Abstract
Medicinal plants serve as vital resources for preventing and treating diseases, with their flowers, fruits, leaves, roots, or entire plants being utilized in the pharmaceutical industry or as direct therapeutic agents. During our investigation of microfungi associated with medicinal plants in Guizhou and Sichuan Provinces, China, several asexual and sexual fungal morphs were collected. Multi-locus phylogenetic analysis based on combined ITS, LSU, SSU and TEF1-α datasets revealed that these taxa are related to the family Dictyosporiaceae. Morphological characteristics, along with multi-locus phylogenetic analysis, supported the establishment of Dictyocheirospora alangii sp. nov. and Pseudocoleophoma rosae sp. nov., as well as the introduction of a novel genus Neoxylochrysis, which accommodates Neoxylochrysis typhicola comb. nov. (≡Pseudocoleophoma typhicola). In addition, four new host records are introduced for Aquadictyospora lignicola from Periploca forrestii, Dendryphiella eucalyptorum from Leonurus japonicus, Ophiopogon japonicus and Sambucus javanica, D. vinosa from Phytolacca americana, and Dictyocheirospora rotunda from Euonymus japonicus and Prinsepia utilis. Detailed descriptions, micrographs of the new taxa and a phylogenetic tree are provided.
1. Introduction
The family Dictyosporiaceae, originally proposed as Dictyosporaceae by Liu et al. [1], was formally introduced by Boonmee et al. [2] within the order Pleosporales and class Dothideomycetes. The type genus of Dictyosporiaceae is Dictyosporium Corda, with D. elegans designated as the type species. This holomorphic family is globally distributed and consists of 20 genera. Most asexual morphs in this family are hyphomycetous, viz., Aquadictyospora, Aquaticheirospora, Cheirosporium, Dendryphiella, Dictyocheirospora, Dictyopalmispora, Dictyosporium, Digitodesmium, Jalapriya, Neodendryphiella, Neodigitodesmium, Pseudodictyosporium and Vikalpa, characterized by the production of cheiroid (digitate), multi-septate, palmate or dictyosporous, and pale brown to brown conidia [2,3,4,5,6,7]. A few genera, viz., Immotthia, Pseudocoleophoma, Pseudoconiothyrium, Pseudocyclothyriella, Sajamaea and Verrucoccum have coelomycetous asexual morphs [8,9,10,11,12,13]. Among these, only five genera viz., Dictyosporium, Gregarithecium, Immotthia, Pseudocoleophoma and Verrucoccum, are known to produce sexual morphs, which are characterized by brown to black ascomata, bitunicate, cylindric-clavate asci with a short ocular chamber, and septate, hyaline to brown, sheathed ascospores [8,9,12]. Notably, Gregarithecium lacks an asexual morph [9].
The genus Pseudocoleophoma, typified by P. calamagrostidis Kaz. Tanaka and K. Hiray., was established by Tanaka et al. [9] to accommodate two species, P. calamagrostidis and P. polygonicola. Fourteen Pseudocoleophoma species are listed in Index Fungorum (accessed September 2024), viz., P. bauhiniae, P. calamagrostidis, P. clematidis, P. flavescens, P. guizhouensis, P. heteropanacicola, P. paraphysoidea, P. polygonicola, P. puerensis, P. rhapidis, P. rusci, P. typhicola, P. yunnanensis and P. zingiberacearum [13,14]. However, P. clematidis was transferred to Pseudocyclothyriella based on morphology and multi-locus phylogenetic analysis. by Jiang et al. [13]. The sexual morph of Pseudocoleophoma is characterized by cylindrical to clavate asci and hyaline, fusiform, septate ascospores with an apparent sheath [15,16], while the asexual morph is characterized by hyaline, aseptate, cylindrical or oblong, smooth-walled conidia with obtuse ends [17,18,19,20].
Aquadictyospora was introduced by Li et al. [5] to accommodate the species, A. lignicola, collected from submerged decaying wood in China. Phukhamsakda et al. [21] introduced the second species, A. clematidis, based on morphology and phylogenetic analyses. Aquadictyospora is an asexual morph genus, with no sexual morph linked to it. The genus is characterized by superficial, compact, scattered, globose or subglobose, dark brown to black conidiomata, micronematous conidiophores with monoblastic conidiogenous cells, and uniformly medium brown conidia with a broadly ovate to subglobose, hyaline cell in the lower half [5,21].
Dendryphiella was introduced by Ranojevic [22] with the type species D. interseminata Bubák. This asexual genus has 20 epithets in Index Fungorum (accessed September 2024). It is characterized by macronematous, fasciculate, septate conidiophores with polytretic, verrucose conidiogenous cells enlarged at the apex, and solitary to catenate, aseptate or septate, hyaline to pale brown, thick-walled, verrucose conidia [23,24]. It is worth noting that the mode of conidiogenous cell development of Dendryphiella members is tretic, while other asexual morphs in the family are blastic [2,6,23]. Boonmee et al. [2] established Dictyocheirospora to accommodate three species, D. bannica, D. rotunda (type species) and D. vinaya, and transferred four species of Dictyosporium to Dictyocheirospora. Currently, 29 epithets of Dictyocheirospora are listed in Index Fungorum (accessed September 2024). Dictyocheirospora is similar to Dictyosporium except for the conidial arms, which are arranged differently [2,25,26,27]. The sexual stage of Dictyocheirospora has never been reported.
In this study, we explored the diversity and taxonomy of the family Dictyosporiaceae, focusing on species associated with medicinal plants. The taxa of Dictyosporiaceae have been subject to frequent taxonomic revisions and phylogenetic realignments, reflecting the complexity of their evolutionary relationships. By examining Dictyosporiaceae species collected from medicinal plant, we aimed to expand the understanding of their biodiversity, refine their taxonomy through multi-locus phylogenetic analyses, and assess their ecological roles in these specialized habitats. This study also contributes to the understanding of classification and biodiversity of this diverse fungal family.
2. Materials and Methods
2.1. Specimen Collection, Examination, and Single Spore Isolation
The dead or decaying leaves and twigs of medicinal plants (Alangium chinense, Euonymus japonicus, Leonurus japonicus, Ophiopogon japonicus, Periploca forrestii, Phytolacca americana, Prinsepia utilis, Rhaphiolepis indica, Rosa roxbunghii and Sambucus javanica, Figure 1) were collected from Guizhou and Sichuan Provinces, China (detailed information about the collection sites is provided in the ‘Materials examined’ subsection of Section 3.2). The samples were kept in paper envelopes and brought to the laboratory following the method described in Senanayake et al. [28]. Morphological observations of fungal structures were made using a Nikon SMZ745 dissecting microscope (Nikon Corporation, Tokyo, Japan), following the method described in Chomnunti et al. [29]. Photomicrographs of the fungal specimens were captured using a Nikon Eclipse Ni-U compound microscope fitted with a Nikon DS-Ri2 digital camera (Nikon Corporation, Tokyo, Japan). Macro-morphological structures were photographed with a Nikon SMZ800N stereo microscope fitted with a Nikon DS-Fi3 microscope camera (Nikon Corporation, Tokyo, Japan). All measurements were made with the Tarosoft Image Frame Work program v. 0.97, and the photo-plates were made with Adobe Photoshop CC extended version 21.1.2. Single spore isolations were conducted in accordance with the methods described in Senanayake et al. [28]. Germinated spores were transferred to fresh potato dextrose agar (PDA), incubated at 25 °C, and the colony characteristics were observed and recorded after one week, following the method described in Rayner [30].

Figure 1.
Photos of medicinal plant hosts in this study (a) Alangium chinense (Cornaceae). (b) Euonymus japonicus (Celastraceae). (c) Leonurus japonicus (Lamiaceae). (d) Ophiopogon japonicus (Asparagaceae). (e) Periploca forrestii (Apocynaceae). (f,g) Phytolacca americana (Phytolaccaceae). (h) Prinsepia utilis (Rosaceae). (i) Rhaphiolepis indica (Rosaceae). (j,k) Rosa roxburghii (Rosaceae). (l) Sambucus javanica (Viburnaceae).
