Genome-Wide Identification and Functional Characterization of GATA Transcription Factor Gene Family in Alternaria alternata
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Targeted Gene Disruption and Complementation
2.3. Stress Adaptation Assays
2.4. Pathogenicity Assays
2.5. Transcriptome Analysis
2.6. Gene Expression Analysis
2.7. Statistical Analysis
3. Results
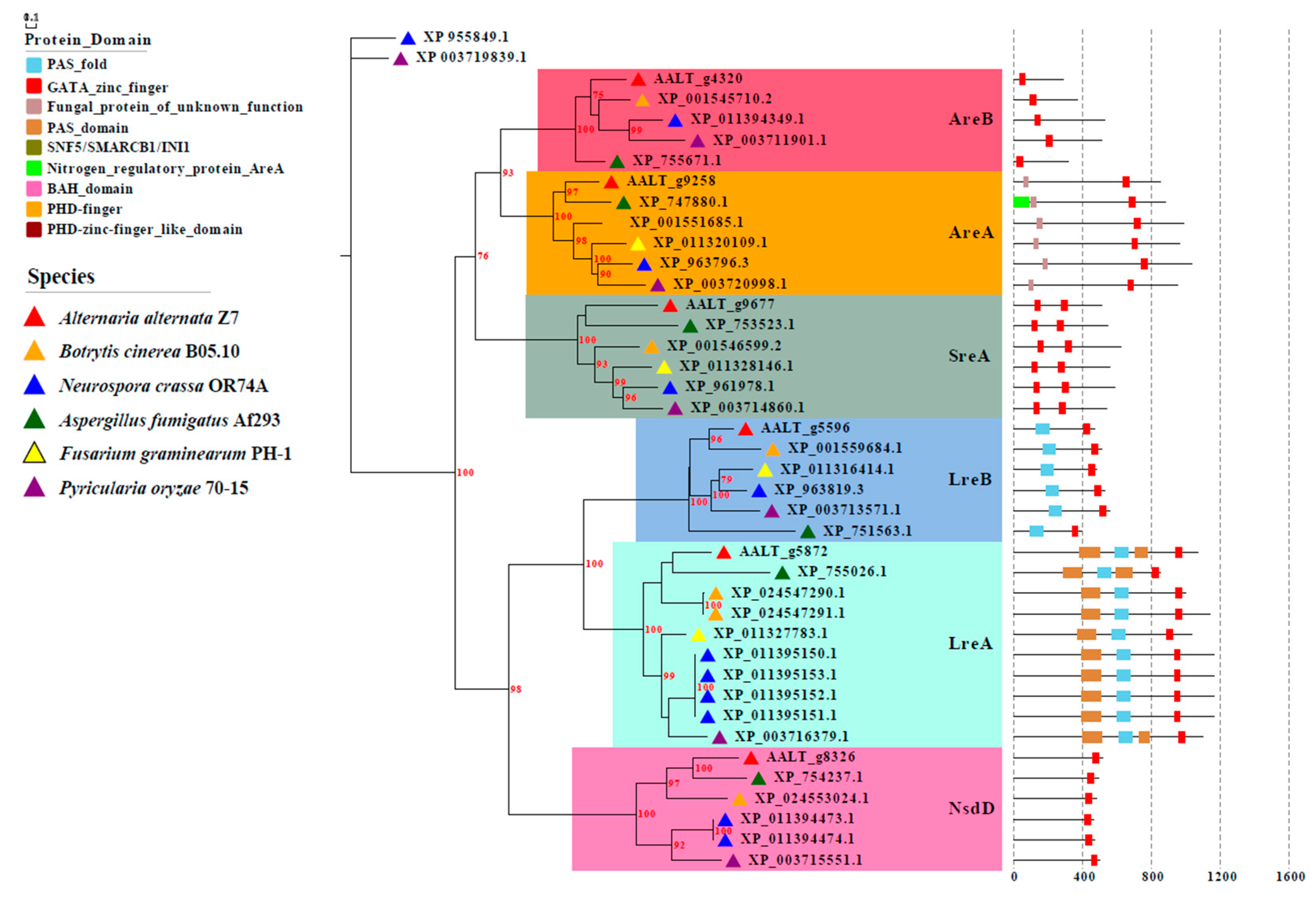
3.1. Identification of GATA Proteins in A. alternata
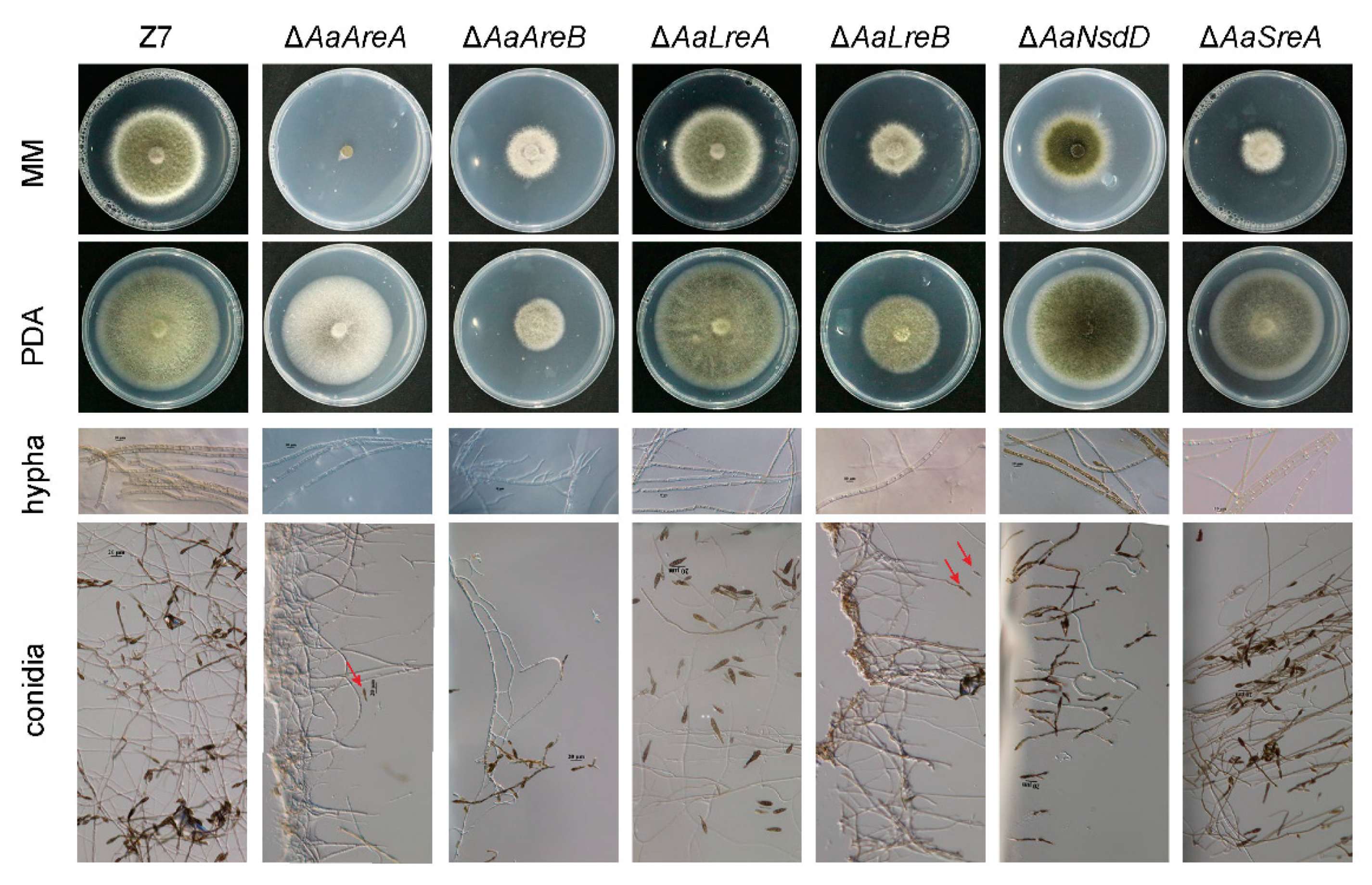
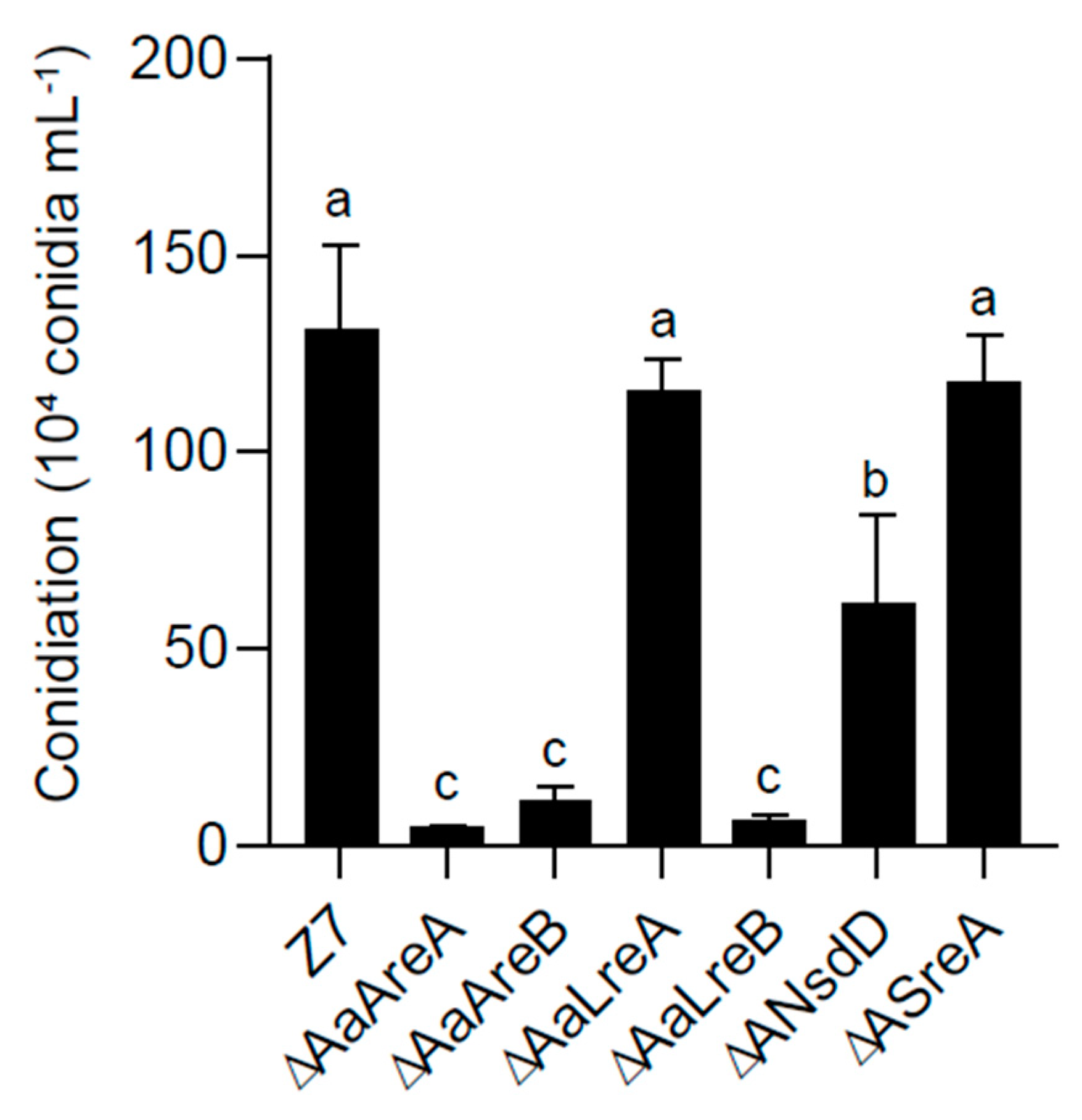
3.2. AaGATA Proteins Are Required for Vegetative Growth and Conidiation
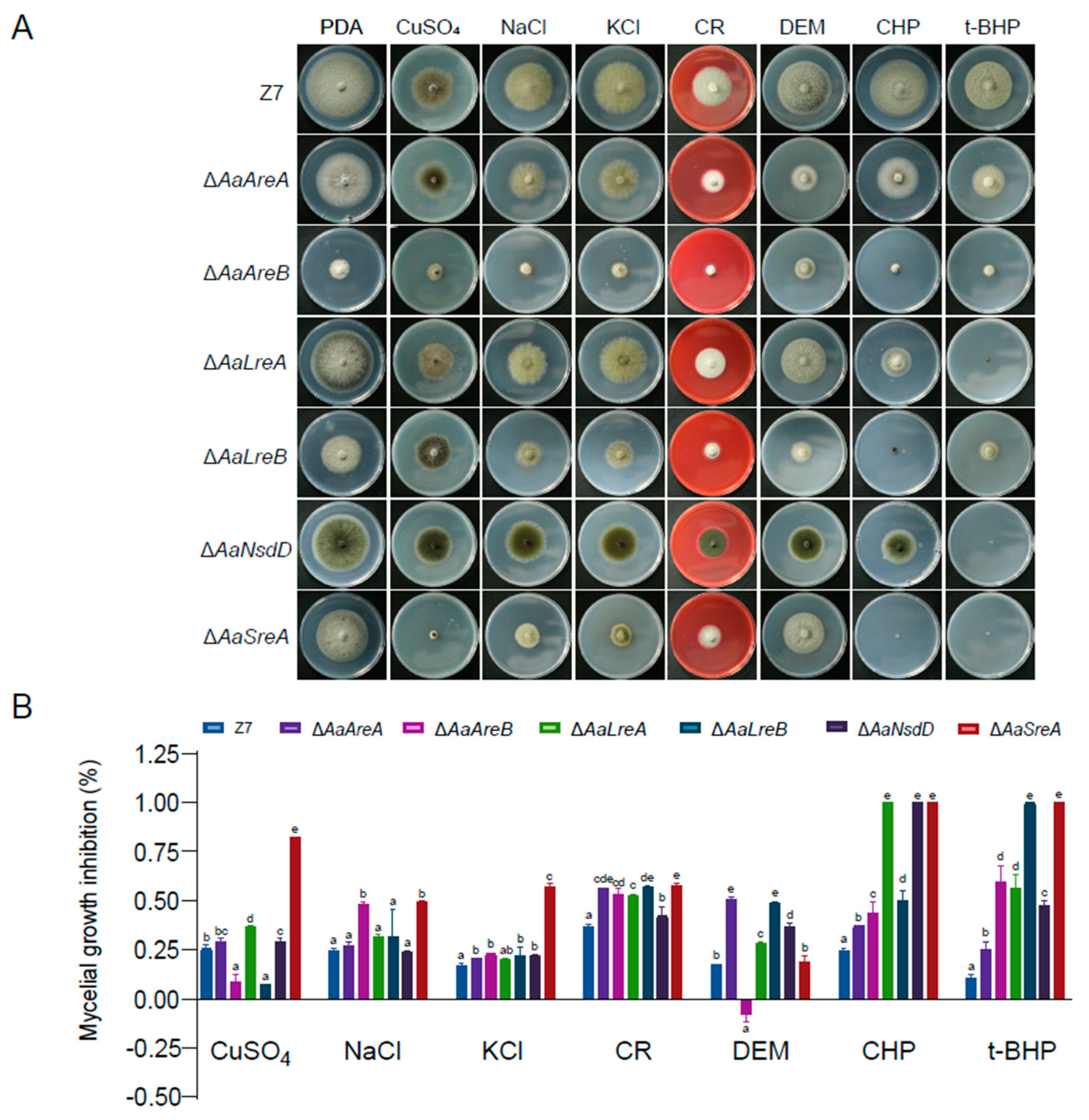
3.3. AaGATA Proteins Are Involved in Resistance to Multiple Stresses
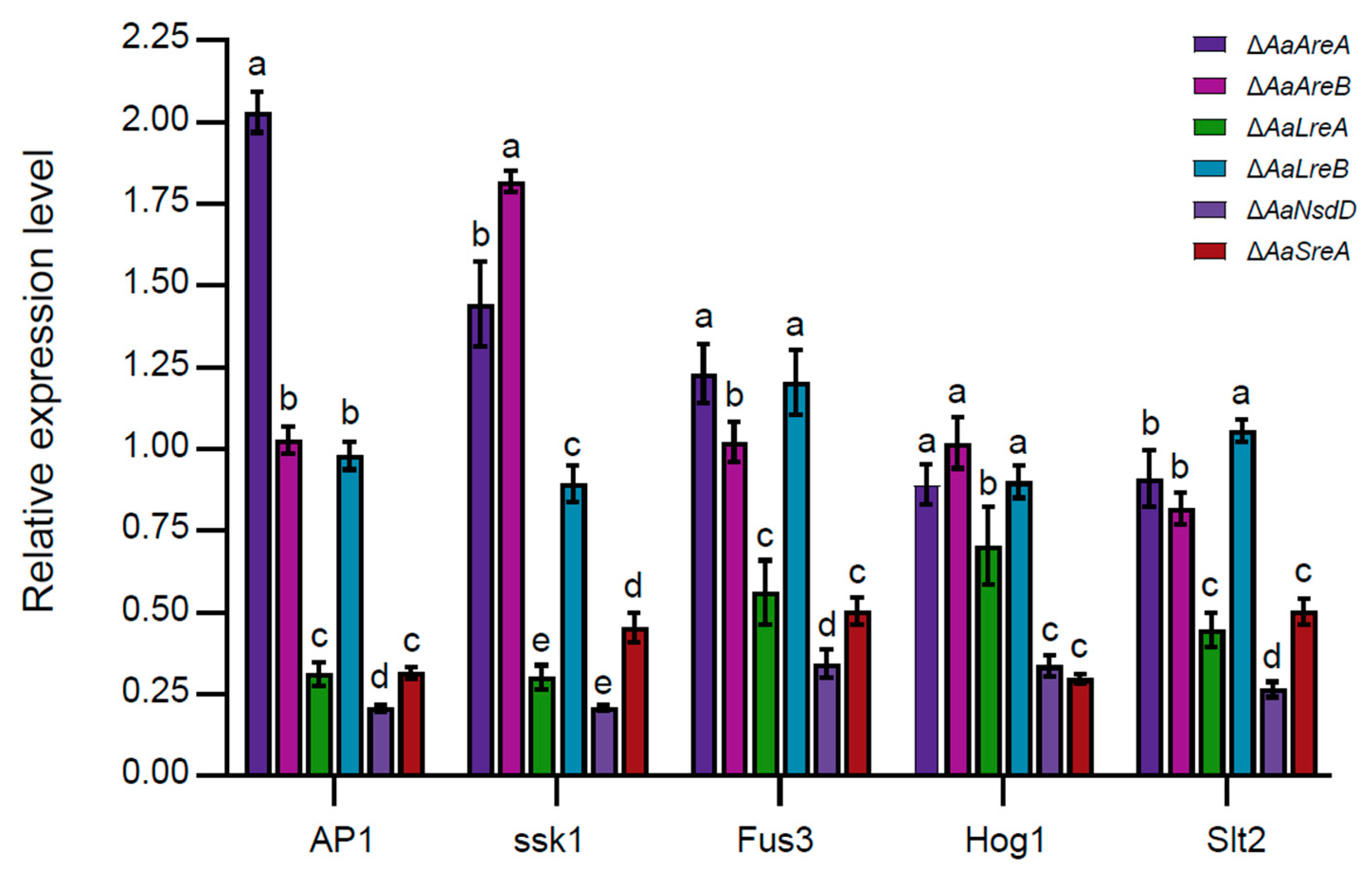
3.4. AaLreA, AaNsdD, and AaSreA Play Negative Roles in Gene Expression
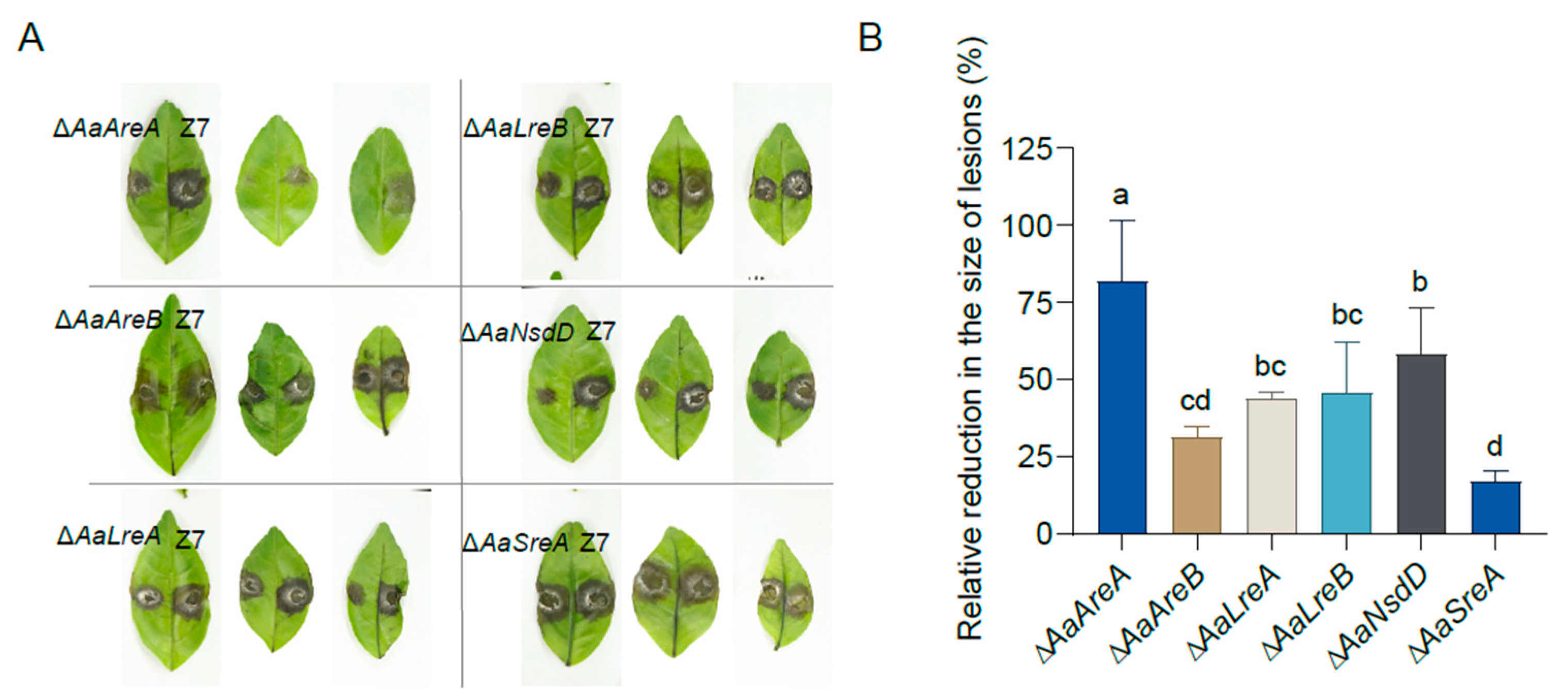
3.5. AaGATA Proteins Are Required for Full Virulence
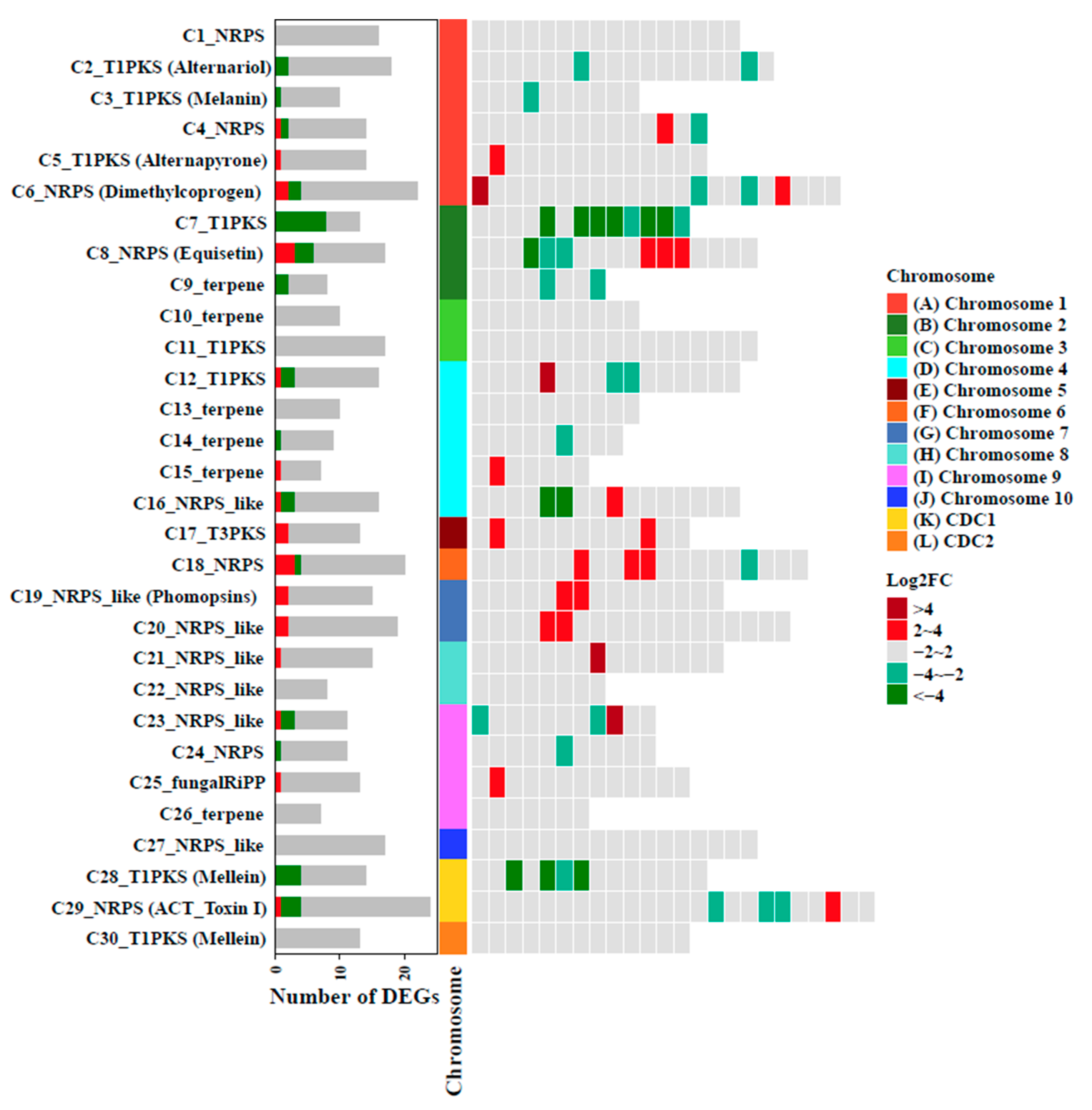
3.6. Transcriptome Analysis Defines the Global Regulatory Role of AaAreA in A. alternata
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wong, K.C. DNA motif recognition modeling from protein sequences. Iscience 2018, 7, 198–211. [Google Scholar] [CrossRef] [Green Version]
- Ko, L.J.; Engel, J.D. DNA-binding specificities of the GATA transcription factor family. Mol. Cell. Biol. 1993, 13, 4011–4022. [Google Scholar]
- Lowry, J.A.; Atchley, W.R. Molecular evolution of the GATA family of transcription factors: Conservation within the DNA-binding domain. J. Mol. Evol. 2000, 50, 103–115. [Google Scholar] [CrossRef]
