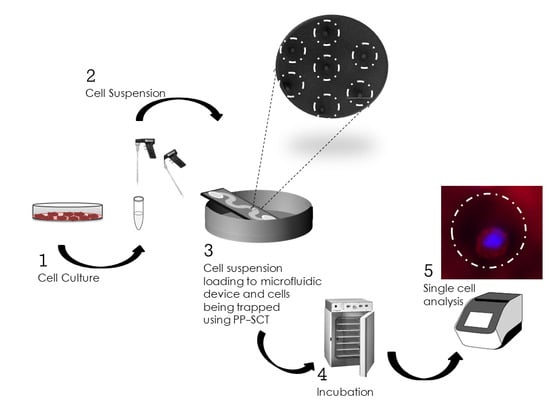
Pipette Petri Dish Single-Cell Trapping (PP-SCT) in Microfluidic Platforms: A Passive Hydrodynamic Technique
Abstract
:1. Introduction
1.1. Microfluidics
1.2. Hydrodynamic Trapping
2. Methods
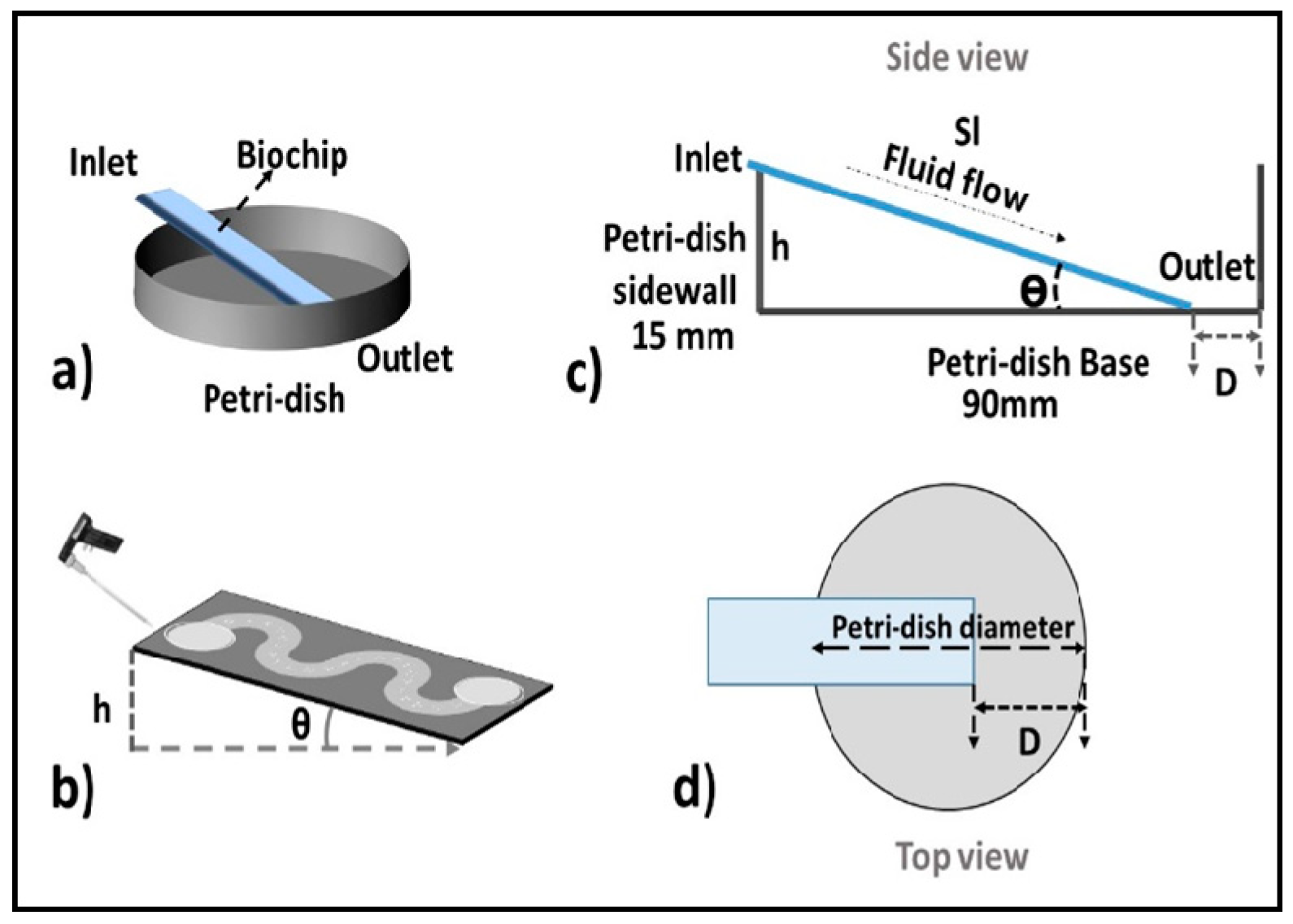
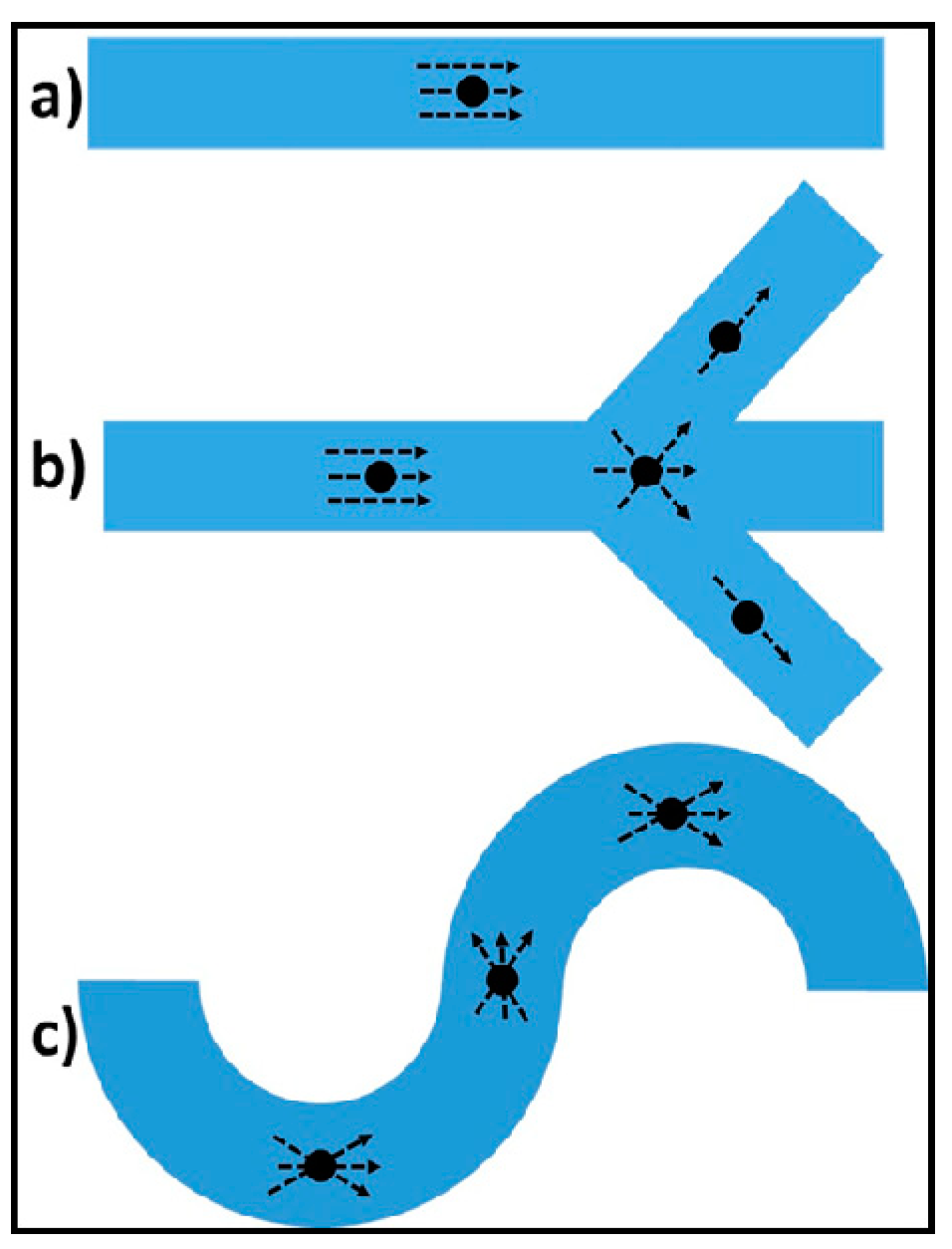
2.1. Concept and Design Criteria
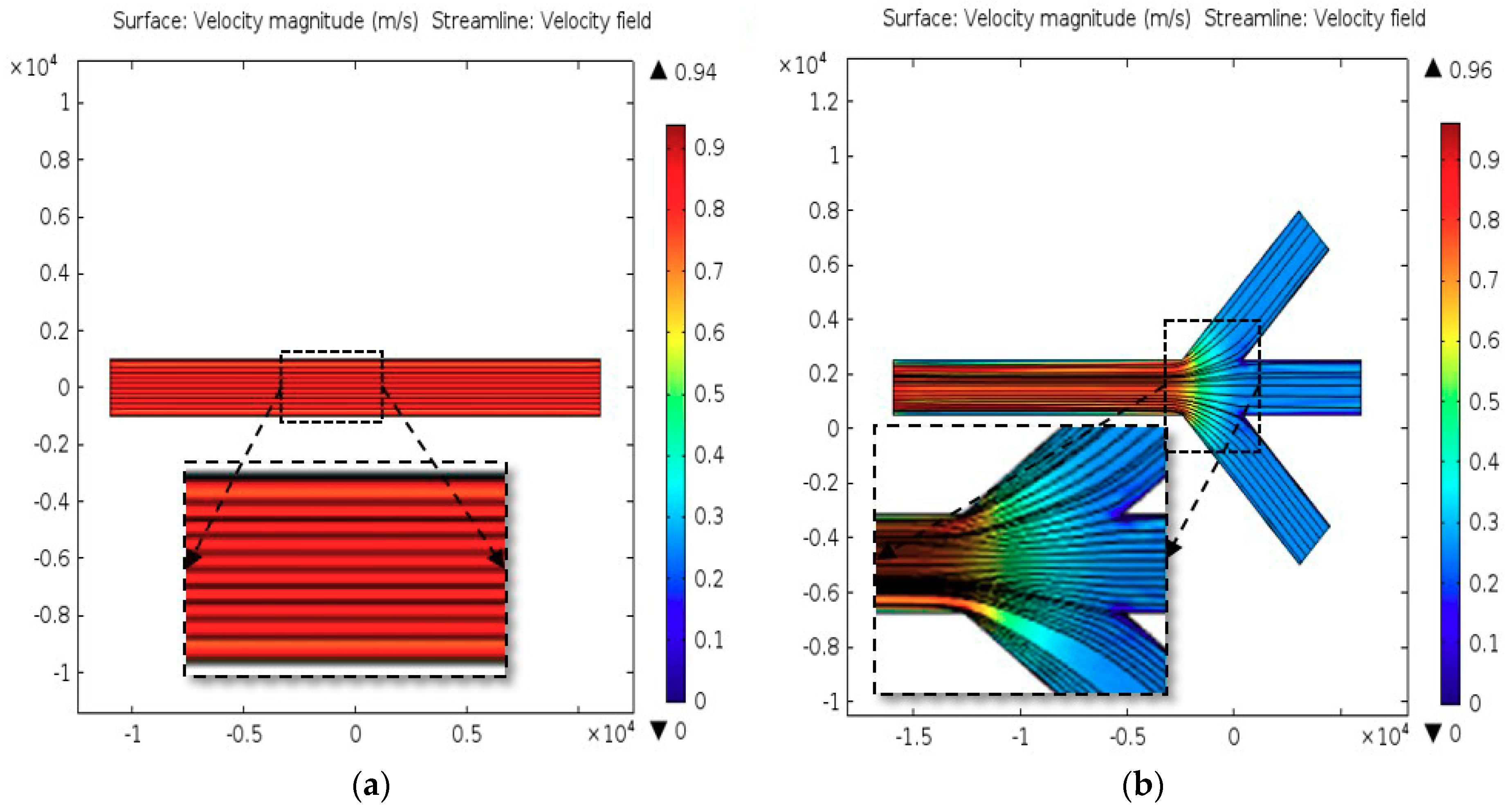
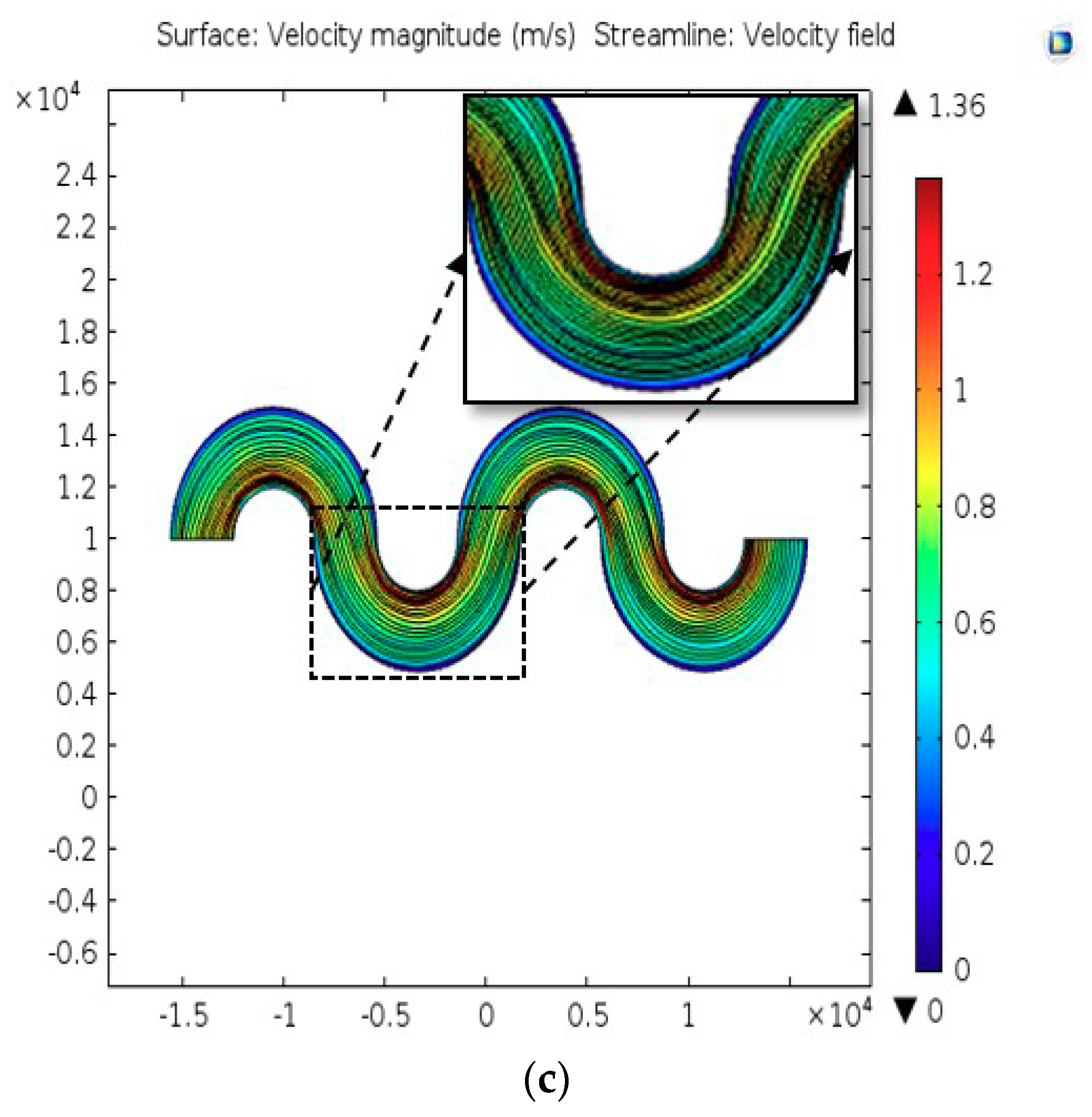
2.2. Computational Analysis
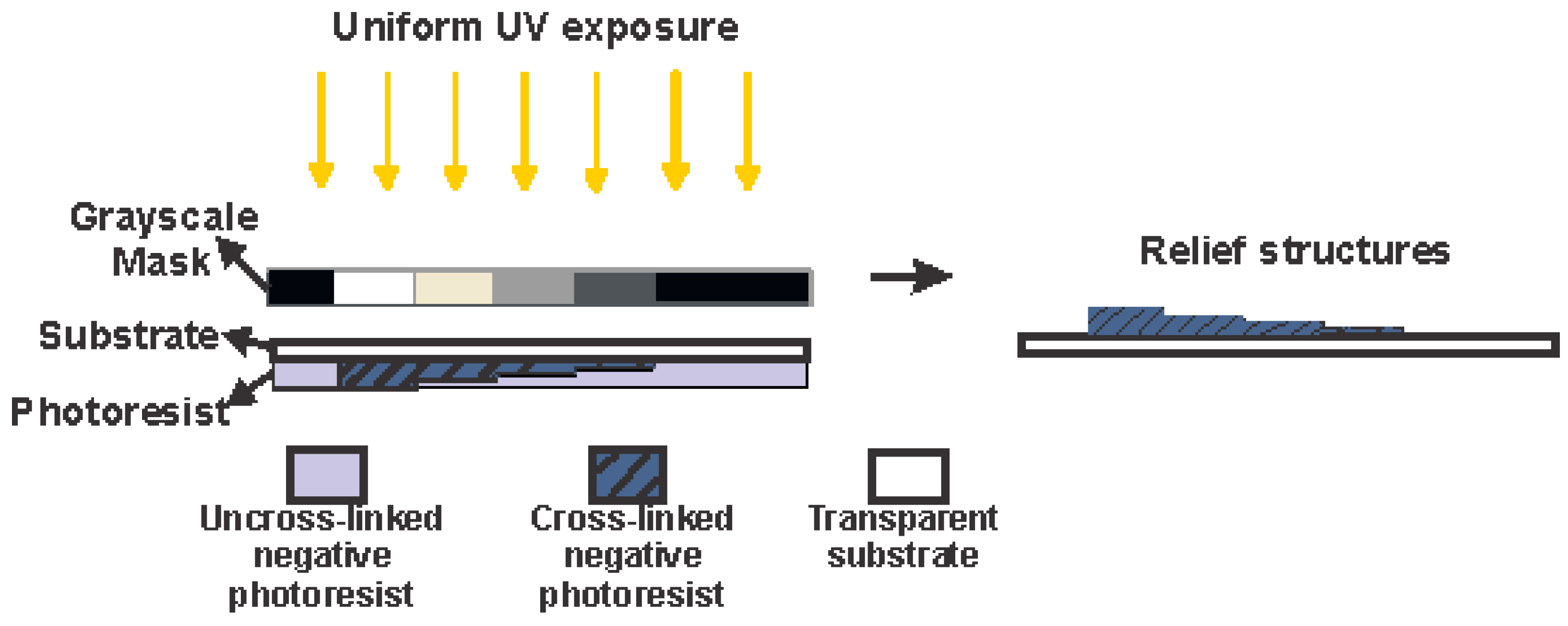
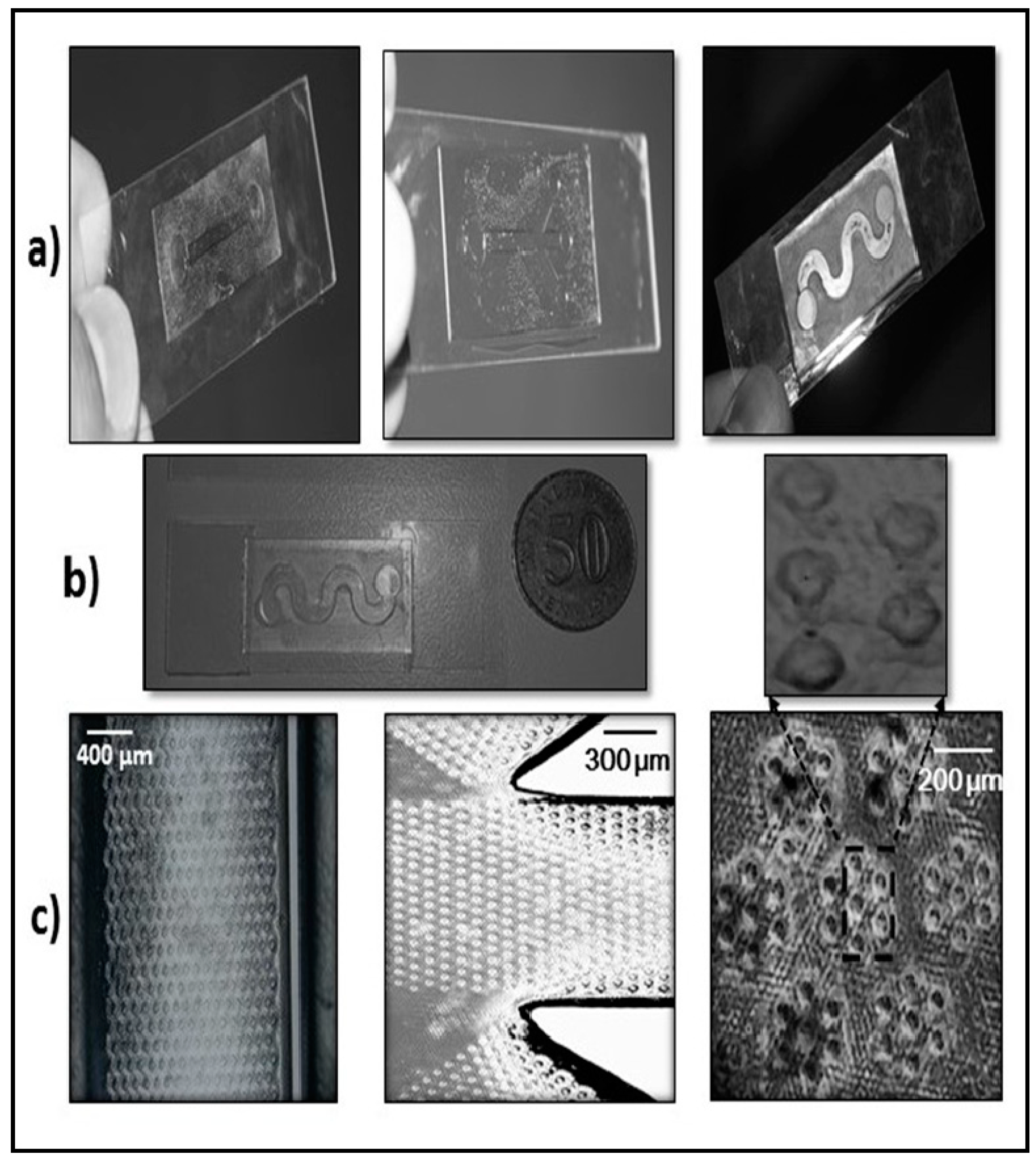
2.3. Biochip Fabrication
2.4. Cell Culture
2.5. Single-Cell Trapping and Cell Viability Tests
2.6. Single-Cell Fluorescent Measurement
3. Results
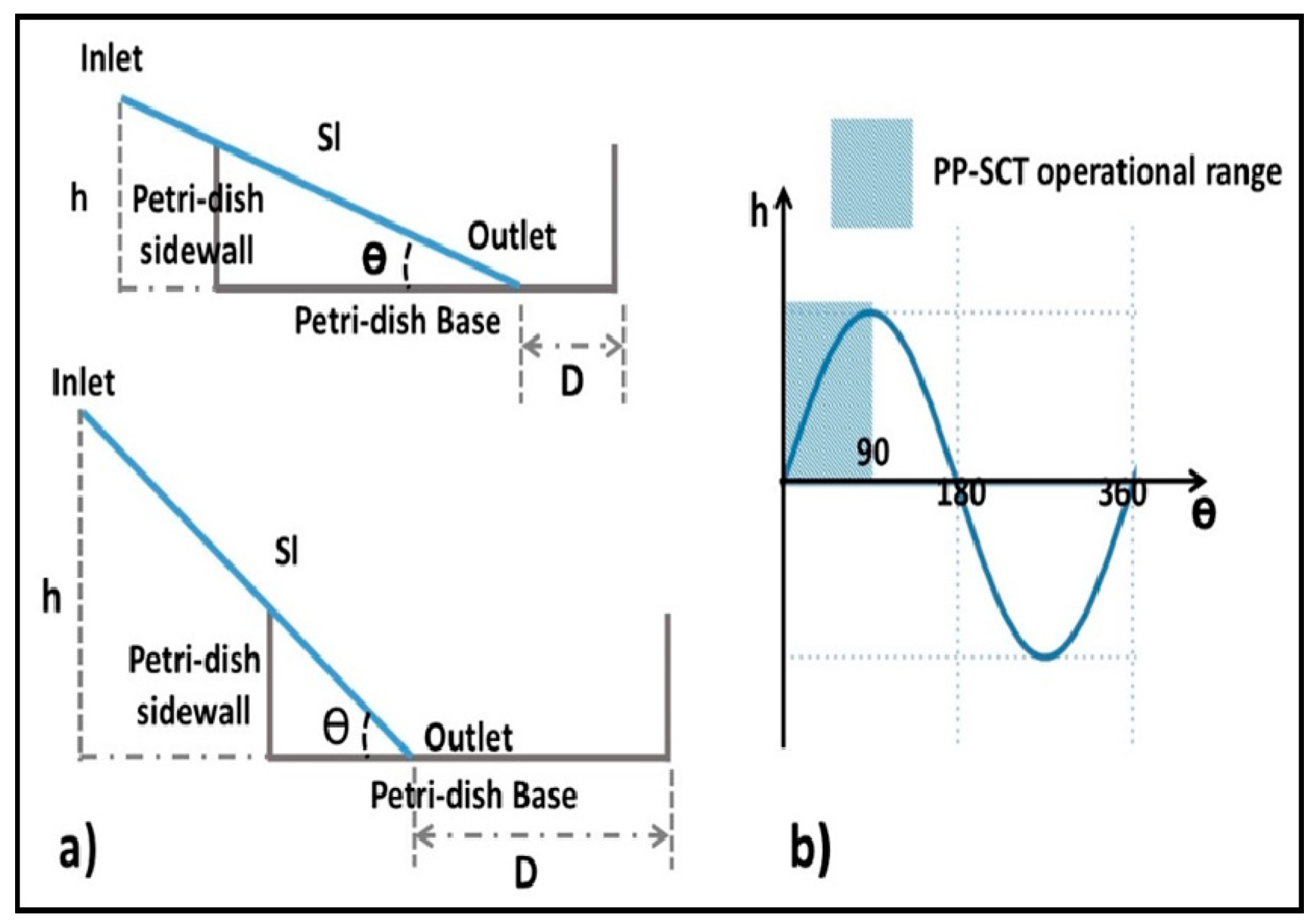
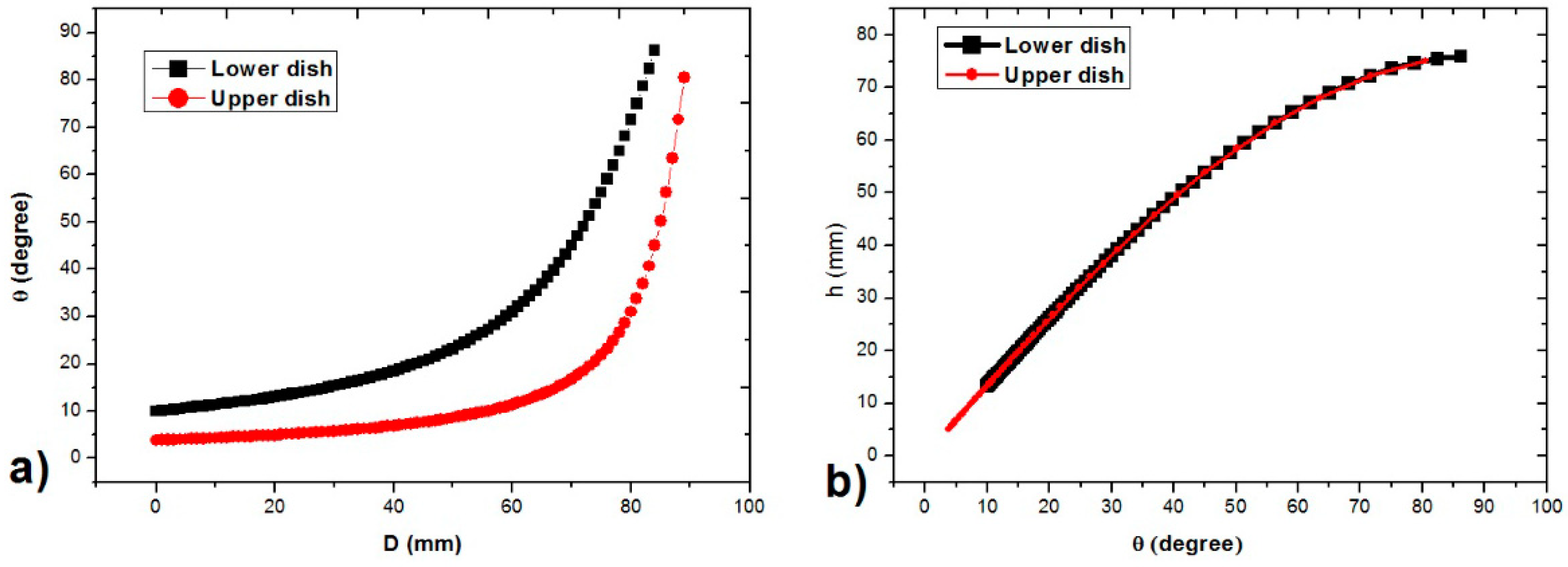
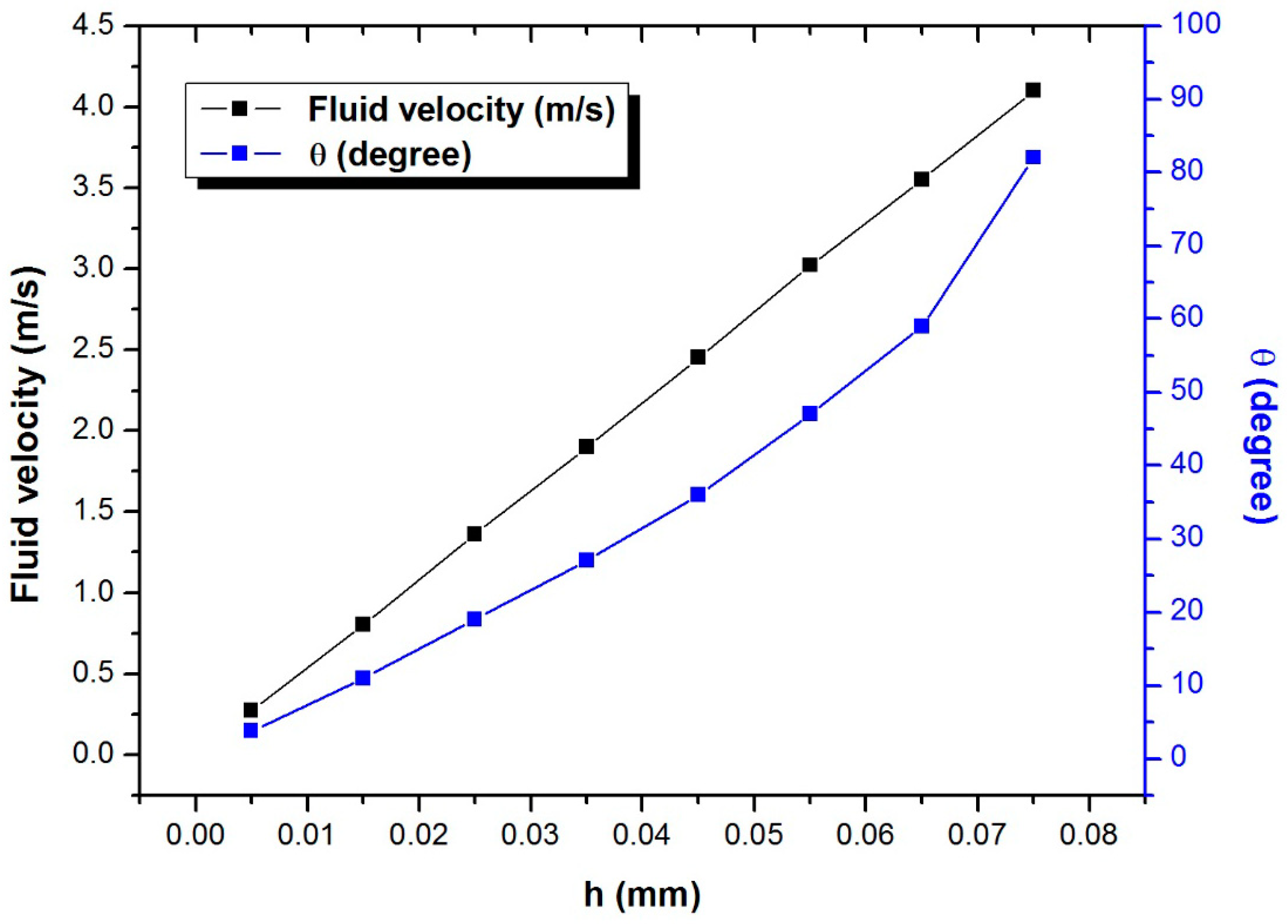
3.1. Characteristics of PP-SCT
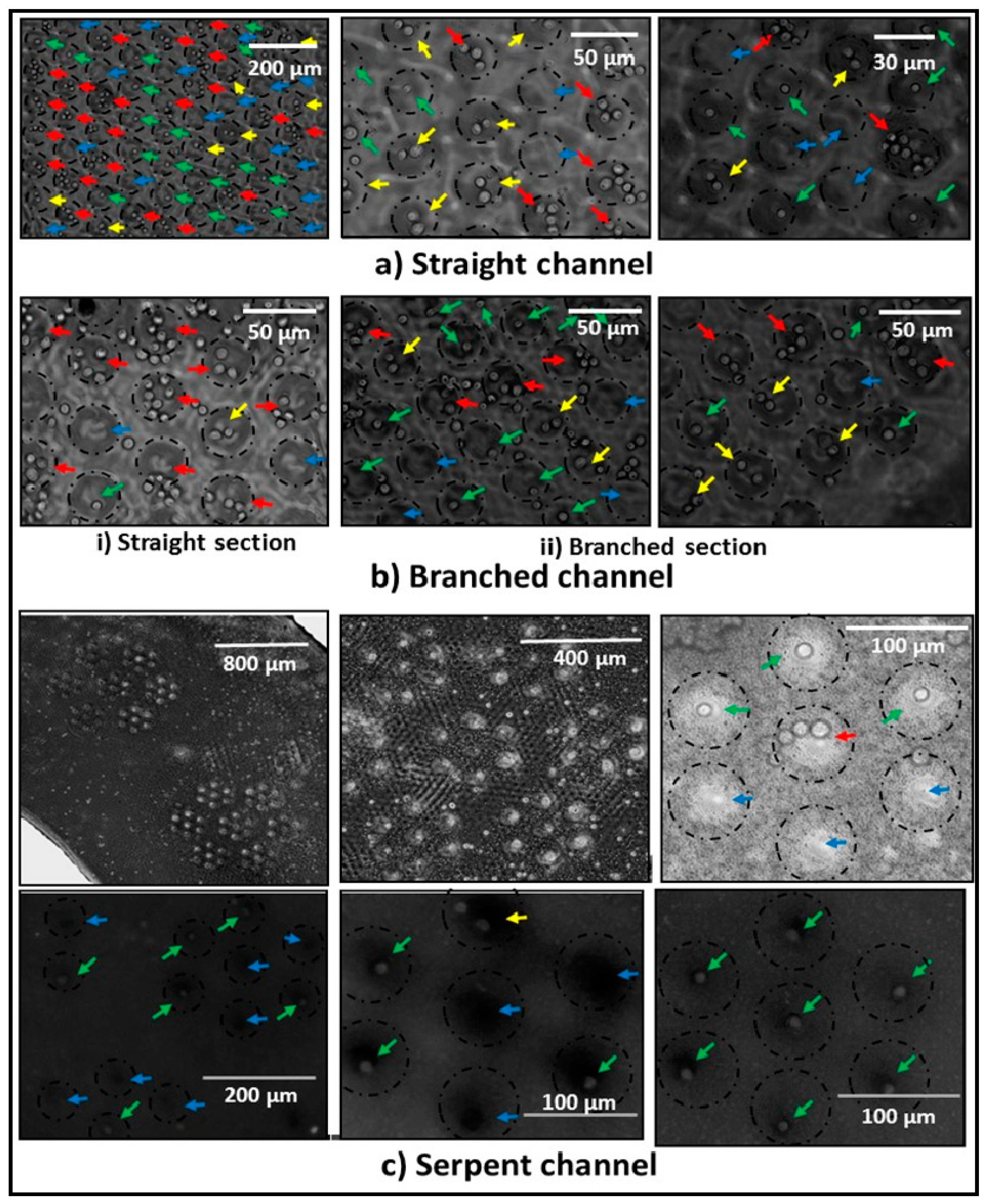
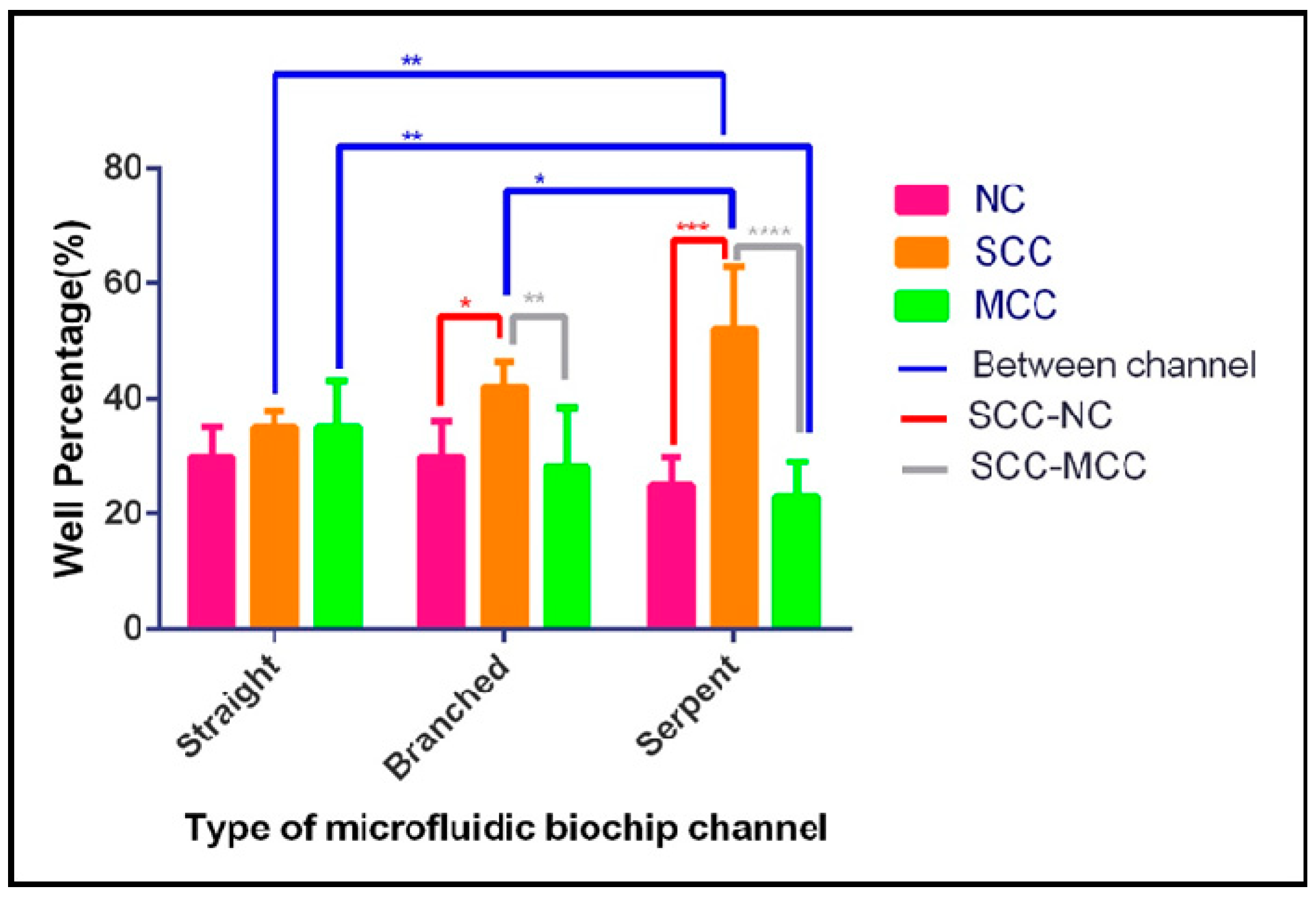
3.2. Single-Cell Trapping and Design Analysis
3.3. Cell Viability and Single-Cell Fluorescent Measurements
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviation
| PP-SCT | Pipette Petri dish single-cell trapping |
| SCC | Single-cell capture |
| MCC | Multiple-cell capture |
| NC | No capture |
| UD | Upper dish |
| LD | Lower dish |
| SCA | Single cell analysis |
| PET | Polyethylene terephthalate |