Herbarium specimens were deposited in the herbarium of Cryptogams, Kunming Institute of Botany, Chinese Academy Sciences (KUN-HKAS), Kunming, China and the Herbarium of University of Electronic Science and Technology (HUEST), Chengdu, China. The pure cultures were deposited in the China General Microbiological Culture Collection Centre (CGMCC), Beijing, China and the University of Electronic Science and Technology Culture Collection (UESTCC), Chengdu, China. Faces of Fungi and Index Fungorum numbers were provided for the new taxa (Index Fungorum 2024) [31].
2.2. DNA Extraction, PCR Amplification and Sequencing
Fresh mycelia (about 50–100 mg) were scraped using a sterilized toothpick from the margin of a colony on a PDA plate, which had been incubated at 25 °C for two to three weeks [32], and stored in 1.5 mL sterilized micro-centrifuge tubes and maintained at −20 °C for long term storage. The TreliefTM Plant Genomic DNA Kit (TSINGKE Biotech, Shanghai, China) was used to extract DNA according to the manufacturer’s instructions. The obtained genomic DNA was stored in two tubes, one at 4 °C for polymerase chain reaction (PCR) amplification and the other at −20 °C for long-term storage. The internal transcribed spacer (ITS), the partial 28S large subunit rRNA (LSU), the partial 18S small subunit rRNA (SSU), and the partial translation elongation factor 1-alpha (TEF1-α) regions were amplified using the following primers: ITS5 and ITS4 [33] for ITS, LR0R and LR5 [34] for LSU, NS1 and NS4 [33] for SSU, and EF1-983F and EF1-2218R [35] for TEF1-α. The final volume (25 μL) contained 2 μL DNA, 12.5 μL PCR mix, 8.5 μL distilled water and 1 μL of each primer.
The PCR thermal cycle program of ITS, LSU, and SSU loci used the following conditions: initial denaturation at 94 °C for 3 min, followed by 40 cycles of denaturation at 94 °C for 45 s, annealing at 56 °C for 50 s, elongation at 72 °C for 1 min, and a final extension at 72 °C for 10 min. The TEF1-α amplification followed these conditions: initial denaturation at 94 °C for 5 min, followed by 34 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 50 s, elongation at 72 °C for 1 min, and a final extension at 72 °C for 5 min. The products were visualized on 1% agarose gel under UV light in a Gel DocTM XR and sequenced at Sangon Biotechnology Co. (Chengdu, China).
2.3. Sequence Alignment and Phylogenetic Analysis
Sequences generated in this study were checked and assembled using BioEdit v. 17.0.1 [36] to ensure sequence quality. Through the BLASTn search tool on NCBI [37] (https://blast.ncbi.nlm.nih.gov/, accessed on 20 August 2023), based on newly generated ITS and LSU sequence data, we found out that our taxa were related to Dictyosporiaceae. According to the BLAST results and previous literature, appropriate sequences were downloaded from GenBank to construct phylogenetic analyses. Two isolates of Periconia igniaria (CBS 379.86 and CBS 845.96) were selected as the outgroup. Details of the isolates used in this study are listed in Table 1. The sequences were aligned using MAFFT v.7 online [38] (https://mafft.cbrc.jp/alignment/server/, accessed on 28 August 2023) and AliView [39], and the results were checked using BioEdit v. 17.0.1 [36] and manually edited where necessary. The concatenation of the genes was conducted using SequenceMatrix 1.8 [40]. The Nexus and Phylip files for phylogenetic analyses were obtained using AliView [39]. Phylogenetic analyses of the combined sequence data were performed using maximum likelihood (ML) and Bayesian inference (BI) methods, as detailed in Dissanayake et al. [41]. Best-fit models for BI analyses were selected using MrModeltest v. 2.2 [42]. The ML analysis was performed using RAxML GUI v. 1.3.1 [43], and the BI analysis was conducted in MrBayes v 3.2.6 [44]. Phylogenetic trees were visualized with FigTree v.1.4.4 [45] (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 9 October 2023) and further edited in Adobe Illustrator 2020 (Adobe Systems, San Jose, CA, USA). The final alignment was submitted to Figshare [46] (https://figshare.com, at https://doi.org/10.6084/m9.figshare.26085910, accessed on 23 June 2024).

Table 1.
GenBank accession numbers and details of isolates chosen for the phylogenetic studies. The newly generated sequences are indicated in bold and ex-type strains are indicated with T after the strain number. N/A denotes no sequence available.
3. Results
3.1. Phylogenetic Analysis
The analyzed dataset was composed of the combined ITS, LSU, SSU and TEF1-α sequence data from 61 taxa (ingroup) with Periconia igniaria (CBS 379.86 and CBS 845.96) as the outgroup (Figure 2). The aligned dataset comprised 3849 characters (ITS: 574 bp, LSU: 883 bp, SSU: 1436 bp, TEF1-α: 956 bp) including gaps. The RAxML analysis of the combined data set yielded a best-scoring tree (Figure 2) with a final ML optimization likelihood value of −20,233.812942. RAxML and Bayesian analyses were conducted and resulted in generally congruent topologies and the familial assignment is similar to previous work [6,27].
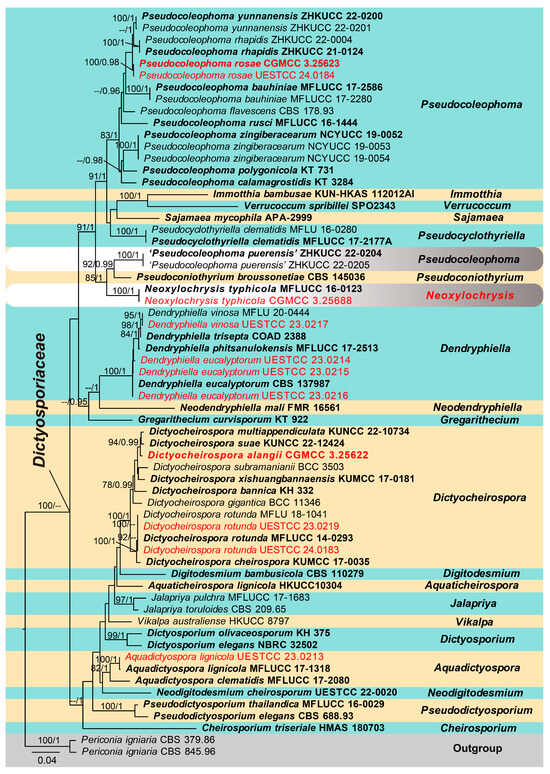
Figure 2.
Phylogenetic tree generated from the maximum likelihood analysis based on the combined ITS, LSU, SSU and TEF1-α sequence data of Dictyosporiaceae. The ML (≥75%) and BI (≥95%) bootstrap supports are given near the nodes, respectively. The new isolates obtained in this study are indicated in red and ex-type strains are in bold. The tree is rooted with Periconia igniaria (CBS 379.86 and CBS 845.96).
The multi-locus phylogenetic analyses showed that eleven isolates obtained in this study were nested within the family Dictyosporiaceae, of which one isolate (UESTCC 23.0213) was identified as Aquadictyospora lignicola. Four isolates (UESTCC 23.0214, UESTCC 23.0215, UESTCC 23.0216 and UESTCC 23.0217) clustered with Dendryphiella eucalyptorum and D. vinosa, respectively. Three isolates belong to the genus Dictyocheirospora, of which UESTCC 23.0219 and UESTCC 24.0183 clustered with Dictyocheirospora rotunda, while the new taxon Dictyocheirospora alangii (CGMCC 3.25622) clustered sister to Di. multiappendiculata (KUNCC 22-10734) and Di. suae (KUNCC 22-12424) with 94% ML bootstrap support (MLBS) and 0.99 Bayesian posterior probability support (BYPP) (Figure 2). Two isolates (CGMCC 3.25623 and UESTCC 24.0184) nested within the genus Pseudocoleophoma, but did not cluster with any previously known species; thus, a novel species, P. rosae, was preliminarily identified. In addition, our isolate CGMCC 3.25688 clustered with Neoxylochrysis typhicola (≡Pseudocoleophoma typhicola) (MFLUCC 16-0123), separated from all taxa of Pseudocoleophoma and formed a clade basal to P. puerensis (ZHKUCC 22-0204, ZHKUCC 22-0205) and Pseudoconiothyrium broussonetiae (CBS 145036). Thus, based on current phylogenetic status, P. typhicola is transferred to the novel genus Neoxylochrysis as N. typhicola.