- Trainor, C.D.; Ghirlando, R.; Simpson, M.A. GATA zinc finger interactions modulate DNA binding and transactivation. J. Biol. Chem. 2000, 275, 28157–28166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, A.H.; Kowalski, K.; King, G.F.; Mackay, J.P.; Crossley, M. Key residues characteristic of GATA N-fingers are recognized by FOG. J. Biol. Chem. 1998, 273, 33595–33603. [Google Scholar] [CrossRef] [Green Version]
- Crispino, J.D.; Horwitz, M.S. GATA factor mutations in hematologic disease. Blood 2017, 129, 2103–2110. [Google Scholar] [CrossRef]
- Romano, O.; Miccio, A. GATA factor transcriptional activity: Insights from genome-wide binding profiles. IUBMB Life 2020, 72, 10–26. [Google Scholar] [CrossRef]
- Daniel-Vedele, F.; Caboche, M. A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol. Gen. Genet. 1993, 240, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004, 134, 1718–1732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, R.; Behringer, C.; Zourelidou, M.; Schwechheimer, C. Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2013, 110, 13192–13197. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Ren, C.; Zou, L.; Wang, Y.; Li, S.; Liang, Z. Characterization of the GATA gene family in Vitis vinifera: Genome-wide analysis, expression profiles, and involvement in light and phytohormone response. Genome 2018, 61, 713–723. [Google Scholar] [CrossRef] [Green Version]
- Teakle, G.R.; Gilmartin, P.M. Two forms of type IV zinc-finger motif and their kingdom-specific distribution between the flora, fauna and fungi. Trends Biochem. Sci. 1998, 23, 100–102. [Google Scholar] [CrossRef]
- Scazzocchio, C. The fungal GATA factors. Curr. Opin. Microbiol. 2000, 3, 126–131. [Google Scholar] [CrossRef]
