Effects of Flow Rate on Mesenchymal Stem Cell Oxygen Consumption Rates in 3D Bone-Tissue-Engineered Constructs Cultured in Perfusion Bioreactor Systems
Abstract
1. Introduction
2. Methods
2.1. Cell Culture
2.2. Scaffold Production and Preparation
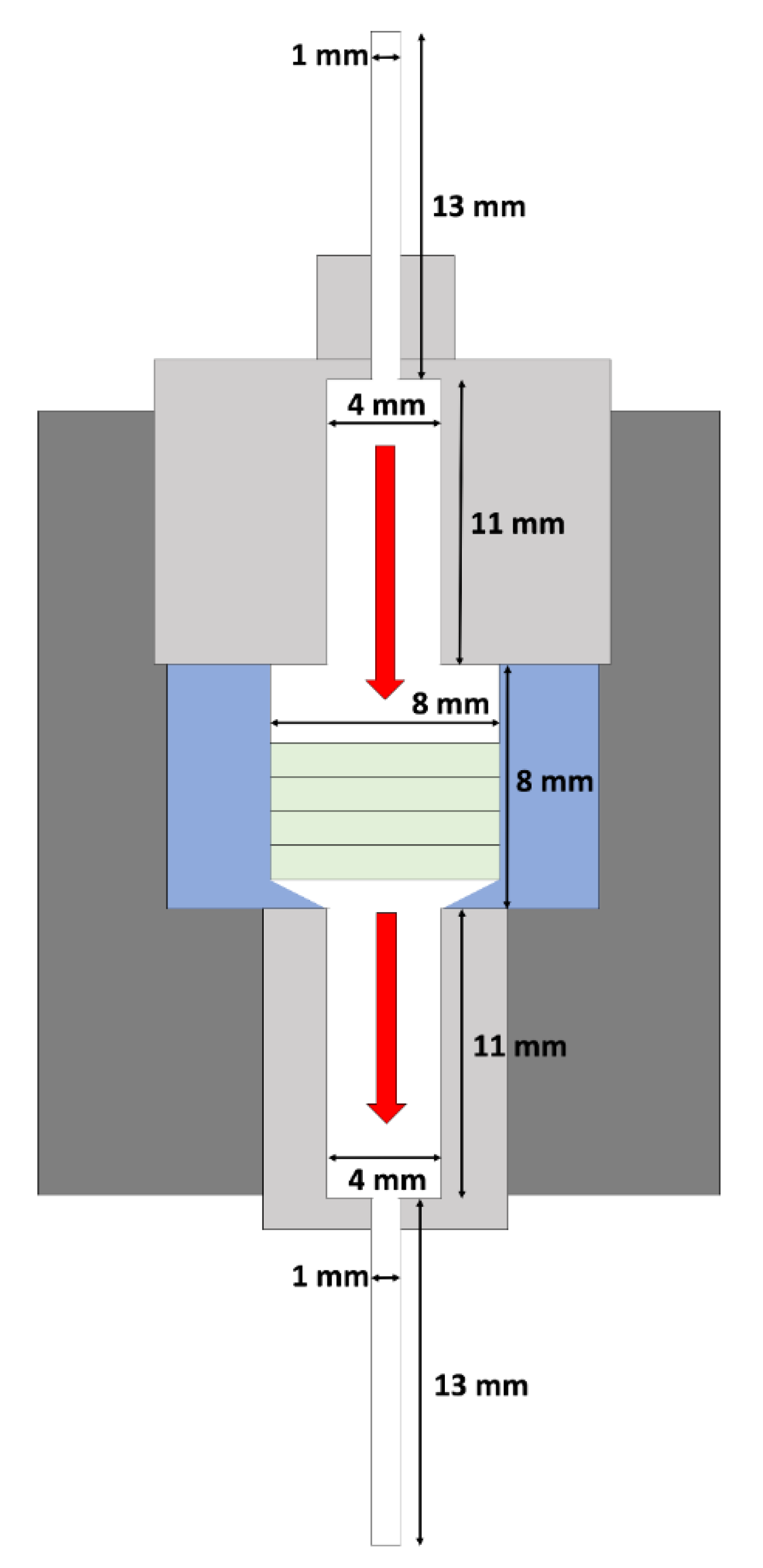
2.3. Bioreactor Setup and Seeding
2.4. Oxygen Data Collection
2.5. Cellularity Quantification
2.6. Residence Time Analysis
2.7. Statistical Analysis
3. Results and Discussion
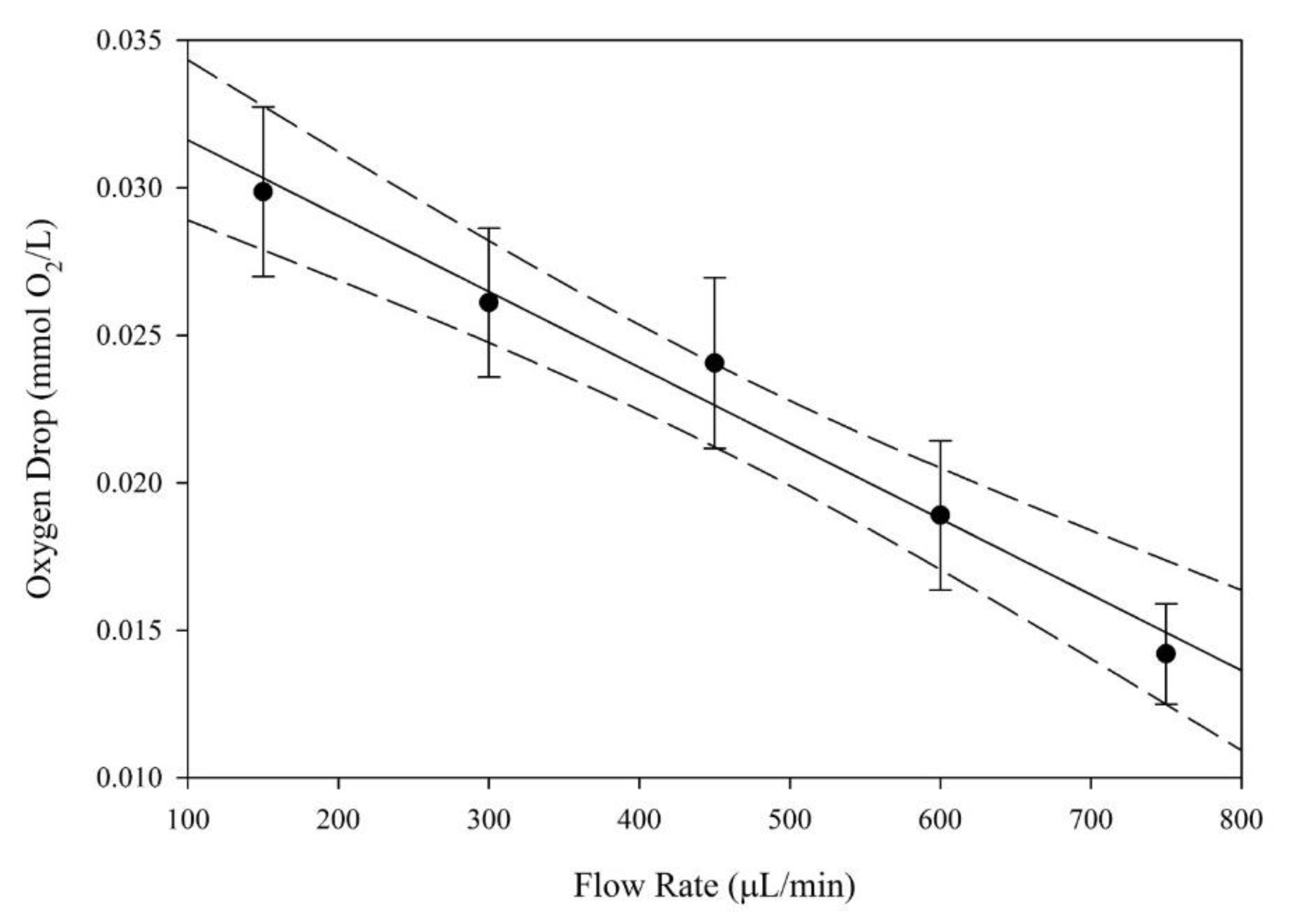
3.1. Oxygen Concentration Drop
3.2. Cellularity Quantification
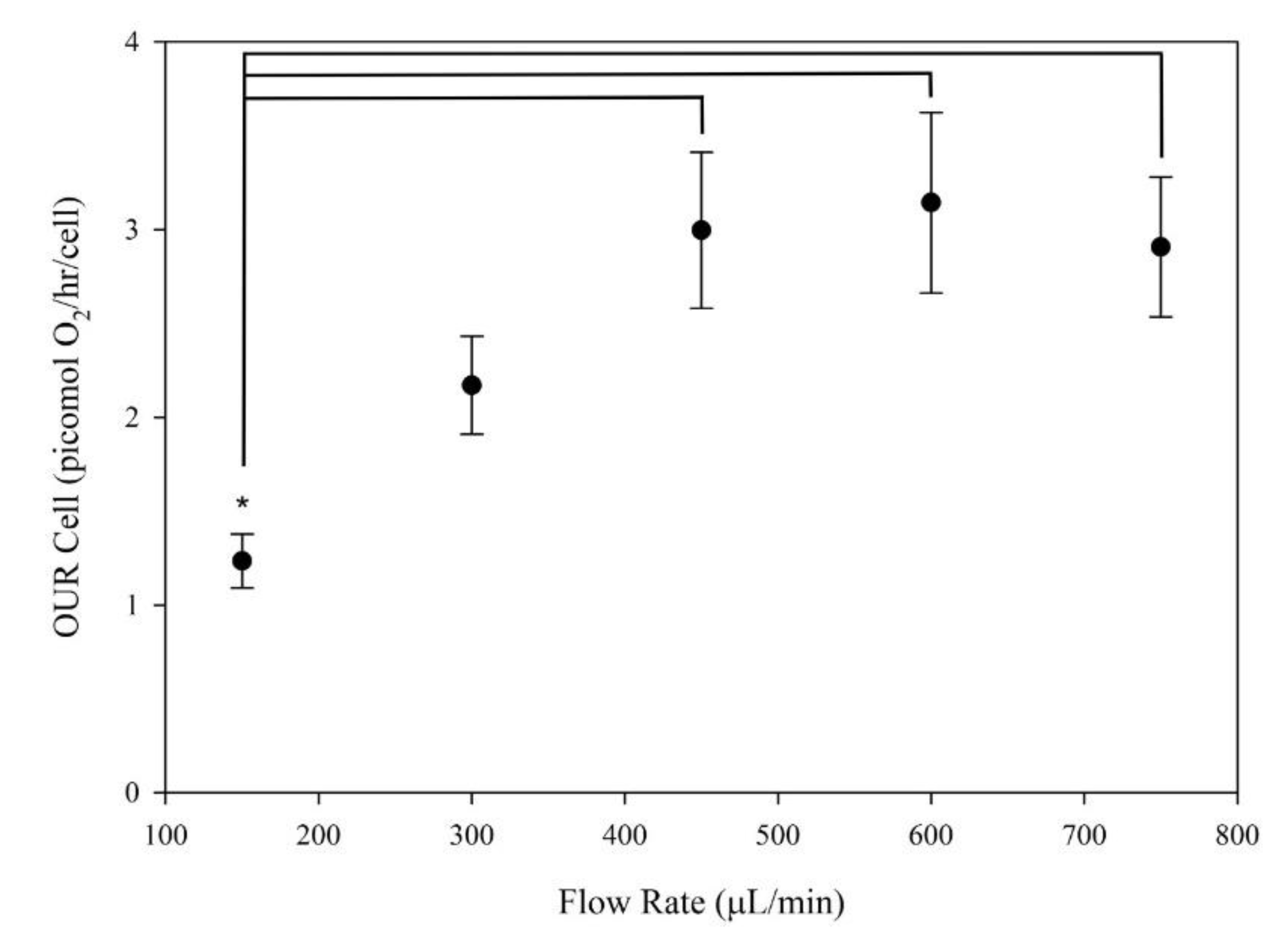
3.3. Oxygen Uptake Rate (OUR)
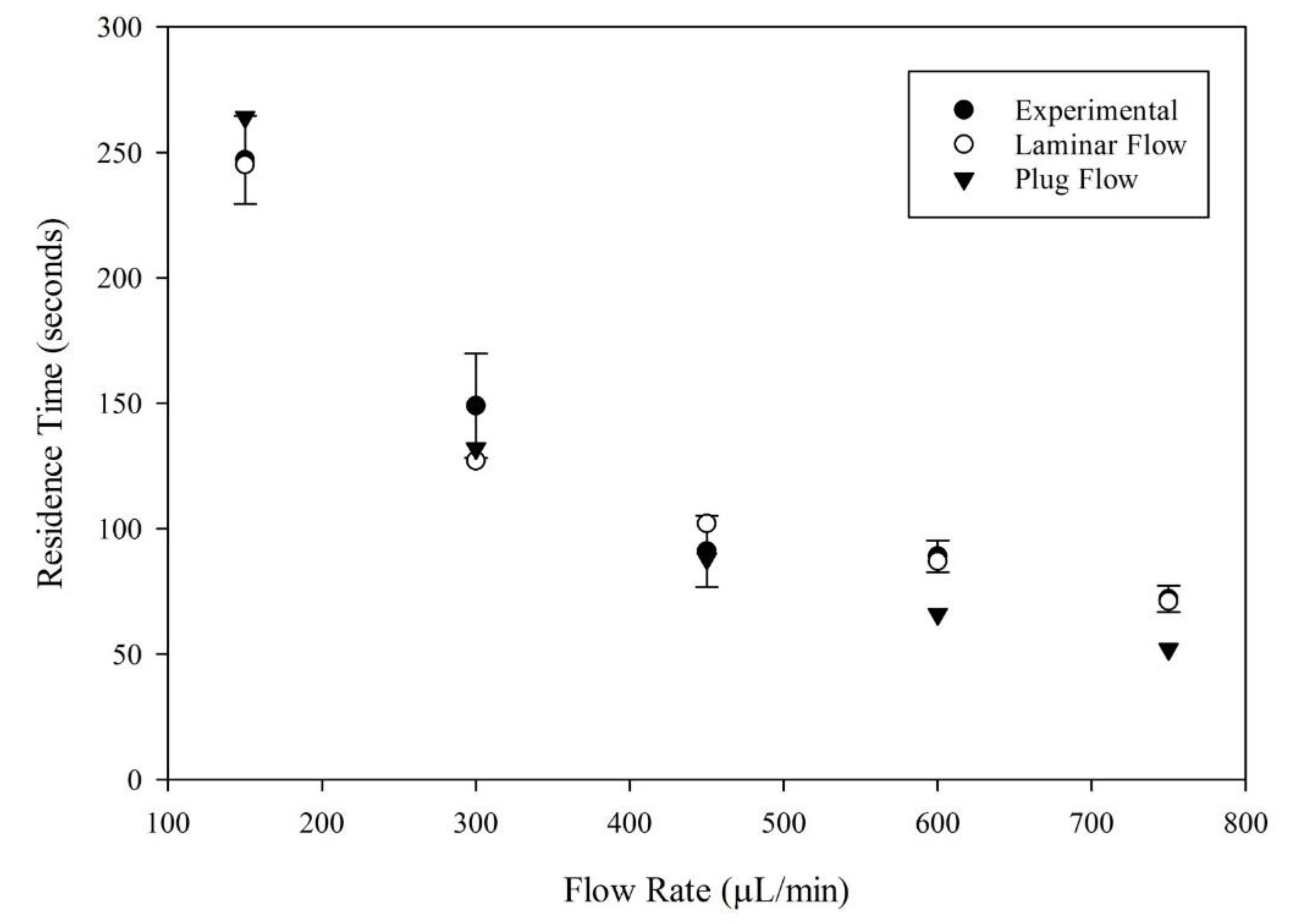
3.4. Residence Time Distribution Analysis
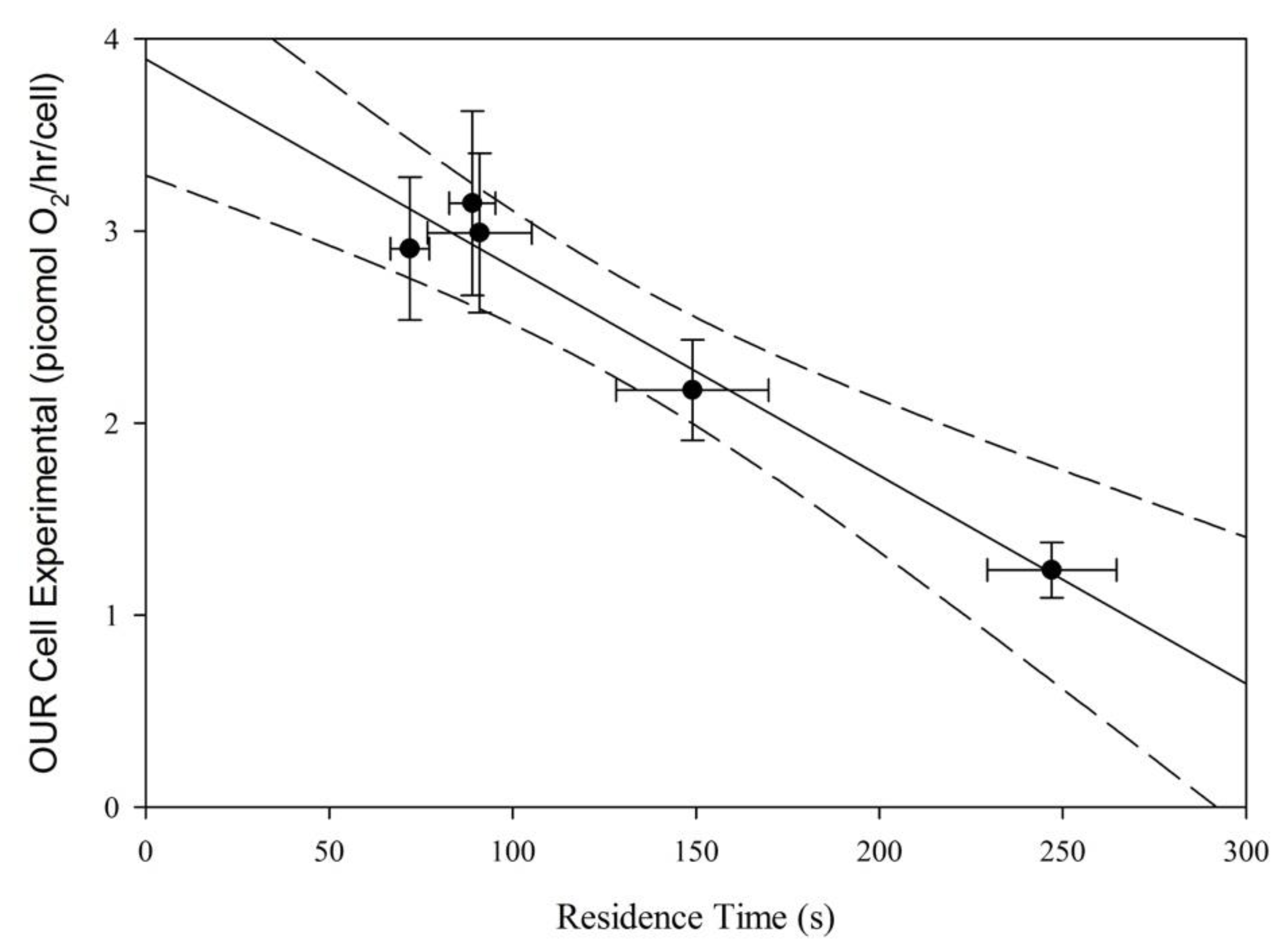
3.5. Oxygen Uptake Rate Relative to Residence Time
3.6. Analysis of Flow Profile in the Bioreactor System