3.2. Taxonomy
Aquadictyospora lignicola Z.L. Luo, W.L. Li, K.D. Hyde & H.Y. Su, Mycosphere 8(10): 1591 [5] (Figure 3).

Figure 3.
Aquadictyospora lignicola (HUEST 23.0213, new host record). (a–c) Colonies on a woody substrate. (d–f) Conidia with partial conidiophores. (g,h) Conidia. (i) Germinated conidium. (j,k) Colonies on PDA, from above (j), from below (k). Scale bars: (d–h) = 20 μm, (i) = 50 μm.
Index Fungorum number: IF553862; Faces of Fungi number: FoF03768
Saprobic on dead twigs of Periploca forrestii Schltr. Sexual morph: Not observed. Asexual morph: Hyphomycetous. Colonies superficial, gregarious, scattered, punctiform, sporodochial, scattered, black, velvety, glistening, orbicular. Mycelium mostly immersed or partly superficial, smooth, with hyaline to pale brown hyphae. Conidiophores micronematous, cylindrical, hyaline to pale brown, smooth-walled, sometimes reduced to conidiogenous cells. Conidiogenous cells holoblastic, monoblastic, integrated, terminal, determinate, hyaline to pale brown, smooth-walled. Conidia 38–52 × 19–33 μm (x = 47 × 26 μm, n = 50), solitary, acrogenous, cheiroid, smooth-walled, yellowish-brown to light brown, with a basal connecting cell, consisting of 5–7 (mostly 6) rows of cells, euseptate, unseparated, each row with 8–9 cells, individual rows discoid, secession schizolytic.
Culture characteristics: Conidia germinated on PDA within 24 h. Germ tubes produced from both ends. Colonies on PDA reaching 20–30 mm diam. after 2 weeks at 25 °C in natural light, circular, wrinkled, with dense white to pale yellow mycelia in the middle, sparser towards the edge; in reverse yellowish brown in the middle, pale yellow at the entire margin.
Material examined: China, Sichuan Province, Chengdu City, Dujiangyan City, Qingcheng Mountain scenic spot, 103°28′36′′ E, 30°55′9′′ N, on dead twigs of medicinal plant Periploca forrestii, 27 March 2021, H.Z. Du, S172 (HUEST 23.0213), living culture UESTCC 23.0213.
Notes: Aquadictyospora lignicola, the type species of Aquadictyospora, was introduced by Li et al. [5] from submerged decaying wood. In this study, an isolate was obtained from Periploca forrestii. Our collection (HUEST 23.0213) is similar to A. lignicola (MFLU 17-1422) in having yellowish-brown to light brown conidia, composed of 4–7 compactly arranged rows of cells, with a basal hyaline cell. Phylogenetic analysis showed that our strain clustered with the type strain of A. lignicola (MFLUCC 17-1318) with ML/BI 100%/1 bootstrap support (Figure 2). Thus, we identified our collection as A. lignicola and report it as a new host record.

Figure 4.
Dendryphiella eucalyptorum (HUEST 23.0214, new host record). (a,b) Host. (c,d) Colonies on a woody substrate. (e,f) Conidiophores. (g–i) Conidiogenous cells and conidia. (j–p) Conidia. (q) Germinated conidium. (r,s) Colonies on PDA, from above (r), from below (s). Scale bars: (e) = 100 μm, (f) = 50 μm, (g–i) = 20 μm, (j–q) = 10 μm.
Index Fungorum number: IF808918; Faces of Fungi number: FoF06712
Saprobic on dead twigs of Sambucus javanica Blume. Sexual morph: Not observed. Asexual morph: Hyphomycetous. Colonies on natural substrate superficial, effuse, dark brown. Mycelium mostly immersed, composed of smooth, septate, branched, brown hyphae. Conidiophores 185–300 μm long, 3–4 μm wide, macronematous, mononematous, brown, wider at the base, slightly paler at the apex, fasciculate, thick-walled, erect, straight or slightly flexuous, smooth or verruculose, septate, unbranched or sometimes branched, wider at the base. Conidiogenous cells 12–30 μm long, 5–7 μm wide (x = 20 × 6 μm, n = 30), polytretic, integrated, terminal and intercalary, later becoming subterminal, proliferating asymmetrically, clavate, brown, enlarged at the vertex. Conidia 12–27 × 5–7 μm (x = 21 × 6 μm, n = 50), subcylindrical, apex obtuse, base bluntly rounded, pale brown, aseptate or 1-septate when young, brown or dark brown, 3-septate when mature, constricted at septa, thick-walled, verruculose to verrucose.
Culture characteristics: Conidia germinated on PDA medium within 24 h. Germ tubes produced from both ends. Colonies on PDA reaching 10–15 mm diam. after two weeks at 25 °C in natural light, circular, cottony, with regular margins, white from above and white to pale grey from below.
Material examined: China, Sichuan Province, Chengdu City, High-tech West District, Yaobo Park, 103°56′21′′ E, 30°43′57′′ N, on dead twigs of medicinal plant Sambucus javanica, 11 August 2021, H.Z. Du, S343 (HUEST 23.0214), living culture UESTCC 23.0214; ibid., Leshan City, Muchuan County, Huangdan Town, 103°41′37′′ E, 29°12′58′ ′N, on dead leaves of medicinal plant Ophiopogon japonicus (L. f.) Ker-Gawl., 31 October 2021, H.Z. Du, S463 (HUEST 23.0215), living culture UESTCC 23.0215; ibid., Yaan City, Mingshan District, Wangu Town, 103°7′57′′ E, 30°10′45′′ N, on dead twigs of medicinal plant Leonurus japonicus Houttuyn, 29 October 2021, H.Z. Du, S459 (HUEST 23.0216), living culture UESTCC 23.0216.
Notes: Dendryphiella eucalyptorum (CBS 137987) was collected from Eucalyptus globulus in Spain [24]. We identified our collections as D. eucalyptorum based on morphology and phylogeny, and report them as new host records from medicinal plants (Leonurus japonicus, Ophiopogon japonicus and Sambucus javanica).
Dendryphiella vinosa (Berk. & M.A. Curtis) Reisinger, Bull. trimest. Soc. mycol. Fr. 84(1): 27 [47] (Figure 5).

Figure 5.
Dendryphiella vinosa (HUEST 23.0217, new host record). (a,b) Host. (c–e) Colonies on a woody substrate. (f,g) Conidiophores. (h–j) Conidiogenous cells and conidia. (k–p) Conidia. (q,r) Colony on PDA, from above (q), from below (r). Scale bars: (f,g) = 50 μm, (h–j) = 20 μm, (k–p) = 10 μm.
Index Fungorum number: IF329796; Faces of Fungi number: FoF08673
Saprobic on the twigs of Phytolacca americana L. Sexual morph: Not observed. Asexual morph: Hyphomycetous. Colonies on natural substrate superficial, effuse, brown to dark brown. Conidiophores 180–230 μm long, 4–7 μm wide, macronematous, mononematous, erect, straight or slightly flexuous, smooth or verruculose, septate, unbranched or sometimes branched, wider at the base. Conidiogenous cells 12–28 μm long, 5–7 μm wide (x = 18 × 6 μm, n = 30), polytretic, terminal, proliferating asymmetrically, brown, verrucose, enlarged at the vertex. Conidia 19–30 × 6–8 μm (x = 25 × 7 μm, n = 50), fusiform to ellipsoidal, pale brown to brown or dark brown, aseptate when young, 3-septate when mature, constricted at septa, thick-walled, smooth or occasionally verruculose.
Culture characteristics: Conidia germinated on PDA medium within 24 h. Germ tubes produced from both ends. Colonies on PDA reaching 40–50 mm diam. after two weeks at 25 °C in natural light. Mycelium superficial, with regular margins, white from above, and brown to yellowish in the middle zone from below, paler toward the margins.
Material examined: China, Guizhou Province, Guiyang City, Huaxi District, 106°39′59′′ E, 26°30′14′′ N, on dead twigs of medicinal plant Phytolacca americana, 24 January 2021, H.Z. Du, S61 (HUEST 23.0217), living culture UESTCC 23.0217.