| IPA | Isopropyl alcohol |
| CGM | Complete growth media |
| PBS | Phosphate-buffered solution |
References
- Khoshnoud, F.; de Silva, C.W. Recent advances in mems sensor technology–biomedical applications. IEEE Instrum. Meas. Mag. 2012, 15, 8–14. [Google Scholar] [CrossRef]
- Kim, C.-S.; Ahn, S.-H.; Jang, D.-Y. Review: Developments in micro/nanoscale fabrication by focused ion beams. Vacuum 2012, 86, 1014–1035. [Google Scholar] [CrossRef]
- Vigneswaran, N.; Samsuri, F.; Ranganathan, B. Recent advances in nano patterning and nano imprint lithography for biological applications. Procedia Eng. 2014, 97, 1387–1398. [Google Scholar] [CrossRef]
- Wang, D.; Bodovitz, S. Single cell analysis: The new frontier in ‘omics’. Trends Biotechnol. 2010, 28, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, P.; Jakubowski, N. Current trends in single cell analysis. Anal. Bioanal. Chem. 2014, 406, 6957–6961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navin, N.; Hicks, J. Future medical applications of single-cell sequencing in cancer. Genome Med. 2011, 3, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Navin, N.E. Advances and applications of single-cell sequencing technologies. Mol. Cell 2015, 58, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Occhetta, P.; Malloggi, C.; Gazaneo, A.; Redaelli, A.; Candiani, G.; Rasponi, M. High-throughput