGD/R in primary microglial cells | Controled inflammatory response through NFκB signaling pathway | [79] |
| Mouse | Mechanical injury | Primary astrocytes | Induced after stretch injury. Upregulates Tnfα expression by sponging miR-466l-3p | [66] | |
| Meg3 | Human | --- | HMEC-1 | Downregulation of MEG3 increased angiogenesis | [76] |
| Rat | Ischemic brain damage | MCAO | Downregulated after ischemic stroke. Silencing of Meg3 improved neurological outcome. | [76] | |
| Neat1 | Mouse | TBI | Controlled cortical impact | Upregulated after injury. Absence of Neat1 increases apoptosis around the impacted area. | [81,82] |
| Mouse | Ischemia | OGD in primary new-born neurons, HT22, and BV2 lines | Upregulated under bexatorene treatment or OGD. Promoted axonal extension in primary neurons. Anti-inflammatory effect via Pidd1. | [81] | |
| Other | Rat | CA-ROSC | Electrically-induced ventricular tachycardia followed by manual chest compression | Dysregulation of 58 lncRNAs and 258 mRNAs in brain cortex of rats. | [80] |
| TBI | Fluid percussion injury | Upregulation of 271 lncRNAs in the hippocampus assessed by microarray, including 4 lncRNAs validated by PCR (Zfas1, Bsr, Gas5, and Snhg6) | [83] | ||
| Stroke | Subarachnoid haemorrhage | Microarray analysis showed 64 upregulated and 144 downregulated lncRNAs between control and haemorrhagic animals. | [84] | ||
| Mouse | TBI | Controlled cortical impact | Alteration of the expression levels of 823 lncRNAs assessed by RNA-Seq 24 h after injury. | [82] | |
| Stroke | Subarachnoid haemorrhage | RNA-Seq analysis identified 103 upregulated and 514 downregulated lncRNAs between injured and control mice. | [85] |
| Specie | Disease | Model | Observation | Ref. |
|---|---|---|---|---|
| Mouse | Ischemic brain damage | MCAO | Microarray analysis after RNAse R treatment | [90] |
| Microarray analysis after RNAse R treatment | [91] | |||
| TBI | Fluid percussion injury | RNA sequencing analysis from brain exosomes. | [93] | |
| Rat | TBI | Fluid percussion injury | Microarray analysis using RNA from ipsilateral hippocampus after RNAse R treatment | [92] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salgado-Somoza, A.; Stefanizzi, F.M.; Stammet, P.; Erlinge, D.; Friberg, H.; Nielsen, N.; Devaux, Y. Non-Coding RNAs to Aid in Neurological Prognosis after Cardiac Arrest. Non-Coding RNA 2018, 4, 42. https://doi.org/10.3390/ncrna4040042
Salgado-Somoza A, Stefanizzi FM, Stammet P, Erlinge D, Friberg H, Nielsen N, Devaux Y. Non-Coding RNAs to Aid in Neurological Prognosis after Cardiac Arrest. Non-Coding RNA. 2018; 4(4):42. https://doi.org/10.3390/ncrna4040042
Chicago/Turabian StyleSalgado-Somoza, Antonio, Francesca Maria Stefanizzi, Pascal Stammet, David Erlinge, Hans Friberg, Niklas Nielsen, and Yvan Devaux. 2018. "Non-Coding RNAs to Aid in Neurological Prognosis after Cardiac Arrest" Non-Coding RNA 4, no. 4: 42. https://doi.org/10.3390/ncrna4040042