- Haas, H.; Zadra, I.; Stöffler, G.; Angermayr, K. The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J. Biol. Chem. 1999, 274, 4613–4619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muro-Pastor, M.I.; Gonzalez, R.; Strauss, J.; Narendja, F.; Scazzocchio, C. The GATA factor AreA is essential for chromatin remodelling in a eukaryotic bidirectional promoter. EMBO J. 1999, 18, 1584–1597. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.H.; Hynes, M.J.; Todd, R.B.; Davis, M.A. Deletion and overexpression of the Aspergillus nidulans GATA factor AreB reveals unexpected pleiotropy. Microbiology 2009, 155, 3868–3880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conlon, H.; Zadra, I.; Haas, H.; Arst, H.N., Jr.; Jones, M.G.; Caddick, M.X. The Aspergillus nidulans GATA transcription factor gene areB encodes at least three proteins and features three classes of mutation. Mol. Microbiol. 2001, 40, 361–375. [Google Scholar] [CrossRef]
- Purschwitz, J.; Müller, S.; Kastner, C.; Schöser, M.; Haas, H.; Espeso, E.A.; Atoui, A.; Calvo, A.M.; Fischer, R. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr. Biol. 2008, 18, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Han, K.H.; Han, K.Y.; Yu, J.H.; Chae, K.S.; Jahng, K.Y.; Han, D.M. The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol. Microbiol. 2001, 41, 299–309. [Google Scholar] [CrossRef]
- Park, J.-S.; Kim, H.-J.; Kim, S.-O.; Kong, S.-H.; Park, J.-J.; Kim, S.-R.; Han, H.-Y.; Park, B.-S.; Jung, K.-Y.; Lee, Y.-H. A comparative genome-wide analysis of GATA transcription factors in fungi. Genom. Inform. 2006, 4, 147–160. [Google Scholar]
- Magasanik, B.; Kaiser, C.A. Nitrogen regulation in Saccharomyces cerevisiae. Gene 2002, 290, 1–18. [Google Scholar] [CrossRef]
- Talora, C.; Franchi, L.; Linden, H.; Ballario, P.; Macino, G. Role of a white collar-1-white collar-2 complex in blue-light signal transduction. EMBO J. 1999, 18, 4961–4968. [Google Scholar] [CrossRef]
- Ballario, P.; Macino, G. White collar proteins: PASsing the light signal in Neurospora crassa. Trends Microbiol. 1997, 5, 458–462. [Google Scholar] [CrossRef]