microfluidic platform for adherent single cells non-viral gene delivery. RSC Adv. 2015, 5, 5087–5095. [Google Scholar] [CrossRef] [Green Version]
- Fujita, H.; Esaki, T.; Masujima, T.; Hotta, A.; Kim, S.H.; Noji, H.; Watanabe, T.M. Comprehensive chemical secretory measurement of single cells trapped in a micro-droplet array with mass spectrometry. RSC Adv. 2015, 5, 16968–16971. [Google Scholar] [CrossRef]
- Sun, H.; Olsen, T.; Zhu, J.; Tao, J.; Ponnaiya, B.; Amundson, S.A.; Brenner, D.J.; Lin, Q. A bead-based microfluidic approach to integrated single-cell gene expression analysis by quantitative rt-pcr. RSC Adv. 2015, 5, 4886–4893. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Rettig, J.R.; Folch, A. Large-scale single-cell trapping and imaging using microwell arrays. Anal. Chem. 2005, 77, 5628–5634. [Google Scholar] [CrossRef] [PubMed]
- Di Carlo, D.; Wu, L.Y.; Lee, L.P. Dynamic single cell culture array. Lab Chip 2006, 6, 1445–1449. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Morgan, M.; Sachs, A.N.; Samorezov, J.; Teller, R.; Shen, Y.; Pienta, K.J.; Takayama, S. Single cell trapping in larger microwells capable of supporting cell spreading and proliferation. Microfluid. Nanofluid. 