Diversity of Lignicolous Freshwater Fungi from Yuanjiang River in Yunnan (China), with the Description of Four New Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Morphology
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Phylogenetic Analyses
3. Results
3.1. Taxonomy
3.1.1. Dothideomycetes O.E. Erikss & Wink
- Pleosporales Luttr. ex M.E. Barr
- Dictyosporiaceae Boonmee & K.D. Hyde
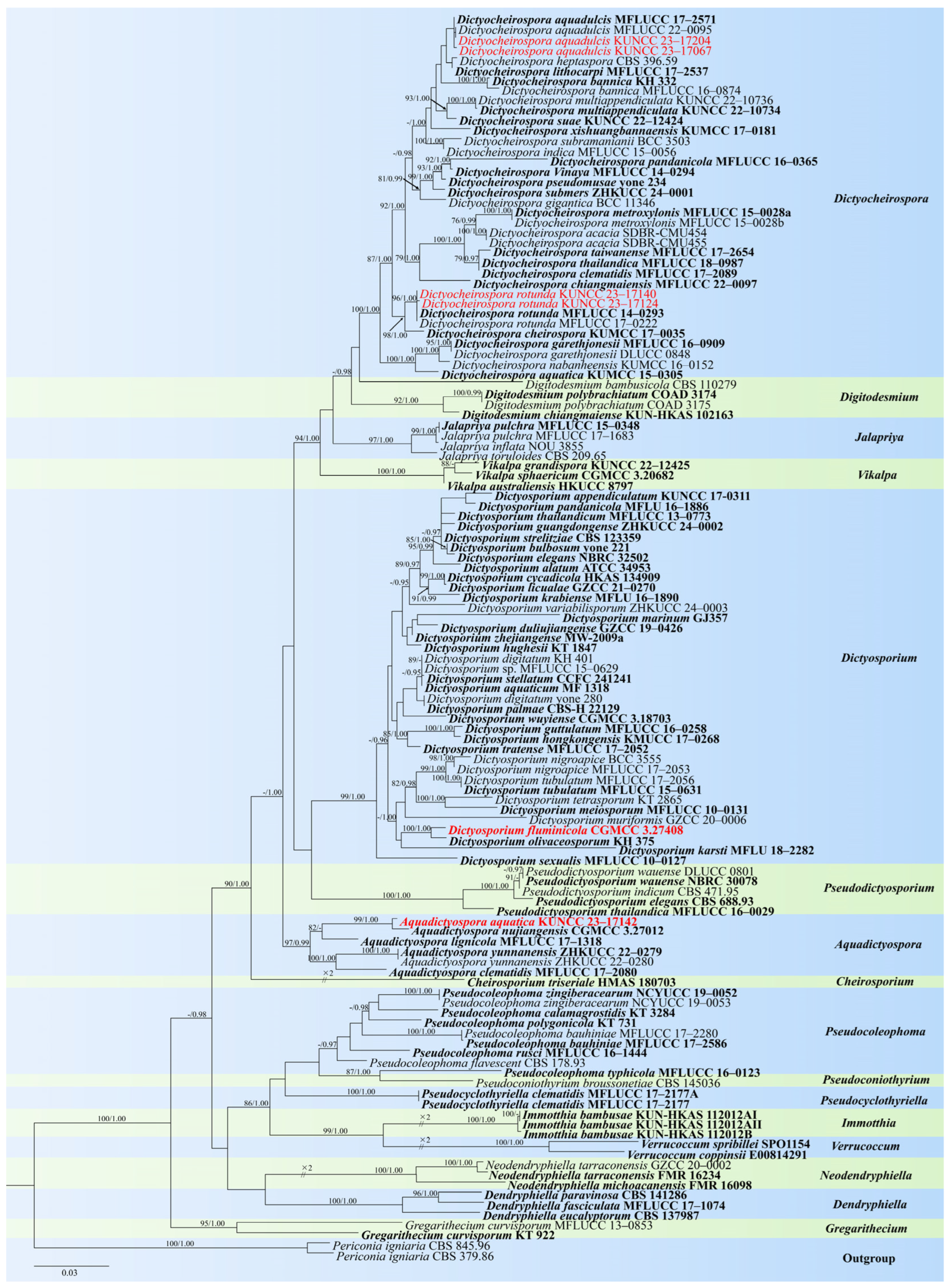
- Aquadictyospora Z.L. Luo, K.D. Hyde & H.Y. Su
- Aquadictyospora aquatica L. Zhang & Z.L. Luo, sp. nov., Figure 1

- Dictyocheirospora D’souza, Boonmee & K.D. Hyde


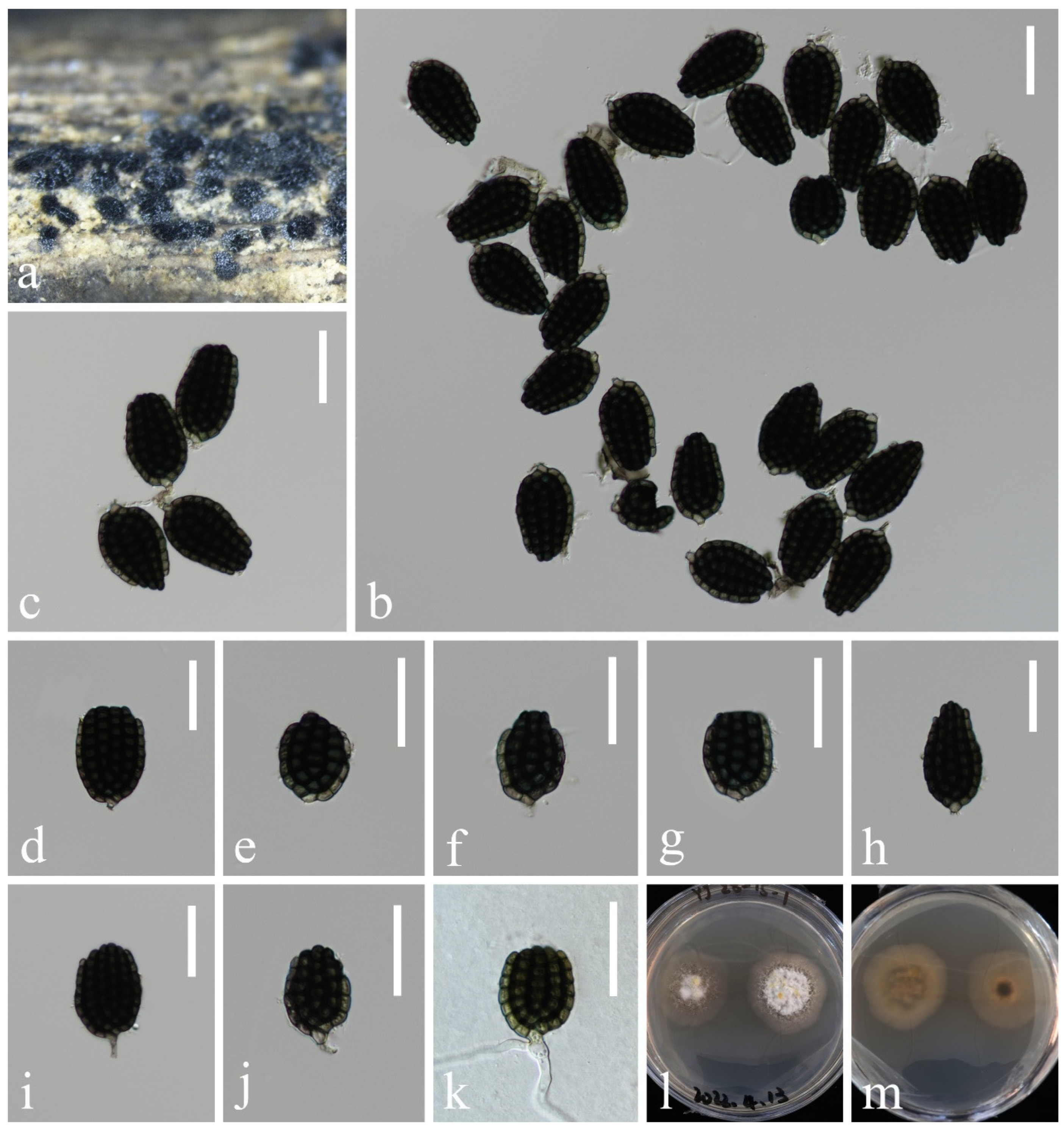
- Dictyocheirospora rotunda D’souza, Bhat & K.D. Hyde, Fungal Diversity 80: 465 (2016), Figure 5

- Dictyosporium Corda
- Dictyosporium fluminicola L. Zhang & Z.L. Luo, sp. nov., Figure 6
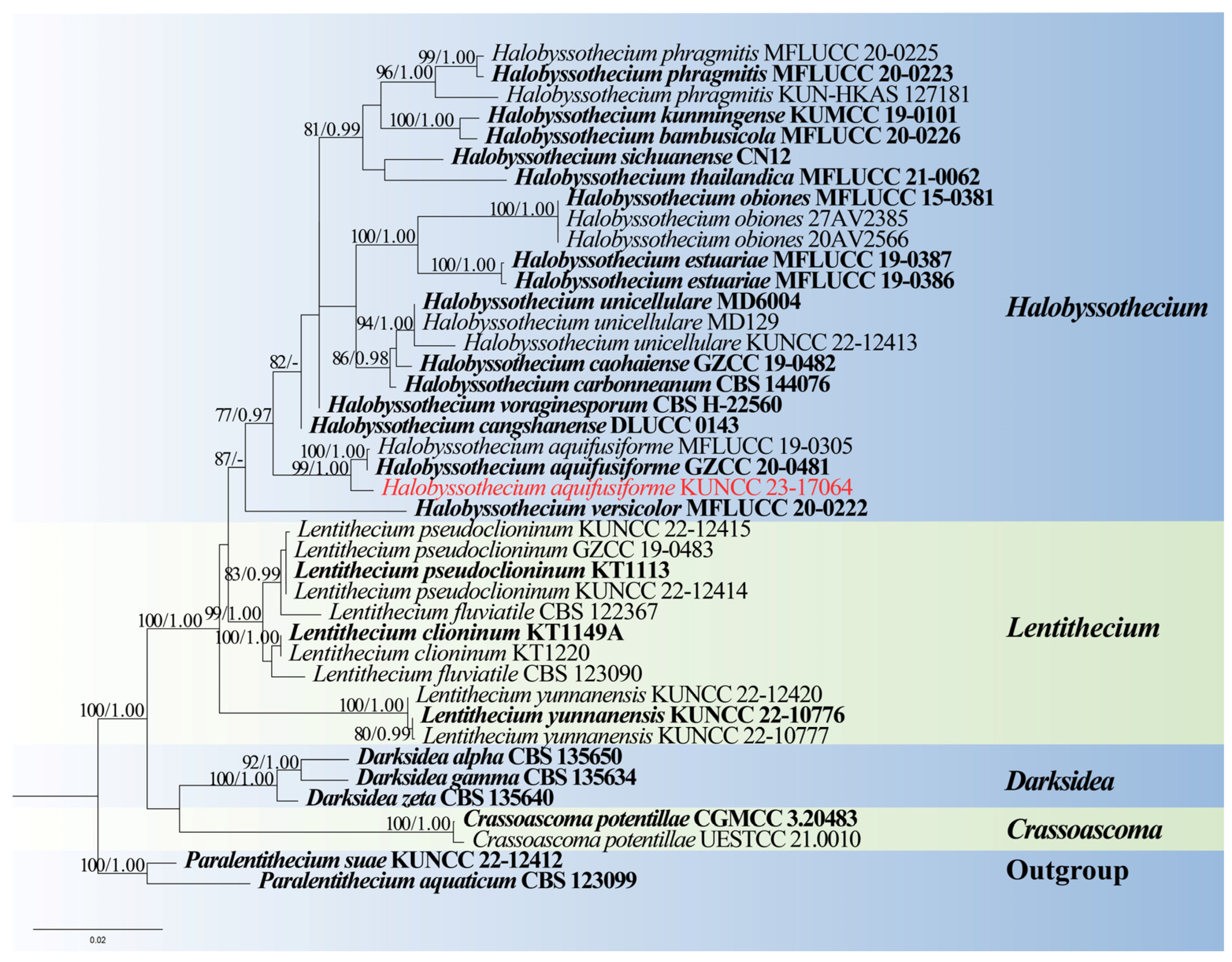
- Lentitheciaceae Y. Zhang, C.L. Schoch, J. Fourn, Crous & K.D. Hyde
- Halobyssothecium Dayarathne, E.B.G. Jones & K.D. Hyde
- Halobyssothecium aquifusiforme J. Yang, J.K. Liu & K.D. Hyde, Fungal Diversity 119: 39 (2023), Figure 7

3.1.2. Sordariomycetes O.E. Erikss. & Winka
- Myrmecridiales Crous
- Myrmecridiaceae Crous
- Myrmecridium Arzanlou, W. Gams & Crous
- Myrmecridium hydei Asghari, Phukhams & E.B.G. Jones, Phytotaxa 625: 265–279 (2023), Figure 9

- Myrmecridium submersum L. Zhang & Z.L. Luo, sp. nov., Figure 11

- Neomyrmecridium Crous
- Neomyrmecridium fusiforme L. Zhang & Z.L. Luo sp. nov., Figure 12

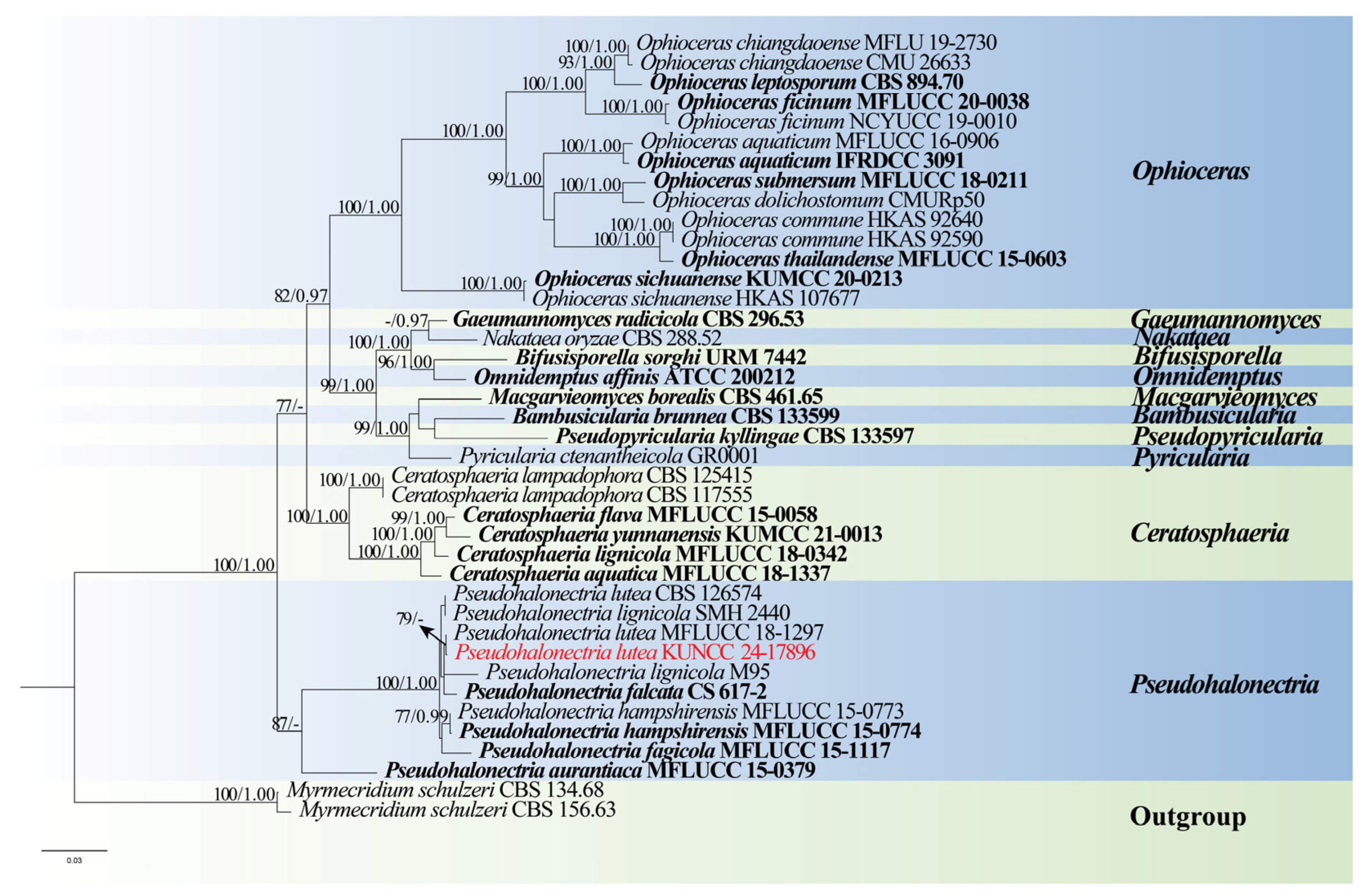
- Magnaporthales Thongk, Vijaykr & K.D. Hyde
- Pseudohalonectriaceae Hongsanan & K.D. Hyde
- Pseudohalonectria Minoura & T. Muroi
- Pseudohalonectria lutea Shearer, Can. J. Bot. 67: 1950 (1989), Figure 13

4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hyde, K.D.; Fryar, S.; Tian, Q.; Bahkali, A.H.; Xu, J.C. Lignicolous freshwater fungi along a northsouth latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecol. 2016, 19, 190–200. [Google Scholar] [CrossRef]