Notes: Dendryphiella vinosa was initially isolated from unidentified rotten leaves by Reisinger [47]. It has also been reported on decomposing leaves in Japan [48] and Dendrobium officinale in China [49]. We identify our isolate as D. vinosa based on morphology and phylogenetic evidence. Therefore, we report D. vinosa from Phytolacca americana as a new host record in China.
Dictyocheirospora alangii H.Z. Du, N. Wu & Jian K. Liu, sp. nov. (Figure 6).

Figure 6.
Dictyocheirospora alangii (HKAS 131314, holotype). (a–d) Colonies on a woody substrate. (e,f) Squash mount of a sporodochium. (g–i) Conidia with partial conidiophores. (j) Conidium. (k) Squashed conidium. (l,m) Arms of conidia. (n,o) Colonies on PDA, from above (n), from below (o). Scale bars: (e,f) = 30 μm, (g–m) = 20 μm.
Index Fungorum number: IF902300; Faces of Fungi number: FoF16017
Etymology: The epithet ‘alangii’ refers to the host genus Alangium from which the fungus was collected.
Holotype: HKAS 131314
Saprobic on dead twigs of Alangium chinense (Lour.) Harms. Sexual morph: Not observed. Asexual morph: Hyphomycetous. Colonies on natural substrate were punctiform, solitary, sporodochial, scattered, dark brown to black. Mycelium immersed, composed of pale brown, smooth, septate, branched hyphae. Conidiophores micronematous, short, branched, hyaline to pale brown. Conidiogenous cells 8–13 μm long, 3–6 μm wide (x = 10 × 5 μm, n = 20), holoblastic, cylindrical, integrated, terminal, pale brown, smooth, thin-walled. Conidia 44–60 × 17–27 μm (x = 49 × 20 μm, n = 50), solitary, cheiroid, ellipsoid to cylindrical, not complanate, pale brown to brown, consisting of 6–7 rows of cells, closely appressed, with rows cylindrical, palmately divergent, each row composed of 8–12 cells, euseptate, slightly constricted at the septa, guttulate, smooth, sometimes with 1–2 hyaline, globose to subglobose appendages which are 10–15 × 7–10 μm, and mostly attached at the central part of two outer arms.
Culture characteristics: Conidia germinated on PDA medium within 12 h. Germ tubes produced from both ends. Colonies on PDA reaching 40–50 mm diam. after one month at 25 °C in natural light. Mycelium superficial, with irregular margins, white from above, and yellowish in the middle zone from below, paler toward the margins.
Material examined: China, Guizhou Province, Guiyang City, Wudang District, Xiangzhigou scenic spot, near freshwater stream, 106°55′22′′ E, 26°46′26′′ N, on dead twigs of medicinal plant Alangium chinense, 24 February 2021, H.Z. Du, S130 (HKAS 131314, holotype); ex-type living culture CGMCC 3.25622 = UESTCC 23.0218.
Notes: The phylogenetic analysis showed that Dictyocheirospora alangii (CGMCC 3.25622) nested within Dictyocheirospora, close to Di. multiappendiculata (KUNCC 22-10734) and Di. suae (KUNCC 22-12424) with good bootstrap support (94% MLBS, 0.99 PP) (Figure 2). Dictyocheirospora alangii, Di. multiappendiculata and Di. suae share similar morphology, having cheiroid, ellipsoid to cylindrical, pale brown to brown conidia with hyaline, globose to subglobose appendages. However, Di. alangii differs from Di. multiappendiculata and Di. suae based on the number and location of appendages. Dictyocheirospora multiappendiculata has 1–6 subapical appendages, Di. suae has 1–3 subapical appendages, and sometimes two ellipsoid appendages are closer together on the same side of the conidia or both sides, whereas Di. alangii has only 1–2 appendages, one on each side of the conidia, and located in the central cells of the outer cell-row rather than near the apex. Therefore, Di. alangii is introduced as a new species based on morphology and phylogeny.

Figure 7.
Dictyocheirospora rotunda (HUEST 23.0219, new host record). (a–c) Colonies on a woody substrate. (d) Squash mount of a sporodochium. (e–h) Conidia with partial conidiophores. (i–l) Conidia. (m) Germinated conidium. (n,o) Colonies on PDA, from above (n), from below (o). Scale bars: (d) = 50 μm, (e–l) = 30 μm, (m) = 20 μm.
Index Fungorum number: IF551581; Faces of Fungi number: FoF01262
Saprobic on dead twigs of Euonymus japonicus Thunb. Sexual morph: Not observed. Asexual morph: Hyphomycetous. Colonies on natural substrate scattered, scattered and dark brown. Mycelium composed of immersed or partly superficial, pale brown, smooth, thin-walled hyphae. Conidiophores micronematous, pale brown, smooth-walled. Conidiogenous cells holoblastic, integrated, terminal, cylindrical to subglobose, hyaline to pale brown, smooth, and thin-walled. Conidia 45–53 × 17–23 μm (x = 50 × 21 μm, n = 50), solitary, acrogenous, cheiroid, pale brown to brown, consisting of 5–7 rows of cells, rows digitate, cylindrical, inwardly curved at the tip, arising from a basal cell, without appendages, with each row composed of 10–12 cells, euseptate.
Culture characteristics: Conidia germinated on PDA medium within 24 h. Germ tubes produced from both ends. Colonies on PDA reaching 10–20 mm diam. after two weeks at 25 °C in natural light. Mycelium superficial, with regular margins, white from above, and brown to yellowish in the middle zone from below, paler toward the margins.
Material examined: China, Guizhou Province, Guiyang City, Huaxi District, 106°39′59′′ E, 26°30′14′′ N, on dead twigs of medicinal plant Euonymus japonicus, 24 January 2021, Y.R. Sun, CL30YR (HUEST 24.0200), living culture UESTCC 24.0183; ibid., Sichuan Province, Chengdu City, Dujiangyan City, Qingcheng Mountain scenic spot, 103°28′36′′ E, 30°55′9′′ N, on dead twigs of medicinal plant Prinsepia utilis, 27 March 2022, R.R. Liang, 123rui (HUEST 23.0219), living culture UESTCC 23.0219.
Notes: Boonmee et al. [2] introduced Dictyocheirospora with D. rotunda as the type species from decaying wood in Thailand. The new strains (UESTCC 23.0219 and UESTCC 24.0183) were collected from Euonymus japonicus and Prinsepia utilis in China. We identified our collections as D. rotunda (Figure 2) and report them as new host records from medicinal plants.
Neoxylochrysis N. Wu & Jian K. Liu, gen. nov.
Index Fungorum number: IF902299; Faces of Fungi number: FoF16018
Etymology: “Neoxylochrysis” refers to the resemblance of its conidial morphology to the genus Xylochrysis.
Saprobic on Typha latifolia. Sexual morph: Not observed. Asexual morph: Conidiomata semi-immersed or immersed, visible as black shiny dots on the host, subcuticular in origin, then becoming erumpent, subglobose, brown to black, uniloculate solitary to scattered, glabrous, ostiolate. Ostiole single, centrally or laterally located. Conidiomatal wall comprising 4–5 layers, hyaline to dark brown, thick-walled cells of textura angularis in the upper part, becoming pale brown, thin-walled at the base. Conidiophores reduced to conidiogenous cells. Conidiogenous cells enteroblastic, phialidic, determinate, discrete, smooth-walled, aseptate, cylindrical to subcylindrical, or ampulliform, hyaline. Conidia hyaline, aseptate or 1-euseptate, without constrictions at the septum, oval or oblong to cylindrical, with rounded or obtuse ends, smooth, thin-walled, often with 2–4 small guttules in each cell, without sheath or appendages.
Type Species: Neoxylochrysis typhicola (Kamolhan, Banmai, Boonmee, E.B.G. Jones and K.D. Hyde) N. Wu and Jian K. Liu.
Life Mode and Known Distribution: Neoxylochrysis is reported as a saprobe on submerged stems of Typha latifolia in freshwater habitat. The genus is presently known from UK [50].