2010, 8, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Kobel, S.; Valero, A.; Latt, J.; Renaud, P.; Lutolf, M. Optimization of microfluidic single cell trapping for long-term on-chip culture. Lab Chip 2010, 10, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shah, P.; Phillips, C.; Sims, C.E.; Allbritton, N.L. Trapping cells on a stretchable microwell array for single-cell analysis. Anal. Bioanal. Chem. 2012, 402, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Banaeiyan, A.A.; Ahmadpour, D.; Adiels, C.B.; Goksör, M. Hydrodynamic cell trapping for high throughput single-cell applications. Micromachines 2013, 4, 414–430. [Google Scholar] [CrossRef]
- Huang, L.; Chen, Y.; Chen, Y.; Wu, H. Centrifugation-assisted single-cell trapping in a truncated cone-shaped microwell array chip for the real-time observation of cellular apoptosis. Anal. Chem. 2015, 87, 12169–12176. [Google Scholar] [CrossRef] [PubMed]
- Ashok, P.C.; Dholakia, K. Optical trapping for analytical biotechnology. Curr. Opin. Biotechnol. 2012, 23, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Singh, A.K. Single-cell protein analysis. Curr. Opin. Biotechnol. 2012, 23, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xu, J.; Romero-Gonzalez, M.; Banwart, S.A.; Huang, W.E. Single cell raman spectroscopy for cell sorting and imaging. Curr. Opin. Biotechnol. 