- Dong, W.; Wang, B.; Hyde, K.D.; McKenzie, E.H.C.; Raja, H.A.; Tanaka, K.; Abdel-Wahab, M.A.; Abdel-Aziz, F.A.; Doilom, M.; Phookamsak, R.; et al. Freshwater Dothideomycetes. Fungal Divers. 2020, 105, 319–575. [Google Scholar] [CrossRef]
- Koske, R.E.; Duncan, I.W. Temperature effects on growth, sporulation, and germination of some “aquatic” hyphomycetes. Can. J. Bot. 1973, 52, 1387–1391. [Google Scholar] [CrossRef]
- Zare-Maivan, H.; Shearer, C.A. Extracellular enzyme production and cell wall degradation by freshwater lignicolous fungi. Mycologia 1988, 80, 365–375. [Google Scholar] [CrossRef]
- Duarte, S.; Fernandes, I.; Nogueira, M.J.; Cássio, F.; Pascoal, C. Temperature alters interspecific relationships among aquatic fungi. Fungal Ecol. 2013, 6, 187–191. [Google Scholar] [CrossRef]
- Hibbett, D.; Abarenkov, K.; Kõljalg, U.; Öpik, M.; Chai, B.; Cole, J.; Wang, Q.; Crous, P.; Robert, V.; Helgason, T.; et al. Sequence-based classification and identification of Fungi. Mycologia 2016, 108, 1049–1068. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Nimalrathna, T.S.; Xian, L.Q.; Faraj, T.K.; Xu, J.C.; Mortimer, P.E. Taxonomic novelties and global biogeography of Montagnula (Ascomycota, Didymosphaeriaceae). MycoKeys 2024, 101, 191–232. [Google Scholar] [CrossRef]
- Calabon, M.S.; Hyde, K.D.; Jones, E.B.G.; Luo, Z.L.; Dong, W.; Hurdeal, V.G.; Gentekaki, E.; Rossi, W.; Leonardi, M.; Thiyagaraja, V.; et al. Freshwater fungal numbers. Fungal Divers. 2022, 114, 3–235. [Google Scholar] [CrossRef]
- Calabon, M.S.; Hyde, K.D.; Jones, E.B.G.; Bao, D.F.; Bhunjun, C.S.; Phukhamsakda, C.; Shen, H.W.; Gentekaki, E.; Al Sharie, A.H.; Barros, J.L.; et al. Freshwater fungal biology. Mycosphere 2023, 14, 195–413. [Google Scholar] [CrossRef]
- Luo, Z.L.; Hyde, K.D.; Bhat, D.J.; Jeewon, R.; Maharachchikumbura, S.S.N.; Bao, D.F.; Li, W.L.; Su, X.J.; Yang, X.Y.; Su, H.Y. Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycol. Prog. 2018, 17, 511–530. [Google Scholar] [CrossRef]