Notes: A new monospecific genus, Neoxylochrysis, is introduced herein to accommodate an asexual species in Dictyosporiaceae, typified by N. typhicola, based on distinct morphology and multi-locus phylogenetic analysis. Neoxylochrysis typhicola was previously described as Pseudocoleophoma typhicola by Hyde et al. [50] from submerged stems of Typha latifolia in freshwater environments from UK. Neoxylochrysis is phylogenetically related to Pseudoconiothyrium in the phylogenetic tree and formed an independent clade separated from Pseudocoleophoma. The asexual morph of Neoxylochrysis differs from the members of Pseudocoleophoma by having small, hyaline, oval-shaped, smooth, thin-walled, septate conidia with rounded or obtuse ends and guttules in each cell [50]. A comparison of the asexual morph of Neoxylochrysis with members of Pseudocoleophoma and intuitive photos of conidia are provided in Table 2. Since the sexual morph of Neoxylochrysis has not been discovered, it is impossible to compare the sexual morphologies of Neoxylochrysis with that of Pseudocoleophoma. Therefore, a new genus, Neoxylochrysis, is introduced in Dictyosporiaceae to accommodate a single coelomycetous species, N. typhicola.

Table 2.
Comparison of asexual morphs in Pseudocoleophoma.
Neoxylochrysis typhicola (Kamolhan, Banmai, Boonmee, E.B.G. Jones and K.D. Hyde) N. Wu and Jian K. Liu, comb. nov. (Figure 8).
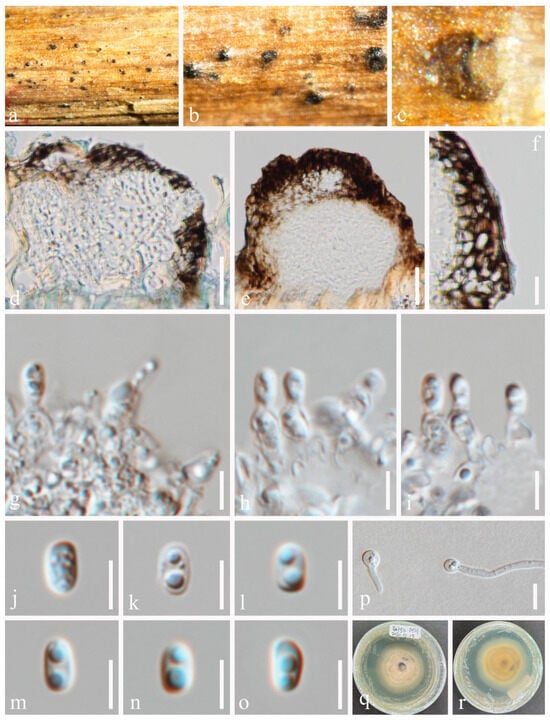
Figure 8.
Neoxylochrysis typhicola (HKAS 131316). (a–c) Appearance of black conidiomata. (d,e) Vertical section of conidiomata. (f) Section of the peridium. (g–i) Conidiogenous cells and developing conidia. (j–o) Conidia. (p) Germinated conidium. (q,r) Colony on PDA, from above (q), from below (r). Scale bars: (d,e) = 20 μm, (f) = 10 μm, (g–o) = 5 μm, (p) = 10 μm.
Index Fungorum number: IF902298; Faces of Fungi number: FoF16019
Basionym: Pseudocoleophoma typhicola Kamolhan, Banmai, Boonmee, E.B.G. Jones and K.D. Hyde, in Hyde et al., Fungal Diversity 80: 34 (2016) [50].
Saprobic on host plants, such as Rhaphiolepis indica and Typha latifolia. Sexual morph: Not observed. Asexual morph: Conidiomata 90–115 μm high × 130–150 μm diam. (x = 105 × 135 μm, n = 10), semi-immersed or immersed, visible as black shiny dots on the host, subcuticular in origin, then becoming erumpent, subglobose, brown to black, unilocular, glabrous, ostiolate. Ostiole 50–75 μm long, 70–100 μm wide, centrally or laterally located. Conidiomatal wall 20–25 μm wide (x = 23 μm, n = 20), comprising 4–5 layers, dark brown, thick-walled cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 2.5–5.5 × 2–3 μm (x = 3.5 × 2.5 μm, n = 30), phialidic, smooth-walled, aseptate, hyaline. Conidia 7.5–11 × 2–3 μm (x = 10 × 2.5 μm, n = 30), hyaline, aseptate, oval, smooth, thin-walled, with 1(–2) guttules in each cell.
Culture characteristics: Conidia germinated on PDA within 12 h. Germ tubes produced from one end. Colonies on PDA reaching 30–35 mm diam. after two weeks at 25 °C in natural light. Mycelium superficial, grayish white from above, pale brown at the center, and yellowish white at the margin from below.
Material examined: UK, Hampshire, Swanick Lakes, on submerged stems of Typha latifolia in freshwater habitat, 28 August 2015, E.B.G. Jones, GJ190 (MFLU 16-0966, holotype; HKAS 94520 isotype), ex-type living culture, MFLUCC 16-0123, KUMCC 16-0007; China, Guizhou Province, Panzhou City, Wetland Park, 104°29′14′′ E, 25°39′33′′ N, on dead twigs of Rhaphiolepis indica in terrestrial habitat, 21 July 2022, Na Wu, YW354 (HKAS 131316), living culture, CGMCC 3.25688 = UESTCC 23.0221.
Notes: In our phylogeny (Figure 2), Neoxylochrysis typhicola (CGMCC 3.25688, MFLUCC 16-0123) clustered with Pseudocoleophoma puerensis (ZHKUCC 22-0204, ZHKUCC 22-0205) and Pseudoconiothyrium broussonetiae (CBS 145036), forming an independent clade in Dictyosporiaceae. This fungus was initially introduced by Hyde et al. [50] from submerged stems of Typha latifolia in freshwater, based on morphology and ITS and LSU sequence data. Neoxylochrysi typhicola can be distinguished from the above closely related species based on ITS and LSU base pair differences from P. puerensis by 76/557 bp (13.6%) in ITS and 17/841 bp (2.0%) in LSU, and P. broussonetiae by 55/557 bp (9.9%) in ITS, 15/841 bp (1.8%) in LSU. Neoxylochrysi is similar to Pseudoconiothyrium, Pseudocyclothyriella and Xylochrysis in their asexual morphs. However, Xylochrysis belongs to Woswasiaceae, whereas Neoxylochrysi belongs to Dictyosporiaceae. The genus Neoxylochrysi differs from Pseudoconiothyrium and Pseudocyclothyriella in having conidia that are hyaline, oval or oblong to cylindrical, rounded or obtuse ends, smooth, thin-walled, often with 2–4 small guttules in each cell, aseptate or 1-euseptate, not constricted at the septum, and without a sheath or appendages. The asexual morph of P. puerensis is undetermined and thus, the asexual morphologies of P. puerensis and N. typhicola could not be compared.
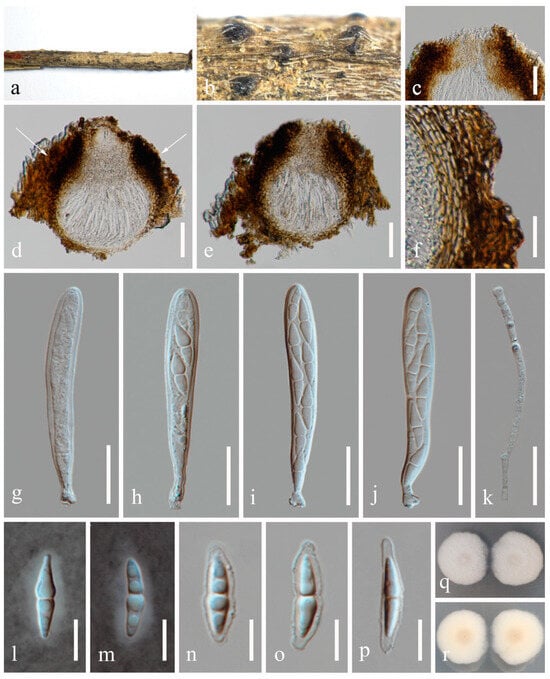
Figure 9.
Sexual morph of Pseudocoleophoma rosae (HKAS 131315, holotype). (a,b) Appearance of thallus and ascomata on the host surface. (c) Vertical section through an ostiole. (d,e) Vertical section through ascomata. (f) Structure of peridium. (g–j) Asci. (k) Pseudoparaphyses. (l–p) Ascospores, (l,m) in Indian ink showing the sheath. (q,r) Colonies on PDA, from above (q), from below (r). Scale bars: (c,f–k) = 20 μm, (d,e) = 50 μm, (l–p) = 10 μm.