2012, 23, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.-H.; Kim, S.-H.; Ahn, K.; Lee, S.-H.; Park, J.Y. Separation and sorting of cells in microsystems using physical principles. J. Micromech. Microeng. 2015, 26, 013003. [Google Scholar] [CrossRef]
- Johann, R.M. Cell trapping in microfluidic chips. Anal. Bioanal. Chem. 2006, 385, 408–412. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Evander, M.; Hammarström, B.; Laurell, T. Review of cell and particle trapping in microfluidic systems. Anal. Chim. Acta 2009, 649, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.-J.; Yao, D.-J. Get to understand more from single-cells: Current studies of microfluidic-based techniques for single-cell analysis. Int. J. Mol. Sci. 2015, 16, 16763–16777. [Google Scholar] [CrossRef] [PubMed]
- Valizadeh, A.; Khosroushahi, A.Y. Single-cell analysis based on lab on a chip fluidic system. Anal. Methods 2015, 7, 8524–8533. [Google Scholar] [CrossRef]
- Shi, Q.; Qin, L.; Wei, W.; Geng, F.; Fan, R.; Shin, Y.S.; Guo, D.; Hood, L.; Mischel, P.S.; Heath, J.R. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc. Natl. Acad. Sci. USA 2012, 109, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Devarenne, T.P.; Han, A. A high-throughput microfluidic single-cell screening platform capable of selective cell extraction. Lab Chip 2015, 15, 2467–2475. [Google Scholar] [CrossRef] [PubMed]