- Bao, D.F.; Hyde, K.D.; McKenzie, E.H.C.; Jeewon, R.; Su, H.Y.; Nalumpang, S.; Luo, Z.L. Biodiversity of lignicolous freshwater hyphomycetes from China and Thailand and description of sixteen species. J. Fungi 2021, 7, 669. [Google Scholar] [CrossRef]
- Bao, D.F.; Bhat, D.J.; Boonmee, S.; Hyde, K.D.; Luo, Z.L.; Nalumpang, S. Lignicolous freshwater ascomycetes from Thailand: Introducing Dematipyriformamuriformis sp. nov., one new combination and two new records in Pleurotheciaceae. MycoKeys 2022, 93, 57–79. [Google Scholar] [CrossRef]
- Bao, D.F.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Perera, R.H.; Thiyagaraja, V.; Hongsanan, S.; Wanasinghe, D.N.; Shen, H.W.; Tian, X.G.; Yang, L.Q.; et al. Taxonomy, phylogeny and evolution of freshwater Hypocreomycetidae (Sordariomycetes). Fungal Divers. 2023, 121, 1–94. [Google Scholar] [CrossRef]
- Li, L.L.; Shen, H.W.; Bao, D.F.; Wanasinghe, D.N.; Lu, Y.Z.; Feng, Y.; Luo, Z.L. The plethora of Tubeufiaceae in lakes of the northwestern Yunnan plateau, China. Front. Microbiol. 2022, 13, 1056669. [Google Scholar] [CrossRef]
- Shen, H.W.; Bao, D.F.; Bhat, D.J.; Su, H.Y.; Luo, Z.L. Lignicolous freshwater fungi in Yunnan Province, China: An overview. Mycology 2022, 13, 119–132. [Google Scholar] [CrossRef]
- Shen, H.W.; Bao, D.F.; Wanasinghe, D.N.; Boonmee, S.; Liu, J.K.; Luo, Z.L. Novel species and records of Dictyosporiaceae from freshwater habitats in China and Thailand. J. Fungi. 2022, 8, 1200. [Google Scholar] [CrossRef]
- Shen, H.W.; Bao, D.F.; Boonmee, S.; Lu, Y.Z.; Su, X.J.; Li, Y.X.; Luo, Z.L. Diversity of Distoseptispora (Distoseptisporaceae) taxa on submerged decaying wood from the Red River in Yunnan, China. MycoKeys 2024, 102, 1–28. [Google Scholar] [CrossRef]
- Zhang, L.; Shen, H.W.; Bao, D.F.; Li, L.L.; Su, X.J.; Luo, Z.L. Wongia suae sp. nov., a lignicolous freshwater fungus from Yuanjiang (Red River) Basin, China. Phytotaxa 2023, 616, 258–268. [Google Scholar] [CrossRef]