- Chung, K.-R.; Wu, P.-C.; Chen, Y.-K.; Yago, J.I. The siderophore repressor SreA maintains growth, hydrogen peroxide resistance, and cell wall integrity in the phytopathogenic fungus Alternaria alternata. Fungal Genet. Biol. 2020, 139, 103384. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P. Alternaria spp.: From general saprophyte to specific parasite. Mol. Plant Pathol. 2003, 4, 225–236. [Google Scholar] [CrossRef]
- Tsuge, T.; Harimoto, Y.; Akimitsu, K.; Ohtani, K.; Kodama, M.; Akagi, Y.; Egusa, M.; Yamamoto, M.; Otani, H. Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 2013, 37, 44–66. [Google Scholar] [CrossRef] [PubMed]
- Akimitsu, K.; Peever, T.L.; Timmer, L.W. Molecular, ecological and evolutionary approaches to understanding Alternaria diseases of citrus. Mol. Plant Pathol. 2003, 4, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, H.; Taga, M.; Kodama, M.; Johnson, R.; Otani, H.; Kohmoto, K. Molecular karyotypes for alternaria plant pathogens known to produce host-specific toxins. Curr. Genet. 1999, 35, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Ajiro, N.; Miyamoto, Y.; Masunaka, A.; Tsuge, T.; Yamamoto, M.; Ohtani, K.; Fukumoto, T.; Gomi, K.; Peever, T.L.; Izumi, Y.; et al. Role of the host-selective act-toxin synthesis gene ACTTS2 encoding an enoyl-reductase in pathogenicity of the tangerine pathotype of Alternaria alternata. Phytopathology 2010, 100, 120–126. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Zhang, B.; Gai, Y.; Sun, X.; Chung, K.R.; Li, H. Cell-wall-degrading enzymes required for virulence in the host selective toxin-producing necrotroph Alternaria alternata of citrus. Front. Microbiol. 2019, 10, 2514. [Google Scholar] [CrossRef]