- Eyer, K.; Kuhn, P.; Hanke, C.; Dittrich, P.S. A microchamber array for single cell isolation and analysis of intracellular biomolecules. Lab Chip 2012, 12, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Shirai, M.; Taniguchi, T.; Kambara, H. Emerging applications of single-cell diagnostics. Top. Curr. Chem. 2012, 336, 99–116. [Google Scholar]
- Yi, C.; Li, C.-W.; Ji, S.; Yang, M. Microfluidics technology for manipulation and analysis of biological cells. Anal. Chim. Acta 2006, 560, 1–23. [Google Scholar] [CrossRef]
- Kim, S.M.; Lee, S.H.; Suh, K.Y. Cell research with physically modified microfluidic channels: A review. Lab Chip 2008, 8, 1015–1023. [Google Scholar] [CrossRef] [PubMed]
- Bhagat, A.A.S.; Bow, H.; Hou, H.W.; Tan, S.J.; Han, J.; Lim, C.T. Microfluidics for cell separation. Med. Biol. Eng. Comput. 2010, 48, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Burger, R.; Ducrée, J. Handling and analysis of cells and bioparticles on centrifugal microfluidic platforms. Expert Rev. Mol. Diagn. 2012, 12, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Cetin, B.; Özer, M.B.; Solmaz, M.E. Microfluidic bio-particle manipulation for biotechnology. Biochem. Eng. J. 2014, 92, 63–82. [Google Scholar] [CrossRef]