- Haruyama, S. Geomorphic Environment of Tonkin Delta; The Study of International Relations; Tsuda College: Tokyo, Japan, 1995; Volume 21, pp. 1–13. [Google Scholar]
- Van den Bergh, G.D.; Boer, W.; Schaapveld, M.A.; Duc, D.M.; Van Weering, T.C. Recent sedimentation and sediment accumulation rates of the Ba Lat prodelta (Red River, Vietnam). J. Asian Earth Sci. 2007, 29, 545–557. [Google Scholar] [CrossRef]
- Gu, Z.; Duan, X.; Shi, Y.; Li, Y.; Pan, X. Spatiotemporal variation in vegetation coverage and its response to climatic factors in the Red River Basin, China. Ecol. Indic. 2018, 93, 54–64. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Wanasinghe, D.N. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Chethana, K.W.T.; Manawasinghe, I.S.; Hurdeal, V.G.; Bhunjun, C.S.; Appadoo, M.A.; Gentekaki, E. What are fungal species and how to delineate them? Fungal Divers 2021, 109, 1–25. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal dna from several cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal rna genes for phylogenetics. PCR Protoc. 1990, 18, 315–322. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. A leaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web-servers. Syst. Biol. 2008, 75, 758–771. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest v2 Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Rannala, B.; Yang, Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996, 43, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.K.; Phookamsak, R.; Doilom, M.; Wikee, S.; Li, Y.M.; Ariyawansha, H.; Boonmee, S.; Chomnunti, P.; Dai, D.Q.; Bhat, D.J.; et al. Towards a natural classification of Botryosphaeriales. Fungal Divers. 2012, 57, 149–210. [Google Scholar] [CrossRef]
- Cai, L.; Jeewon, R.; Hyde, K.D. Phylogenetic investigations of Sordariaceae based on multiple gene sequences and morphology. Mycol. Res. 2006, 110, 137–150. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree 1.4.2. 2014. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 9 July 2014).