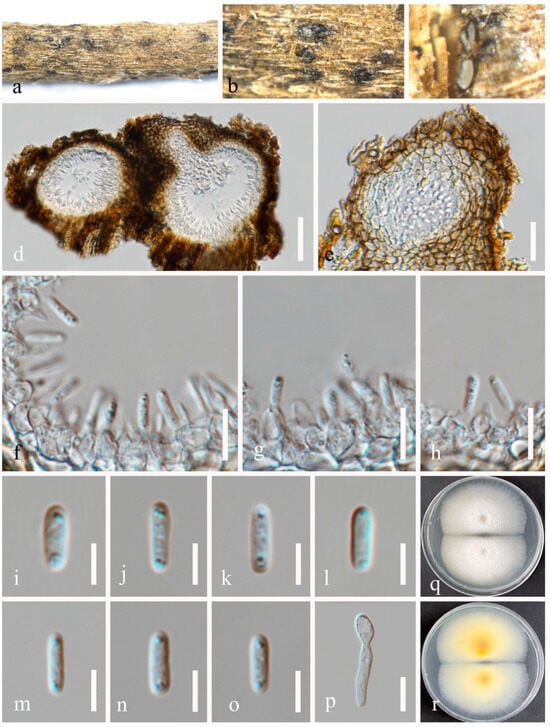
Figure 10.
Asexual morph of Pseudocoleophoma rosae (HUEST 24.0201). (a–c) Conidiomata on the host substrate. (d) Vertical section of conidiomata. (e) Structure of the peridium. (f–h) Conidiogenous cells and developing conidia. (i–o) Conidia. (p) Germinated conidium. (q,r) Colonies on PDA, from above (q), from below (r). Scale bars: (d) = 50 μm, (e) = 20 μm, (f–h) = 10 μm, (i–o) = 5 μm, (p) = 10 μm.
Index Fungorum number: IF902297; Faces of Fungi number: FoF16020
Etymology: Referring to the host genus Rosa from which the fungus was collected.
Holotype: HKAS 131315
Saprobic on dead twigs of Rosa roxbunghii Tratt. Sexual morph: Ascomata 170–210 μm high × 175–220 μm diam. (x = 196 × 188 μm, n = 20), semi-immersed or immersed in the substrate, dark brown, globose to subglobose, unilocular, glabrous, thick-walled, thickened at the apex, visible as black dots or papilla on the host, ostiolate. Ostiole neck central, 59–71 μm long, 49–71 μm wide. Peridium up to 12–35 μm wide, composed of thick-walled, dark brown to pale brown or hyaline cells of textura angularis. Hamathecium up to 2–3 μm wide, hyaline, septate, branched. Asci 55–70 × 7–10 μm (x = 65 × 8 μm, n = 20), 8-spored, cylindrical to clavate, some slightly curved, with an ocular chamber, short-stalked with club-shape pedicel. Ascospores 12–20 × 3–6 μm (x = 16 × 4 μm, n = 50), hyaline, smooth-walled, fusiform with acute ends, 1-septate, slightly constricted at the septum, occasionally 2–4-guttulate when young, surrounded by a mucilaginous sheath; 1–2 μm wide at sides, 2–3 μm at each end. Asexual morph: Coelomycetous. Conidiomata 100–184 μm high × 98–215 μm diam. (x = 140 × 148 μm, n = 10), dark brown to black, visible as black dots covered by epidermal tissues, solitary to gregarious, globose to subglobose, pyriform or irregular in shape, unilocular, glabrous, ostiolate. Ostiole 30–40 μm long, 30–50 μm wide, cylindrical, centrally or laterally located. Conidiomatal wall 8–20 μm wide (x = 15 μm, n = 20), composed of thick-walled, dark brown to hyaline cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells 3–5 × 4–7 μm (x = 4 × 6 μm, n = 30), phialidic, aseptate, smooth-walled, hyaline. Conidia 6–9 × 2–4 μm (x = 8 × 3 μm, n = 50), hyaline, aseptate, oblong to cylindrical, smooth, thin-walled, guttules concentrated to ends.
Culture characteristics: Ascospores and conidia germinated on PDA within 12 h. Colonies on PDA reaching 30–40 mm diam. after two weeks at 25 °C in natural light. Mycelium superficial, with regular margins, slightly raised, fluffy, moderate aerial mycelium on the surface, underneath pale yellow.
Material examined: China, Sichuan Province, Leshan City, Huangdan Town, Muchuan County, 28°49′56′′ N, 103°40′40′′ E, on dead twigs of Rosa roxbunghii, 30 October 2021, Na Wu, H54 (HKAS 131315, holotype), ex-type living culture CGMCC 3.25623 = UESTCC 23.0220; ibid., H245 (HUEST 24.0201), living culture UESTCC 24.0184.
Notes: We isolated the asexual and sexual morphs of Pseudocoleophoma rosae from the same substrate, Rosa roxbunghii. The phylogeny indicates that these two isolates are identical (Figure 2). The multi-locus phylogenetic result (Figure 2) clearly showed that these two isolates are identical. And these two isolates constitute a distinct lineage but clustered close to P. rhapidis and P. yunnanensis. The sexual morph of P. rosae differs from P. yunnanensis by having smaller ascomata (170–210 × 175–220 μm vs. 160–280 × 200–280 μm), asci (55–70 × 7–10 μm vs. 65–90 × 8–11 μm) and ascospores (12–20 × 3–6 μm vs. 16–26 × 4–8 μm). Ascospores of P. rosae have guttules when young that disappear at maturity. The asexual morph of P. rosae differs from P. rhapidis by its smaller conidiomata (100–184 × 98–215 μm vs. 150–225 × 225–300 μm), narrower conidiogenous cells (3–5 × 4–7 μm vs. 7–10 × 13–17 μm), and smaller conidia (6–9 × 2–4 μm vs. 20–25 × 10–15 μm). In addition, the conidia of P. rosae have guttules concentrated at the ends, while P. rhapidis lacks guttules. Therefore, we introduce P. rosae as a new species based on the morphology and phylogeny.
4. Discussion
Pseudocoleophoma is a holomorphic genus distributed in Asia and Europe. Members of the genus have been reported as saprobes on various hosts and substrates in freshwater and terrestrial habitats, with no records as pathogens or endophytes [9,15,16,17,18,19,20,50]. Five Pseudocoleophoma species (P. flavescens, P. paraphysoidea, P. rhapidis, P. rusci and P. zingiberacearum) have been reported as asexual morphs, while four species (P. guizhouensis, P. heteropanacicola, P. puerensis and P. yunnanensis) are described based solely on their sexual morphs. Only P. bauhiniae, P. calamagrostidis, P. polygonicola and P. rosae are holomorphic species in Pseudocoleophoma. Morphologically, P. calamagrostidis and P. polygonicola have longer and narrower conidia than those of P. typhicola, which are wider and more rounded (Table 2) [9]. The location, size and number of the guttules also differ. In addition, the conidia of P. calamagrostidis and P. polygonicola have two small guttules concentrated at the ends of each cell, whereas P. typhicola has 2–4 large guttules filling the interior of the conidia. Also, the conidia of P. typhicola are septate or aseptate, whereas those of P. calamagrostidis and P. polygonicola remain aseptate throughout their entire growth cycle [9,50]. These characteristics have been confirmed in species, viz., P. bauhiniae, P. rusci and P. zingiberacearum, and their conidia are not similar to P. typhicola, but their morphology is similar to the type species, P. calamagrostidis [15,18,19].
When comparing the morphology of asexual species in Pseudocoleophoma, we not only found that the characteristics of Neoxylochrysis typhicola are different from other asexual species of Pseudocoleophoma, but also found that the characteristics of P. flavescens and P. rhapidis are different from P. bauhiniae, P. calamagrostidis, P. polygonicola, P. rusci and P. zingiberacearum [9,15,17,18,19,20,50]. Detailed characteristics and conidial sizes are shown in Table 2. We speculate that these differences are due to host and geographical factors.