- Karimi, A.; Yazdi, S.; Ardekani, A. Hydrodynamic mechanisms of cell and particle trapping in microfluidics. Biomicrofluidics 2013, 7, 021501. [Google Scholar] [CrossRef] [PubMed]
- Batchelor, G.K. An Introduction to Fluid Dynamics; Cambridge University Press: Cambridge, UK, 1967; Volume 515, p. 13. [Google Scholar]
- Stokes, G.G. On the Effect of the Internal Friction of Fluids on the Motion of Pendulums; Pitt Press: Pittsburgh, PA, USA, 1851; Volume 9. [Google Scholar]
- Reynolds, O. An experimental investigation of the circumstances which determine whether the motion of water shall be direct or sinuous, and of the law of resistance in parallel channels. Proc. R. Soc. Lond. 1883, 35, 84–99. [Google Scholar] [CrossRef]
- Lee, T.P.; Mohamed, K. 3D Microfabrication Using Emulsion Mask Grayscale Photolithography Technique; IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2016; p. 012032. [Google Scholar]
- Kuznetsov, S.; Doonan, C.; Wilson, N.; Mohan, S.; Hudson, S.E.; Paulos, E. Diybio things: Open source biology tools as platforms for hybrid knowledge production and scientific participation. In Proceedings of the 33rd Annual ACM Conference on Human Factors in Computing Systems, Seoul, Korea, 18–23 April 2015; ACM: New York, NY, USA, 2015; pp. 4065–4068. [Google Scholar]

| Components | Property | Value/Equation |
|---|---|---|