- Boonmee, S.; D’souza, M.J.; Luo, Z.L.; Pinruan, U.; Tanaka, K.; Su, H.Y.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Taylor, J.E.; et al. Dictyosporiaceae fam. nov. Fungal Divers. 2016, 80, 457–482. [Google Scholar] [CrossRef]
- Kodsueb, R.; Lumyong, S.; Ho, W.H.; Hyde, K.D.; Mckenzie, E.H.; Jeewon, R. Morphological and molecular characterization of Aquaticheirospora and phylogenetics of Massarinaceae (Pleosporales). Bot. J. Linn. Soc. 2007, 155, 283–296. [Google Scholar] [CrossRef]
- Cai, L.; Guo, X.Y.; Hyde, K.D. Morphological and molecular characterisation of a new anamorphic genus Cheirosporium from freshwater in China. Persoonia 2008, 20, 53–58. [Google Scholar] [CrossRef]
- Li, W.L.; Luo, Z.L.; Liu, J.K.; Bhat, D.J.; Bao, D.F.; Su, H.Y.; Hyde, K.D. Lignicolous freshwater fungi from China I: Aquadictyospora lignicola gen. et sp. nov. and new record of Pseudodictyosporium wauense from northwestern Yunnan Province. Mycosphere 2017, 8, 1587–1597. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; McKenzie, E.H.C.; Bhat, D.J.; Phillips, A.J.L.; Wanasinghe, D.N.; Samarakoon, M.C.; Jayawardena, R.S.; Dissanayake, A.J.; Tennakoon, D.S.; et al. Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Divers. 2018, 92, 1–160. [Google Scholar] [CrossRef]
- Yang, J.; Liu, J.K.; Hyde, K.D.; Jones, E.B.G.; Liu, Z.Y. New species in Dictyosporium, new combinations in Dictyocheirospora and an updated backbone tree for Dictyosporiaceae. MycoKeys 2018, 36, 83–105. [Google Scholar] [CrossRef]
- Yang, J.; Liu, L.L.; Jones, E.B.G.; Hyde, K.D.; Liu, Z.Y.; Bao, D.F.; Liu, N.G.; Li, W.L.; Shen, H.W.; Yu, X.D.; et al. Freshwater fungi from karst landscapes in China and Thailand. Fungal Divers. 2023, 119, 1–212. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Atienza, V.; Hawksworth, D.L.; Pérez-Ortega, S. Verrucoccum (Dothideomycetes, Dictyosporiaceae), a new genus of lichenicolous fungi on Lobaria s. lat. for the Dothidea hymeniicola species complex. Mycologia 2021, 113, 1233–1252. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.B.; Jeewon, R.; Karunarathna, S.C.; Phukhamsakda, C.; Doilom, M.; Kakumyan, P.; Suwannarach, N.; Phookamsak, R.; Lumyong, S. Reappraisal of Immotthia in Dictyosporiaceae, Pleosporales: Introducing Immotthia bambusae sp. nov. and Pseudocyclothyriella clematidis comb. et gen. nov. based on morphology and phylogeny. Front. Microbiol. 2021, 12, 656235. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.H.; Chen, Y.P.; Maharachchikumbura, S.S.N. Neodigitodesmium, a novel genus of family Dictyosporiaceae from Sichuan Province, China. Phytotaxa 2022, 559, 176–184. [Google Scholar] [CrossRef]
- Liu, L.L.; Song, L.C.; Gu, X.F.; Wei, Q.Q.; Zhang, M.; Liu, Z.Y.; Gou, J.L. Dictyosporium duliujiangense sp. nov. (Dictyosporiaceae, Pleosporales) from freshwater habitat in Guizhou Province, China. Phytotaxa 2023, 606, 259–272. [Google Scholar] [CrossRef]
- Shu, X.Y.; Doilom, M.; Boonmee, S.; Xu, B.; Dong, W. Three Novel Cheiroid Hyphomycetes in Dictyocheirospora and Dictyosporium (Dictyosporiaceae) from Freshwater Habitats in Guangdong and Guizhou Provinces, China. J. Fungi 2024, 10, 259. [Google Scholar] [CrossRef]
- Phukhamsakda, C.; McKenzie, E.H.C.; Phillips, A.J.L.; Jones, E.B.G.; Jayarama Bhat, D.; Stadler, M.; Bhunjun, C.S.; Wanasinghe, D.N.; Thongbai, B.; Camporesi, E.; et al. Microfungi associated with clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020, 102, 1–203. [Google Scholar] [CrossRef]
- Wang, W.P.; Luan, S.; Shen, H.W.; Liu, N.G.; Li, Y.X.; Luo, Z.L. Two new freshwater dematiaceous hyphomycetes of Pleosporales from Yunnan Province, China. Phytotaxa 2024, 676, 063–074. [Google Scholar] [CrossRef]
- Wang, R.X.; Luo, Z.L.; Hyde, K.D.; Bhat, D.J.; Su, X.J.; Su, H.Y. New species and records of Dictyocheirospora from submerged wood in north-western Yunnan, China. Mycosphere 2016, 7, 1357–1367. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Abreu, V.P.; Bazzicalupo, A.; Chethana, K.W.T.; Clericuzio, M.; Dayarathne, M.C.; Dissanayake, A.J.; Ekanayaka, A.H.; He, M.Q.; et al. Fungal Diversity notes 603–708: Taxonomic and phylogenetic notes on genera and species. Fungal Divers. 2017, 87, 1–235. [Google Scholar] [CrossRef]
- Hyde, K.D.; Danushka, S.; Tennakoon, D.S.; Jeewon, R.; Bhat, D.J.; Maharachchikumbura, S.S.N.; Rossi, W.; Leonardi, M.; Lee, H.B.; Mun, H.Y.; et al. Fungal Diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers 2019, 96, 1–242. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Jeewon, R.; Phillips, A.J.L.; Bhat, D.J.; Wanasinghe, D.N.; Liu, J.K.; Lu, Y.Z.; et al. Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 2019, 10, 1–186. [Google Scholar] [CrossRef]
- Phookamsak, R.; Hyde, K.D.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Raspé, O.; Karunarathna, S.C.; Wanasinghe, D.N.; Hongsanan, S.; et al. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019, 95, 1–273. [Google Scholar] [CrossRef]
- Sushma; Verma, R.K.; Prasher, I.B.; Gautam, A.K.; Rajeshkumar, K.C.; Castaneda-Ruiz, R.F. Dictyocheirospora himachalensis sp. nov. from Himachal Pradesh, India. Mycotaxon 2022, 137, 455–463. [Google Scholar] [CrossRef]