- Ma, H.; Wang, M.; Gai, Y.; Fu, H.; Zhang, B.; Ruan, R.; Chung, K.R.; Li, H. Thioredoxin and glutaredoxin systems required for oxidative stress resistance, fungicide sensitivity, and virulence of Alternaria alternata. Appl. Environ. Microbiol. 2018, 84, e00086-18. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Yang, X.; Ruan, R.; Fu, H.; Li, H. Csn5 is required for the conidiogenesis and pathogenesis of the Alternaria alternata tangerine pathotype. Front. Microbiol. 2018, 9, 508. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.R. Stress response and pathogenicity of the necrotrophic fungal pathogen Alternaria alternata. Scientifica 2012, 2012, 635431. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.; Fu, Y.; Nie, D.; Stewart, J.E.; Peever, T.L.; Li, H. Identification of a novel phylogenetic lineage of Alternaria alternata causing citrus brown spot in China. Fungal Biol. 2015, 119, 320–330. [Google Scholar] [CrossRef]
- Wang, M.; Sun, X.; Yu, D.; Xu, J.; Chung, K.; Li, H. Genomic and transcriptomic analyses of the tangerine pathotype of Alternaria alternata in response to oxidative stress. Sci. Rep. 2016, 6, 32437. [Google Scholar] [CrossRef] [Green Version]
- Gai, Y.; Ma, H.; Chen, Y.; Li, L.; Cao, Y.; Wang, M.; Sun, X.; Jiao, C.; Riely, B.K.; Li, H. Chromosome-scale genome sequence of Alternaria alternata causing Alternaria Brown Spot of Citrus. Mol. Plant Microbe Interact. 2021, 34, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterprofiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de Los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. Antismash 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.; Srivastava, A.; Ohm, R.A.; Lawrence, C.B.; Wang, K.-H.; Grigoriev, I.V.; Marahatta, S.P. Transcription factor Amr1 induces melanin biosynthesis and suppresses virulence in Alternaria brassicicola. PLoS Pathog. 