Pseudocyclothyriella clematidis was initially introduced into Pseudocoleophoma by Phukhamsakda et al. [21] and placed between P. calamagrostidis (KT 3284) and Neoxylochrysis typhicola based on phylogenetic analysis. Jiang et al. [13] transferred P. clematidis to Pseudocyclothyriella based on morphology and multi-locus phylogenetic analysis. We observed that N. typhicola (MFLUCC 16-0123) and Pseudoconiothyrium broussonetiae (CBS 145036) also clustered in phylogenetic trees presented in two previous publications [6,27], separated from Pseudocoleophoma. As there were no fresh specimens, previous studies did not address the phylogenetic issues with N. typhicola. The results mentioned above coincide with our research. Fortunately, we re-collected N. typhicola from Rhaphiolepis indica (Rosaceae) in a terrestrial habitat. In morphology, our sample fits with the description of the N. typhicola holotype provided by Hyde et al. [50]. And our collection is phylogenetically identical to N. typhicola. The addition of fresh samples also allowed us to analyze the genus Pseudocoleophoma again, which confirmed that N. typhicola and Pse. broussonetiae clustered and solved issues related to the phylogeny of N. typhicola.
Pseudocoleophoma puerensis was introduced by Lu et al. [16] from a decaying branch of Coffea arabica var. catimor in China. Phylogenetic analysis showed that two strains of P. puerensis formed a distinct clade basal to all Pseudocoleophoma members, and it clustered with Neoxylochrysis typhicola [16]. Pseudocoleophoma puerensis only has a sexual morph, and it differs from P. bauhiniae, P. calamagrostidis, P. polygonicola and P. yunnanensis (Table 3). Pseudocoleophoma puerensis does not belong to Pseudocoleophoma, but as no fresh samples of this species were collected, we retained it within the genus Pseudocoleophoma until new specimens are found.

Table 3.
Comparison of sexual morphs in Pseudocoleophoma.
The Dictyosporiaceae species investigated in this study were collected from ten different medicinal plants in Guizhou and Sichuan Provinces, China. These regions are characterized by a sub-tropical climate with favorable temperature and humidity conditions that support the growth of medicinal plants [51,52]. Fungal interactions with medicinal plants, including those involving endophytes [53,54], pathogens [55,56], and saprobes [57,58], are known to influence plant health and development. All of the taxa identified in this study were saprophytic fungi, and there is no evidence to indicate they negatively affect their host plants. In addition, our study suggests that the abundance of medicinal plants not only provides favorable conditions for the discovery of more microfungi, but also contributes to the diversity of microfungi associated with medicinal plants, offering a valuable reference for future studies on how fungi affect the potential medicinal value of these plants.
Author Contributions
Conceptualization, N.W. and J.-K.L.; methodology, N.W. and H.-Z.D.; formal analysis, N.W. and H.-Z.D.; resources, N.W.; data curation, N.W.; writing—original draft preparation, N.W., K.W.T.C. and H.-Z.D.; writing—review and editing, K.W.T.C., S.S.N.M., K.K., J.-K.L. and K.D.H.; supervision, J.-K.L.; project administration, J.-K.L.; funding acquisition, J.-K.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Science and Technology Fundamental Resources Investigation Program (Grant No. 2021FY100906).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequence data are available in NCBI GenBank with the accession numbers given in the manuscript.
Acknowledgments
Shaun Pennycook (Manaaki Whenua-Landcare Research, New Zealand) is thanked for corrections to the Latin names of the novel taxa. Na Wu thanks Rui-Ru Liang and Ya-Ru Sun for their help with sample collections. Na Wu also acknowledges Mae Fah Luang University for financial support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Liu, J.K.; Hyde, K.D.; Jones, E.B.G.; Ariyawansa, H.A.; Bhat, D.J.; Boonmee, S.; Maharachchikumbura, S.S.N.; McKenzie, E.H.C.; Phookamsak, R.; Phukhamsakda, C.; et al. Fungal diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 2015, 72, 1–197. [Google Scholar] [CrossRef]
- Boonmee, S.; D’souza, M.J.; Luo, Z.L.; Pinruan, U.; Tanaka, K.; Su, H.Y.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Taylor, J.E.; et al. Dictyosporiaceae fam. nov. Fungal Divers. 2016, 80, 457–482. [Google Scholar] [CrossRef]
- Kirk, P.M. New or interesting microfungi II. Dematiaceous hyphomycetes from Esher Common, Surrey. Trans. Br. Mycol. Soc. 1981, 77, 279–297. [Google Scholar] [CrossRef]
- Cai, L.; Guo, X.Y.; Hyde, K.D. Morphological and molecular characterisation of a new anamorphic genus Cheirosporium from freshwater in China. Persoonia 2008, 20, 53–58. [Google Scholar] [CrossRef]
- Li, W.L.; Luo, Z.L.; Liu, J.K.; Bhat, D.J.; Bao, D.F.; Su, H.Y.; Hyde, K.D. Lignicolous freshwater fungi from China I: Aquadictyospora lignicola gen. et sp. nov. and new record of Pseudodictyosporium wauense from northwestern Yunnan Province. Mycosphere 2017, 8, 1587–1597. [Google Scholar] [CrossRef]
- Shen, H.W.; Bao, D.F.; Wanasinghe, D.N.; Boonmee, S.; Liu, J.K.; Luo, Z.L. Novel species and records of Dictyosporiaceae from freshwater habitats in China and Thailand. J. Fungi 2022, 8, 1200. [Google Scholar] [CrossRef]
- Tian, W.H.; Chen, Y.P.; Maharachchikumbura, S.S.N. Neodigitodesmium, a novel genus of family Dictyosporiaceae from Sichuan Province, China. Phytotaxa 2022, 559, 176–184. [Google Scholar] [CrossRef]
- Barr, M.E. New taxa and combinations in the Loculoascomycetes. Mycotaxon 1987, 29, 501–505. [Google Scholar]
- Tanaka, K.; Hirayama, K.; Yonezawa, H.; Sato, G.; Toriyabe, A.; Kudo, H.; Hashimoto, A.; Matsumura, M.; Harada, Y.; Kurihara, Y.; et al. Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud. Mycol. 2015, 82, 75–136. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Schumache, R.K.; Akulov, A.; Thangavel, R.; Hernández-Restrepo, M.; Carnegie, A.J.; Cheewangkoon, R.; Wingfeld, M.J.; Summerel, B.A.; Quaedvlieg, W.; et al. New and interesting fungi. 2. Fungal Syst. Evol. 2019, 3, 57–134. [Google Scholar] [CrossRef]
- Piątek, M.; Rodriguez-Flakus, P.; Domic, A.; Palabral-Aguilera, A.N.; Gómez, M.I.; Flakus, A. Phylogenetic placement of Leptosphaeria polylepidis, a pathogen of Andean endemic Polylepis tarapacana, and its newly discovered mycoparasite Sajamaea mycophila gen. et sp. nov. Mycol. Prog. 2020, 19, 1–14. [Google Scholar] [CrossRef]
- Atienza, V.; Hawksworth, D.L.; Pérez-Ortega, S. Verrucoccum (Dothideomycetes, Dictyosporiaceae), a new genus of lichenicolous fungi on Lobaria s. lat. for the Dothidea hymeniicola species complex. Mycologia 2021, 113, 1233–1252. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.B.; Jeewon, R.; Karunarathna, S.C.; Phukhamsakda, C.; Doilom, M.; Kakumyan, P.; Suwannarach, N.; Phookamsak, R.; Lumyong, S. Reappraisal of Immotthia in Dictyosporiaceae, Pleosporales: Introducing Immotthia bambusae sp. nov. and Pseudocyclothyriella clematidis comb. et gen. nov. based on morphology and phylogeny. Front. Microbiol. 2021, 12, 656235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.F.; Liu, J.K.; Hyde, K.D.; Chen, Y.Y.; Ran, H.Y.; Liu, Z.Y. Ascomycetes from karst landscapes of Guizhou Province, China. Fungal Divers. 2023, 122, 1–160. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Jeewon, R.; Phillips, A.J.L.; Bhat, D.J.; Wanasinghe, D.N.; Liu, J.K.; Lu, Y.Z.; et al. Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 2019, 10, 1–186. [Google Scholar] [CrossRef]