- Tennakoon, D.S.; de Silva, N.I.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Kumla, J.; Suwannarach, N.; Lumyong, S. Exploring more on Dictyosporiaceae: The species geographical distribution and intriguing novel additions from plant litter. Diversity 2023, 15, 410. [Google Scholar] [CrossRef]
- Long, C.S.; Wu, Y.P.; Zhang, X.; Lin, Y.; Shen, X.C.; Ma, J.; Li, Q.R. Additions to hyphomycetes from Yungui Plateau, China with three new species (Ascomycota, Sordariomycetes). Biodivers. Data J. 2023, 11, e101629. [Google Scholar] [CrossRef]
- Corda, A.J.C. Mykologische Beobachtungen. In Beitrage zur Gesammtem Natur-und Heilwissenschaften, 1st ed.; Weitenweber, W.R., Ed.; Commission bei Kronberger und Weber: Prague, Czech, 1836; Volume 1, pp. 80–88. [Google Scholar]
- Goh, T.; Hyde, K.; Ho, W.; Yanna. A revision of the genus Dictyosporium, with descriptions of three new species. Fungal Divers. 1999, 2, 65–100. [Google Scholar]
- Cai, L.; Zhang, K.Q.; McKenzie, E.H.C.; Hyde, K.D. Freshwater fungi from bamboo and wood submerged in the Liput River in the Philippines. Fungal Divers. 2003, 13, 1–12. [Google Scholar] [CrossRef]
- Whitton, S.R.; McKenzie, E.H.C.; Hyde, K.D. Fungi associated with Pandanaceae. Fungal Divers. Res. Ser. 2012, 21, 278–280. [Google Scholar]
- Silva, C.R.; Gusmão, L.F.P.; Castañeda-Ruiz, R.F. Dictyosporium amoenum sp. nov. from Chapada Diamantina, Bahia, Brazil. Mycotaxon 2015, 130, 1125–1133. [Google Scholar] [CrossRef]
- Dubey, R. Dictyosporium matherense sp. nov.: A new-fangled cheirosporous fungal species described from the Western Ghats of India. Asian J. 2022, 6, 1–8. [Google Scholar] [CrossRef]
- Zhang, J.F.; Liu, J.K.; Hyde, K.D.; Chen, Y.Y.; Ran, H.Y.; Liu, Z.Y. Ascomycetes from karst landscapes of Guizhou Province, China. Fungal Divers. 2023, 122, 1–160. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, H.K.; Fournier, J.; Crous, P.W.; Jeewon, R.; Pointing, S.B.; Hyde, K.D. Towards a phylogenetic clarification of Lophiostoma/Massarina and morphologically similar genera in the Pleosporales. Fungal Divers. 2009, 38, 225–251. [Google Scholar]
- Zhang, Y.; Crous, P.W.; Schoch, C.L.; Hyde, K.D. Pleosporales . Fungal Divers. 2012, 53, 1–221. [Google Scholar] [CrossRef]
- Hyde, K.D.; Jones, E.B.G.; Liu, J.K.; Ariyawansa, H.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.Q.; et al. Families of Dothideomycetes. Fungal Divers. 2013, 63, 1–313. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Jones, E.B.G.; Camporesi, E.; Boonmee, S.; Ariyawansa, H.A.; Wijayawardene, N.N.; Mortimer, P.E.; Xu, J.; Yang, J.B.; Hyde, K.D. An exciting novel member of Lentitheciaceae in Italy from Clematis vitalba. Cryptogam. Mycol. 2014, 35, 323–337. [Google Scholar] [CrossRef]
- Calabon, M.S.; Jones, E.B.G.; Hyde, K.D.; Boonmee, S.; Tibell, S.; Tibell, L.; Pang, K.L.; Phookamsak, R. Phylogenetic assessment and taxonomic revision of Halobyssothecium and Lentithecium (Lentitheciaceae, Pleosporales). Mycol. Prog. 2021, 20, 701–720. [Google Scholar] [CrossRef]
- Wijayawardene, N.; Hyde, K.; Dai, D.; Sánchez-García, M.; Goto, B.; Saxena, R.; Erdoğdu, M.; Selçuk, F.; Rajeshkumar, K.; Aptroot, A.; et al. Outline of fungi and fungus-like taxa—2021. Mycosphere 2022, 13, 53–453. [Google Scholar] [CrossRef]
- Liu, Z.P.; Zhang, S.N.; Cheewangkoon, R.; Zhao, Q.; Liu, J.K. Crassoascoma gen. nov. (Lentitheciaceae, Pleosporales): Unrevealing microfungi from the Qinghai-Tibet plateau in China. Diversity 2022, 14, 15. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, S.N.; Yu, X.D.; Liu, J.K. Pseudokeissleriella bambusicola gen. et sp. nov. (Lentitheciaceae, Pleosporales) from bamboos in Sichuan Province, China. Phytotaxa 2022, 560, 263–273. [Google Scholar] [CrossRef]
- Rajeshkumar, K.C.; Varma, R.K.; Sruthi, O.P.; Gautam, A.K.; Crous, P.W. Groenewaldia (Lentitheciaceae), a new corticolous fungal genus from India. Mycol. Prog. 2023, 22, 43. [Google Scholar] [CrossRef]
- Dayarathne, M.C.; Wanasinghe, D.N.; Jones, E.B.G.; Chomnunti, P.; Hyde, K.D. A novel marine genus, Halobyssothecium (Lentitheciaceae) and epitypifcation of Halobyssothecium obiones comb. nov. Mycol. Prog. 2018, 17, 1161–1171. [Google Scholar] [CrossRef]
- Hyde, K.D.; Suwannarach, N.; Jayawardena, R.S.; Manawasinghe, I.S.; Liao, C.; Doilom, M.; Cai, L.; Zhao, P.; Buyck, B.; Phukhamsakda, C.; et al. Mycosphere notes 325–344—Novel species and records of fungal taxa from around the world. Mycosphere 2021, 12, 1101–1156. [Google Scholar] [CrossRef]
- Arzanlou, M.; Groenewald, J.Z.; Gams, W.; Braun, U.; Shin, H.D.; Crous, P.W. Phylogenetic and morpho-taxonomic revision of Ramichloridium and allied genera. Stud. Mycol. 2007, 58, 57–93. [Google Scholar] [CrossRef]
- Crous, P.W.; Luangsa-ard, J.J.; Wingfield, M.J.; Carnegie, A.J.; Hernández-Restrepo, M.; Lombard, L.; Roux, J.; Barreto, R.W.; Baseia, I.G.; Cano-Lira, J.F.; et al. Fungal Planet description sheets: 785–867. Persoonia 2018, 41, 238–417. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfeld, M.J.; Guarro, J.; Hernández-Restrepo, M.; Sutton, D.A.; Acharya, K.; Barber, P.A.; Boekhout, T.; Dimitrov, R.A.; Dueñas, M.; et al. Fungal Planet description sheets: 320–370. Persoonia 2015, 34, 167–266. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Hardy, G.E.; Crane, C.; Barrett, S.; Cano-Lira, J.F.; Le Roux, J.J.; Thangavel, R.; Guarro, J.; et al. Fungal Planet description sheets: 469–557. Persoonia 2016, 37, 218–403. [Google Scholar] [CrossRef]