| Fluid (water) | Dynamic Viscosity (mu) | 8.90 × 10−4 (Pa·s) |
| Density (rho) | 997 (kg/m3) | |
| Channel | Channel Surface Roughness | Glass (0.0015 mm) |
| Initial Values | Pressure | 101,325 (Pa) |
| Tangential Velocity | 0 m/s | |
| Volume Force | x | 0 (N/m3) |
| y | (−g_const) * pfl.rho (N/m3) | |
| Pressure | Inlet | 101,325 (Pa) |
| Outlet | 101,325 (Pa) + (5(mm)) * g_const * pfl.rho |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narayanamurthy, V.; Lee, T.P.; Khan, A.Y.F.; Samsuri, F.; Mohamed, K.; Hamzah, H.A.; Baharom, M.B. Pipette Petri Dish Single-Cell Trapping (PP-SCT) in Microfluidic Platforms: A Passive Hydrodynamic Technique. Fluids 2018, 3, 51. https://doi.org/10.3390/fluids3030051
Narayanamurthy V, Lee TP, Khan AYF, Samsuri F, Mohamed K, Hamzah HA, Baharom MB. Pipette Petri Dish Single-Cell Trapping (PP-SCT) in Microfluidic Platforms: A Passive Hydrodynamic Technique. Fluids. 2018; 3(3):51. https://doi.org/10.3390/fluids3030051
Chicago/Turabian StyleNarayanamurthy, Vigneswaran, Tze Pin Lee, Al’aina Yuhainis Firus Khan, Fahmi Samsuri, Khairudin Mohamed, Hairul Aini Hamzah, and Madia Baizura Baharom. 2018. "Pipette Petri Dish Single-Cell Trapping (PP-SCT) in Microfluidic Platforms: A Passive Hydrodynamic Technique" Fluids 3, no. 3: 51. https://doi.org/10.3390/fluids3030051