- Lu, L.; Karunarathna, S.C.; Dai, D.Q.; Xiong, Y.R.; Suwannarach, N.; Stephenson, S.L.; Elgorban, A.M.; Al-Rejaie, S.; Jayawardena, R.S.; Tibpromma, S. Description of four novel species in Pleosporales associated with coffee in Yunnan, China. J. Fungi 2022, 8, 1113. [Google Scholar] [CrossRef] [PubMed]
- Boerema, G.H.; de Gruyter, J.; Noordeloos, M.E.; Hamers, M.E.C. Phoma Identification Manual: Differentiation of Specific and Infra-specific Taxa in Culture; CABI: Wallingford, UK, 2004; p. 470. [Google Scholar]
- Tennakoon, D.S.; Bhat, D.J.; Kuo, C.H.; Hyde, K.D. Leaf litter saprobic Dictyosporiaceae (Pleosporales, Dothideomycetes): Pseudocoleophoma zingiberacearum sp. nov. from Hedychium coronarium. Kavaka 2019, 53, 1–7. [Google Scholar] [CrossRef]
- Li, W.J.; McKenzie, E.H.C.; Liu, J.K.; Bhat, D.J.; Dai, D.Q.; Camporesi, E.; Tian, Q.; Maharachchikumbura, S.S.N.; Luo, Z.L.; Shang, Q.J.; et al. Taxonomy and phylogeny of hyaline-spored coelomycetes. Fungal Divers. 2020, 100, 279–801. [Google Scholar] [CrossRef]
- Kularathnage, N.D.; Wanasinghe, D.N.; Senanayake, I.C.; Yang, Y.H.; Manawasinghe, I.S.; Phillips, A.J.L.; Hyde, K.D.; Dong, W.; Song, J.G. Microfungi associated with ornamental palms: Byssosphaeria phoenicis sp. nov. (Melanommataceae) and Pseudocoleophoma rhapidis sp. nov. (Dictyosporiaceae) from south China. Phytotaxa 2022, 568, 149–169. [Google Scholar] [CrossRef]
- Phukhamsakda, C.; McKenzie, E.H.C.; Phillips, A.J.L.; Gareth Jones, E.B.; Jayarama Bhat, D.; Stadler, M.; Bhunjun, C.S.; Wanasinghe, D.N.; Thongbai, B.; Camporesi, E.; et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020, 102, 1–203. [Google Scholar] [CrossRef]
- Ranojevic, N. Dritter Beitrag zur Pilzflora Serbiens. Ann. Mycol. 1914, 12, 393–421. [Google Scholar]
- Ellis, M.B. Dematiaceous Hyphomycetes; CABI: Wallingford, UK, 1971. [Google Scholar]
- Crous, P.W.; Shivas, R.G.; Quaedvlieg, W.; van der Bank, M.; Zhang, Y.; Summerell, B.A.; Guarro, J.; Wingfield, M.J.; Wood, A.R.; Alfenas, A.C.; et al. Fungal Planet description sheets: 214–280. Persoonia 2014, 32, 184–306. [Google Scholar] [CrossRef]
- Prasher, I.B.; Verma, R.K. Two new species of Dictyosporium from India. Phytotaxa 2015, 204, 193–202. [Google Scholar] [CrossRef]
- Silva, C.R.; Gusmão, L.F.P.; Castaneda-Ruiz, R.F. Dictyosporium amoenum sp. nov. from Chapada Diamantina, Bahia, Brazil. Mycotaxon 2016, 130, 1125–1133. [Google Scholar] [CrossRef]
- Tennakoon, D.S.; de Silva, N.I.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Kumla, J.; Suwannarach, N.; Lumyong, S. Exploring more on Dictyosporiaceae: The species geographical distribution and intriguing novel additions from plant litter. Diversity 2023, 15, 410. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Chomnunti, P.; Hongsanan, S.; Hudson, B.A.; Tian, Q.; Peršoh, D.; Dhami, M.K.; Alias, A.S.; Xu, J.C.; Liu, X.Z.; Stadler, M.; et al. The sooty moulds. Fungal Divers. 2014, 66, 1–36. [Google Scholar] [CrossRef]
- Rayner, R.W. A Mycological Colour Chart; Commonwealth Mycological Institute and British Mycological Society: Kew, UK, 1970. [Google Scholar]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Wu, Z.H.; Wang, T.H.; Huang, W.; Qu, Y.B. A simplified method for chromosome DNA preparation from filamentous fungi. Mycosystema 2001, 20, 575–577. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- GenBank. Available online: http://www.ncbi.nlm.nih.gov (accessed on 12 July 2023).
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, A.J.; Bhunjun, C.S.; Maharachchikumbura, S.S.N.; Liu, J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 2020, 11, 2653–2677. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest (Version 2.2); Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Silvestro, D.; Michalak, I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2012, 12, 335–337. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree 1.4.4. 2014. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 11 July 2024).
- Figshare. Available online: https://figshare.com/account/home#/data (accessed on 23 June 2024).
- Reisinger, O. Remarques sur les genres Dendryphiella et Dendryphion. Bull. De La Société Mycol. De Fr. 1968, 84, 27–51. [Google Scholar]
- Dela Cruz, T.; Edison, E. Marine Dendryphiella Species from Different Geographical Locations: An Integrated, Polyphasic Approach to Its Taxonomy and Physioecology. 2006. Available online: https://nbn-resolving.org/urn:nbn:de:gbv:084-10363 (accessed on 14 October 2020).
- Chethana, K.W.T.; Niranjan, M.; Dong, W.; Samarakoon, M.C.; Bao, D.F.; Calabon, M.S.; Chaiwan, N.; Chuankid, B.; Dayarathne, M.C.; de Silva, N.I.; et al. AJOM new records and collections of fungi: 101–150. Asian J. Mycol. 2021, 4, 113–260. [Google Scholar] [CrossRef]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, M.; Bussmann, W.R.; Liu, H.M.; Liu, Y.Y.; Peng, Y.D.; Zu, K.L.; Zhao, Y.M.; Liu, Z.B.; Yu, S.X. Species richness and conservation gap analysis of karst areas: A case study of vascular plants from Guizhou, China. Glob. Ecol. Conserv. 2018, 16, e00460. [Google Scholar] [CrossRef]
- Shan, Z.J.; Ye, J.F.; Hao, D.C.; Xiao, P.G.; Chen, Z.D.; Lu, A.M. Distribution patterns and industry planning of commonly used traditional Chinese medicinal plants in China. Plant Divers. 2022, 44, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Z.; Huang, X.L.; Tong, B.L.; Wang, D.; Liu, J.M.; Liao, X.F.; Sun, Q.W. Effects of rhizosphere fungi on the chemical composition of fruits of the medicinal plant Cinnamomum migao endemic to southwestern China. BMC Microbiol. 2021, 21, 206. [Google Scholar] [CrossRef]
- Du, T.Y.; Karunarathna, S.C.; Zhang, X.; Dai, D.Q.; Mapook, A.; Suwannarach, N.; Xu, J.C.; Stephenson, S.L.; Elgorban, A.M.; Al-Rejaie, S.; et al. Endophytic fungi associated with Aquilaria sinensis (Agarwood) from China show antagonism against bacterial and fungal pathogens. J. Fungi 2022, 8, 1197. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.P.; Gao, H.; Qi, J.M.; Chen, M.C.; Tao, A.; Xu, J.T.; Dai, Z.G.; Su, J.G. Colletotrichum species associated with jute (Corchorus capsularis L.) anthracnose in southeastern China. Sci. Rep. 2016, 6, 25179. [Google Scholar] [CrossRef] [PubMed]
- Petrasch, S.; Knapp, S.J.; van Kan, J.A.L.; Blanco-Ulate, B. Grey mould of strawberry, a devastating disease caused by the ubiquitous necrotrophic fungal pathogen Botrytis cinerea. Mol. Plant Pathol. 2019, 20, 877–892. [Google Scholar] [CrossRef]
- Yang, H.Z.; He, R.Y.; Cui, Y.; Li, Y.; Ge, X. Saprophytic Bacillus accelerates the release of effective components in agarwood by degrading cellulose. Molecules 2022, 27, 1428. [Google Scholar] [CrossRef]
- Du, H.Z.; Lu, Y.H.; Cheewangkoon, R.; Liu, J.K. Morpho-phylogenetic evidence reveals novel species and new records of Nigrograna(Nigrogranaceae) associated with medicinal plants in Southwestern China. MycoKeys 2024, 110, 1–33. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).