- Peintner, U.; Knapp, M.; Fleischer, V.; Walch, G.; Dresch, P. Myrmecridium hiemale sp. nov. from snow-covered alpine soil is the first eurypsychrophile in this genus of anamorphic fungi. Int. J. Syst. Evol. Microbiol. 2016, 66, 2592–2598. [Google Scholar] [CrossRef]
- Réblová, M.; Fournier, J.; Štepánek, V. Two new lineages of aquatic ascomycetes: Atractospora gen. nov. and Rubellisphaeria gen. et sp. nov., and a sexual morph of Myrmecridium montsegurinum sp. nov. Mycol. Prog. 2016, 15, 1–39. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; Jeewon, R.; Maharachchikumbura, S.S.N.; Liu, J.K.; Bhat, D.J.; Jones, E.B.G.; McKenzie, E.H.C.; Camporesi, E.; Bulgakov, T.S.; et al. Fungal diversity notes 491–602: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2017, 83, 1–261. [Google Scholar] [CrossRef]
- Jie, C.Y.; Zhou, Q.X.; Zhao, W.S.; Jiang, Y.L.; Hyde, K.D.; McKenzie, E.H.C.; Wang, Y. A new Myrmecridium species from Guizhou, China. Mycotaxon 2013, 124, 1–8. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Chooi, Y.H.; Gilchrist, C.L.M.; Lacey, E.; Pitt, J.I.; Roets, F.; Swart, W.J.; Cano-Lira, J.F.; ValenzuelaLopez, N.; et al. Fungal Planet description sheets: 1042–1111. Persoonia 2020, 44, 301–459. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Osieck, E.R.; Jurjevicì, Z.; Boers, J.; Van Iperen, A.L.; Starink-Willemse, M.; Dima, B.; Balashov, S.; Bulgakov, T.S.; Johnston, P.R.; et al. Fungal Planet description sheets: 1284–1382. Persoonia 2021, 47, 178–374. [Google Scholar] [CrossRef]
- Crous, P.W.; Boers, J.; Holdom, D.; Osieck; Steinrucken, T.V.; Tan, Y.P.; Vitelli, J.S.; Shivas, R.G.; Barrett, M.; Boxshall, A.G.; et al. Fungal Planet description sheets: 1383–1435. Persoonia 2022, 48, 261–371. [Google Scholar] [CrossRef]
- Serrano, L.; Sosa, D.; Magdama, F.; Espinoza, F.; Quevedo, A.; Vera, M.; Villavicencio, M.; Maridueña, G.; Pérez-Martinez, S.; Malosso, E.; et al. Neomyrmecridium asymmetricum sp. nov. from Ecuador. Mycotaxon 2020, 135, 151–165. [Google Scholar] [CrossRef]
- Asghari, R.; Phukhamsakda, C.; Karimi, O.; Apurillo, C.C.S.; Jones, E.B.G. Myrmecridium hydei, a novel marine species from Thailand. Phytotaxa 2023, 625, 265–279. [Google Scholar] [CrossRef]
- Tian, X.G.; Bao, D.F.; Karunarathna, S.C.; Jayawardena, R.S.; Hyde, K.D.; Bhat, D.J.; Luo, Z.L.; Elgorban, A.M.; Hongsanan, S.; Rajeshkumar, K.C.; et al. Taxonomy and phylogeny of ascomycetes associated with selected economically important monocotyledons in China and Thailand. Mycosphere 2024, 15, 1–274. [Google Scholar] [CrossRef]
- Tan, Y.P.; Steinrucken, T. Index of Australian Fungi No. 36; ResearchGate: Berlin, Germany, 2024. [Google Scholar] [CrossRef]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.; et al. Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef]
- Xu, R.J.; Dong, W.; Wei, D.P.; Zhao, Q.; Boonmee, S. Two new species in Neomyrmecridium and two new records in Myrmecridium (Myrmecridiaceae, Myrmecridiales) from the Tibetan Plateau, China. Curr. Res. Environ. Appl. Mycol. (J. Fungal Biol.) 2023, 13, 489–504. [Google Scholar] [CrossRef]
- Luo, Z.L.; Hyde, K.D.; Liu, J.K.; Maharachchikumbura, S.S.N.; Jeewon, R.; Bao, D.F.; Bhat, D.J.; Lin, C.G.; Li, W.L.; Yang, J.; et al. Freshwater Sordariomycetes. Fungal Divers. 2019, 99, 451–660. [Google Scholar] [CrossRef]
- Hongsanan, S.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Samarakoon, M.C.; Jeewon, R.; Zhao, Q.; Al-Sadi, A.M.; Bahkali, A.H. An updated phylogeny of Sordariomycetes based on phylogenetic and molecular clock evidence. Fungal Divers. 2017, 84, 25–41. [Google Scholar] [CrossRef]
- Asthana, A.; Shearer, C.A. Antagonistic activity of Pseudohalonectria and Ophioceras. Mycologia 1990, 82, 554–561. [Google Scholar] [CrossRef]
- Foremska, E.; Kostecki, M.; Chelkowski, J. Biosynthesis, preparation and properties of ascochitine. Acta Biotechnol. 1992, 12, 461–465. [Google Scholar] [CrossRef]
- Dong, J.Y.; Zhao, Z.X.; Cai, L.; Liu, S.Q.; Zhang, H.R.; Duan, M.; Zhang, K.Q. Nematicidal efect of freshwater fungal cultures against the pine-wood nematode, Bursaphelenchus xylophilus. Fungal Divers. 2004, 15, 123–133. [Google Scholar]
- Dong, J.Y.; Zhou, Y.P.; Li, R.; Zhou, W.; Li, L.; Zhu, Y.H.; Huang, R.; Zhang, K.Q. Newnematicidal azaphilones fromtheaquatic fungus Pseudohalonectria adversaria YMF1.01019. FEMS Microbiol. Lett. 2006, 264, 65–69. [Google Scholar] [CrossRef]
- Park, J.H.; Choi, G.J.; Jang, K.S.; Lim, H.K.; Kim, H.T.; Cho, K.Y.; Kim, J.C. Antifungal activity against plant pathogenic fungi of chaetoviridins isolated from Chaetomium globosum. FEMS Microbiol. Lett. 2005, 252, 309–313. [Google Scholar] [CrossRef]
- Minoura, K.; Muroi, T. Some freshwater ascomycetes from Japan. Trans. Mycol. Soc. Jpn. 1978, 19, 129–134. [Google Scholar]