2012, 8, e1002974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, Y.; Kim, K.H.; La Rota, M.; Scott, D.; Santopietro, G.; Callihan, M.; Mitchell, T.K.; Lawrence, C.B. Identification of novel virulence factors associated with signal transduction pathways in Alternaria brassicicola. Mol. Microbiol. 2009, 72, 1316–1333. [Google Scholar] [CrossRef] [PubMed]
- John, E.; Singh, K.B.; Oliver, R.P.; Tan, K.C. Transcription factor control of virulence in phytopathogenic fungi. Mol. Plant Pathol. 2021, 22, 858–881. [Google Scholar] [CrossRef]
- Chandarlapaty, S.; Errede, B. Ash1, a daughter cell-specific protein, is required for pseudohyphal growth of Saccharomyces cerevisiae. Mol. Cell. Biol. 1998, 18, 2884–2891. [Google Scholar] [CrossRef] [Green Version]
- Long, R.M.; Singer, R.H.; Meng, X.; Gonzalez, I.; Nasmyth, K.; Jansen, R.P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 1997, 277, 383–387. [Google Scholar] [CrossRef] [Green Version]
- Jung, W.H.; Sham, A.; White, R.; Kronstad, J.W. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 2006, 4, e410. [Google Scholar] [CrossRef] [Green Version]
- Jung, W.H.; Kronstad, J.W. The iron-responsive, GATA-type transcription factor Cir1 influences mating in Cryptococcus neoformans. Mol. Cells 2011, 31, 73–77. [Google Scholar] [CrossRef] [Green Version]
- He, Q.; Cheng, P.; Yang, Y.; Wang, L.; Gardner, K.H.; Liu, Y. White collar-1, a DNA binding transcription factor and a light sensor. Science 2002, 297, 840–843. [Google Scholar] [CrossRef]
- Todd, R.B.; Fraser, J.A.; Wong, K.H.; Davis, M.A.; Hynes, M.J. Nuclear accumulation of the GATA factor AreA in response to complete nitrogen starvation by regulation of nuclear export. Eukaryot. Cell 2005, 4, 1646–1653. [Google Scholar] [CrossRef] [Green Version]