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Simmons, A.D.; Sikavitsas, V.I. Monitoring Bone Tissue Engineered (BTE) Constructs Based on the Shifting Metabolism of Differentiating Stem Cells. Ann. Biomed. Eng. 2018, 46, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Janssen, F.W.; Hofland, I.; van Oorschot, A.; Oostra, J.; Peters, H.; van Blitterswijk, C.A. Online measurement of oxygen consumption by goat bone marrow stromal cells in a combined cell-seeding and proliferation perfusion bioreactor. J. Biomed. Mater. Res. A 2006, 79, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Starly, B. Real time in vitro measurement of oxygen uptake rates for HEPG2 liver cells encapsulated in alginate matrices. Microfluid. Nanofluidics 2009, 6, 373–381. [Google Scholar] [CrossRef]
- Super, A.; Jaccard, N.; Cardoso Marques, M.P.; Macown, R.J.; Griffin, L.D.; Veraitch, F.S.; Szita, N. Real-time monitoring of specific oxygen uptake rates of embryonic stem cells in a microfluidic cell culture device. Biotechnol. J. 2016, 11, 1179–1189. [Google Scholar] [CrossRef]
- Simmons, A.D.; Williams, C., 3rd; Degoix, A.; Sikavitsas, V.I. Sensing metabolites for the monitoring of tissue engineered construct cellularity in perfusion bioreactors. Biosens. Bioelectron. 2017, 90, 443–449. [Google Scholar] [CrossRef]
- Kellner, K.; Liebsch, G.; Klimant, I.; Wolfbeis, O.S.; Blunk, T.; Schulz, M.B.; Gopferich, A. Determination of oxygen gradients in engineered tissue using a fluorescent sensor. Biotechnol. Bioeng. 2002, 80, 73–83. [Google Scholar] [CrossRef]
- Santoro, R.; Krause, C.; Martin, I.; Wendt, D. On-line monitoring of oxygen as a non-destructive method to quantify cells in engineered 3D tissue constructs. J. Tissue Eng. Regen Med. 2012, 6, 696–701. [Google Scholar] [CrossRef]
- Curcio, E.; Piscioneri, A.; Morelli, S.; Salerno, S.; Macchiarini, P.; De Bartolo, L. Kinetics of oxygen uptake by cells potentially used in a tissue engineered trachea. Biomaterials 2014, 35, 6829–6837. [Google Scholar] [CrossRef]