- Shearer, C.A. Pseudohalonectria (Lasiosphaeriaceae), an antagonistic genus from wood in freshwater. Can. J. Bot. 1989, 67, 1944–1955. [Google Scholar] [CrossRef]
- Perera, R.H.; Maharachchikumbura, S.S.N.; Ariyawansa, H.; Bahkali, A.H.; Jones, E.B.G.; Al-Sadi, A.M.; Hyde, K.D.; Liu, Z.Y. Two new Pseudohalonectria species on beech cupules (Fagus sylvatica) and a new genus to accommodate P. suthepensis. Phytotaxa 2016, 278, 115–131. [Google Scholar] [CrossRef]
- Ono, Y.; Kobayashi, T. Notes on new and noteworthy plant-inhabiting fungi from Japan (1). Mycoscience 2001, 42, 439–446. [Google Scholar] [CrossRef]
- Huhndorf, S.M.; Greif, M.; Miller, A.N. Two new genera in the Magnaporthaceae, a new addition to Ceratosphaeria and two new species of Lentomitella. Mycologia 2008, 100, 940–955. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Goh, T.K. Fungi on submerged wood from the River Coln, England. Mycol. Res. 1999, 103, 1561–1574. [Google Scholar] [CrossRef]
- Ho, W.H.; Hyde, K.D.; Hodgkiss, I.J. Fungal communities on submerged wood from streams in Brunei, Hong Kong and Malaysia. Mycol. Res. 2001, 105, 1492–1501. [Google Scholar] [CrossRef]
- Ho, W.H.; Yanna; Hyde, K.D.; Hodgkiss, I.J. Seasonality and sequential occurrence of fungi on wood submerged in Tai Po Kau Forest Stream, Hong Kong. Fungal Divers. 2002, 10, 21–43. [Google Scholar]
- Hyde, K.D.; Taylor, J.E.; Fröhlich, J. Two new species of Pseudohalonectria from palms. Mycologia 1999, 91, 520–524. [Google Scholar] [CrossRef]
- Kohlmeyer, J.; Volkmann-Kohlmeyer, B.; Tsui, C.K.M. Fungi on Juncus roemerianus. 17. New ascomycetes and the hyphomycete genus Kolletes gen. nov. Bot. Mar. 2005, 48, 306–317. [Google Scholar] [CrossRef]
- Stern, D.L. The genetic causes of convergent evolution. Nat. Rev. Genet. 2013, 14, 751–764. [Google Scholar] [CrossRef]
- O’Brien, P.J. Catalytic promiscuity and the divergent evolution of DNA repair enzymes. Chem. Rev. 2006, 106, 720–752. [Google Scholar] [CrossRef]
- Hu, X.; Xiao, G.; Zheng, P.; Shang, Y.; Su, Y.; Zhang, X.; Liu, X.; Zhan, S.; St Leger, R.J.; Wang, C. Trajectory and genomic determinants of fungal-patho gen speciation and host adaptation. Proc. Natl. Acad. Sci. USA 2014, 111, 16796–16801. [Google Scholar] [CrossRef]
- Takaoka, S.; Kurata, M.; Harimoto, Y.; Hatta, R.; Yamamoto, M.; Akimitsu, K.; Tsuge, T. Complex regulation of secondary metabolism controlling pathogenicity in the phytopathogenic fungus Alternaria alternata. New Phytol. 2014, 202, 1297–1309. [Google Scholar] [CrossRef]

| Species | Conidiophores | Conidiogenous Cells | Conidia | Host | Distribution | References |
|---|---|---|---|---|---|---|
| Neomyrmecridium aquaticum | Macronematous, mononematous, erect, unbranched, multi-septate, 211–308 × 5–7 μm | Holoblastic, polyblastic, integrated, terminal | Obovoid, 3-septate, 14–16 × 4–6 μm | On decaying wood in freshwater habitat | China | [97] |
| N. fusiforme | Macronematous, mononematous, solitary, erect, unbranched, 214–258 × 4–5 μm | Polyblastic, terminal, integrated, subcylindrical | Fusiform, navicular to tapering and pointed at both ends, (0–)1–(3–)septate, 29–34 × 4–6 μm | On decaying wood in freshwater habitat | China | This study |
| N. guizhouense | Macronematous, mononematous, solitary, erect, unbranched, 75–140 × 2–4.5 μm | Polyblastic, terminal, integrated, subcylindrical, 2.2–4.3 μm | Fusoid-ellipsoid, (2–)3-septate, 8.9–12.7 × 2.8–4.8 μm | On decaying wood in freshwater habitat | China | [95] |
| N. naviculare | Macronematous, mononematous, erect, unbranched, 100–200 × 4–5.6 µm | Polyblastic, integrated, terminal, sympodial, denticulate | Navicular to fusiform, tapering to a hilum towards the base, (1–)3–septate, 16–24 × 5.5–7.5 µm, with a thin mucilaginous sheath | On decaying submerged wood in freshwater habitats | China | [45] |
| N. septatum | Solitary, erect, straight, unbranched, 1–4-septate, 40–70 × 4–5 µm | Polyblastic, terminal, integrated, denticles, 30–40 × 4–5 µm | Fusoid-ellipsoid, 1–3-septate, 12–20 × 3.5–5 µm, mucoid sheath | On leaves of unidentified vine | Thailand | [81] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Bao, D.-F.; Shen, H.-W.; Luo, Z.-L. Diversity of Lignicolous Freshwater Fungi from Yuanjiang River in Yunnan (China), with the Description of Four New Species. J. Fungi 2024, 10, 881. https://doi.org/10.3390/jof10120881
Zhang L, Bao D-F, Shen H-W, Luo Z-L. Diversity of Lignicolous Freshwater Fungi from Yuanjiang River in Yunnan (China), with the Description of Four New Species. Journal of Fungi. 2024; 10(12):881. https://doi.org/10.3390/jof10120881
Chicago/Turabian StyleZhang, Liang, Dan-Feng Bao, Hong-Wei Shen, and Zong-Long Luo. 2024. "Diversity of Lignicolous Freshwater Fungi from Yuanjiang River in Yunnan (China), with the Description of Four New Species" Journal of Fungi 10, no. 12: 881. https://doi.org/10.3390/jof10120881
APA StyleZhang, L., Bao, D.-F., Shen, H.-W., & Luo, Z.-L. (2024). Diversity of Lignicolous Freshwater Fungi from Yuanjiang River in Yunnan (China), with the Description of Four New Species. Journal of Fungi, 10(12), 881. https://doi.org/10.3390/jof10120881