- Macios, M.; Caddick, M.X.; Weglenski, P.; Scazzocchio, C.; Dzikowska, A. The GATA factors AREA and AREB together with the co-repressor NMRA, negatively regulate arginine catabolism in Aspergillus nidulans in response to nitrogen and carbon source. Fungal Genet. Biol. 2012, 49, 189–198. [Google Scholar] [CrossRef]
- Michielse, C.B.; Pfannmüller, A.; Macios, M.; Rengers, P.; Dzikowska, A.; Tudzynski, B. The interplay between the GATA transcription factors AreA, the global nitrogen regulator and AreB in Fusarium fujikuroi. Mol. Microbiol. 2014, 91, 472–493. [Google Scholar] [CrossRef]
- Marroquin-Guzman, M.; Wilson, R.A. GATA-dependent glutaminolysis drives appressorium formation in Magnaporthe oryzae by suppressing TOR inhibition of cAMP/PKA signaling. PLoS Pathog. 2015, 11, e1004851. [Google Scholar] [CrossRef]
- Wilson, R.A.; Gibson, R.P.; Quispe, C.F.; Littlechild, J.A.; Talbot, N.J. An NADPH-dependent genetic switch regulates plant infection by the rice blast fungus. Proc. Natl. Acad. Sci. USA 2010, 107, 21902–21907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crosthwaite, S.K.; Dunlap, J.C.; Loros, J.J. Neurospora wc-1 and wc-2: Transcription, photoresponses, and the origins of circadian rhythmicity. Science 1997, 276, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Yang, Y.; Gardner, K.H.; Liu, Y. PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady-state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol. Cell. Biol. 2002, 22, 517–524. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.K.; Kwon, N.J.; Choi, J.M.; Lee, I.S.; Jung, S.; Yu, J.H. NsdD is a key repressor of asexual development in Aspergillus nidulans. Genetics 2014, 197, 159–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schumacher, J.; Simon, A.; Cohrs, K.C.; Viaud, M.; Tudzynski, P. The transcription factor BcLTF1 regulates virulence and light responses in the necrotrophic plant pathogen Botrytis cinerea. PLoS Genet. 