- Guarino, R.D.; Dike, L.E.; Haq, T.A.; Rowley, J.A.; Pitner, J.B.; Timmins, M.R. Method for determining oxygen consumption rates of static cultures from microplate measurements of pericellular dissolved oxygen concentration. Biotechnol. Bioeng. 2004, 86, 775–787. [Google Scholar] [CrossRef]
- Lewis, M.C.; Macarthur, B.D.; Malda, J.; Pettet, G.; Please, C.P. Heterogeneous proliferation within engineered cartilaginous tissue: The role of oxygen tension. Biotechnol. Bioeng. 2005, 91, 607–615. [Google Scholar] [CrossRef]
- Bancroft, G.N.; Sikavitsas, I.; Mikos, A.G. Design of a flow perfusion bioreactor system for bone tissue-engineering applications. Tissue Eng. 2003, 9, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Kasper, F.K.; Liao, J.; Kretlow, J.D.; Sikavitsas, V.I.; Mikos, A.G. Flow perfusion culture of mesenchymal stem cells for bone tissue engineering. In StemBook; Harvard Stem Cell Institute: Cambridge, MA, USA, 2008. [Google Scholar]
- Gharravi, A.M.; Orazizadeh, M.; Hashemitabar, M. Fluid-induced low shear stress improves cartilage like tissue fabrication by encapsulating chondrocytes. Cell Tissue Bank. 2016, 17, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Tang, T.; Lu, J.; Dai, K. Effects of flow shear stress and mass transport on the construction of a large-scale tissue-engineered bone in a perfusion bioreactor. Tissue Eng. Part. A 2009, 15, 2773–2783. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, A.S.; Juarez, T.M.; Helmke, C.D.; Gustin, M.C.; Mikos, A.G. Effect of convection on osteoblastic cell growth and function in biodegradable polymer foam scaffolds. Biomaterials 2001, 22, 1279–1288. [Google Scholar] [CrossRef]
- Mauney, J.R.; Sjostorm, S.; Blumberg, J.; Horan, R.; O’Leary, J.P.; Vunjak-Novakovic, G.; Volloch, V.; Kaplan, D.L. Mechanical stimulation promotes osteogenic differentiation of human bone marrow stromal cells on 3-D partially demineralized bone scaffolds in vitro. Calcif. Tissue Int. 2004, 74, 458–468. [Google Scholar] [CrossRef]
- Bacabac, R.G.; Smit, T.H.; Mullender, M.G.; Dijcks, S.J.; Van Loon, J.J.; Klein-Nulend, J. Nitric oxide production by bone cells is fluid shear stress rate dependent. Biochem. Biophys. Res. Commun. 2004, 315, 823–829. [Google Scholar] [CrossRef]
- VanGordon, S.B.; Voronov, R.S.; Blue, T.B.; Shambaugh, R.L.; Papavassiliou, D.V.; Sikavitsas, V.I. Effects of Scaffold Architecture on Preosteoblastic Cultures under Continuous Fluid Shear. Ind. Eng. Chem. Res. 2011, 50, 620–629. [Google Scholar] [CrossRef]
- Ocean Optics Oxygen Sensing. 2018. Available online: http://www.oceaninsight.com/products/systems/benchtop/sensors/ (accessed on 8 August 2018).
- Simmons, A.D. The Development of a Combined approach for the Real-Time, Non-Destructive Monitoring of In Vitro Bone Tissue Engineered Constructs Utilizing Physio-Metabolic Markers, in School of Chemical, Biological, and Materials Engineering; University of Oklahoma: Norman, OK, USA, 2016; p. 120. [Google Scholar]
- Coker, A.K.; Ludwig, E.E. Ludwig’s Applied Process Design for Chemical and Petrochemical Plants, 4th ed.; Gulf Professional Publishing: Houston, TX, USA, 2007. [Google Scholar]
- Fogler, H.S. Elements of Chemical Reaction Engineering, 5th ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2016. [Google Scholar]
- Portner, R.; Koop, M. A model for oxygen supply in fixed bed reactors with immobilized hybridoma cells. Bioprocess. Eng. 1997, 17, 269–275. [Google Scholar] [CrossRef]
- Kumar, S.; Vaidya, M. Hypoxia inhibits mesenchymal stem cell proliferation through HIF1alpha-dependent regulation of P27. Mol. Cell Biochem. 2016, 415, 29–38. [Google Scholar] [CrossRef]
- Pattappa, G.; Heywood, H.K.; de Bruijn, J.D.; Lee, D.A. The metabolism of human mesenchymal stem cells during proliferation and differentiation. J. Cell Physiol. 2011, 226, 2562–2570. [Google Scholar] [CrossRef]
- Schop, D.; van Dijkhuizen-Radersma, R.; Borgart, E.; Janssen, F.W.; Rozemuller, H.; Prins, H.J.; de Bruijn, J.D. Expansion of human mesenchymal stromal cells on microcarriers: Growth and metabolism. J. Tissue Eng. Regen Med. 2010, 4, 131–140. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Felder, M.L.; Simmons, A.D.; Shambaugh, R.L.; Sikavitsas, V.I. Effects of Flow Rate on Mesenchymal Stem Cell Oxygen Consumption Rates in 3D Bone-Tissue-Engineered Constructs Cultured in Perfusion Bioreactor Systems. Fluids 2020, 5, 30. https://doi.org/10.3390/fluids5010030
Felder ML, Simmons AD, Shambaugh RL, Sikavitsas VI. Effects of Flow Rate on Mesenchymal Stem Cell Oxygen Consumption Rates in 3D Bone-Tissue-Engineered Constructs Cultured in Perfusion Bioreactor Systems. Fluids. 2020; 5(1):30. https://doi.org/10.3390/fluids5010030
Chicago/Turabian StyleFelder, Michael L., Aaron D. Simmons, Robert L. Shambaugh, and Vassilios I. Sikavitsas. 2020. "Effects of Flow Rate on Mesenchymal Stem Cell Oxygen Consumption Rates in 3D Bone-Tissue-Engineered Constructs Cultured in Perfusion Bioreactor Systems" Fluids 5, no. 1: 30. https://doi.org/10.3390/fluids5010030
APA StyleFelder, M. L., Simmons, A. D., Shambaugh, R. L., & Sikavitsas, V. I. (2020). Effects of Flow Rate on Mesenchymal Stem Cell Oxygen Consumption Rates in 3D Bone-Tissue-Engineered Constructs Cultured in Perfusion Bioreactor Systems. Fluids, 5(1), 30. https://doi.org/10.3390/fluids5010030