2014, 10, e1004040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerwien, F.; Skrahina, V.; Kasper, L.; Hube, B.; Brunke, S. Metals in fungal virulence. FEMS Microbiol. Rev. 2018, 42, fux050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenzo-Gutiérrez, D.; Gómez-Gil, L. Cu transporter protein CrpF protects against Cu-induced toxicity in Fusarium oxysporum. Virulence 2020, 11, 1108–1121. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.L.; Chen, L.H.; Chung, K.R. How the pathogenic fungus fungus Alternaria alternata copes with stress via the response regulators ssk1 and sho1. PLoS ONE 2016, 11, e0149153. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.H.; Chung, K.R. Specialized and shared functions of the histidine kinase- and HOG1 MAP kinase-mediated signaling pathways in Alternaria alternata, a filamentous fungal pathogen of citrus. Fungal Genet. Biol. 2010, 47, 818–827. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Q.; Sun, Y.; Zhang, Y.; Zhang, X.; Liu, J.; Yu, G.; Pan, H. Sssfh1, a gene encoding a putative component of the RSC chromatin remodeling complex, is involved in hyphal growth, reactive oxygen species accumulation, and pathogenicity in Sclerotinia sclerotiorum. Front. Microbiol. 2018, 9, 1828. [Google Scholar] [CrossRef]
- Niehaus, E.M.; Schumacher, J.; Burkhardt, I.; Rabe, P.; Spitzer, E.; Münsterkötter, M.; Güldener, U.; Sieber, C.M.K.; Dickschat, J.S.; Tudzynski, B. The GATA-type transcription factor Csm1 regulates conidiation and secondary metabolism in Fusarium fujikuroi. Front. Microbiol. 2017, 8, 1175. [Google Scholar] [CrossRef]
- Motoyama, T. Secondary metabolites of the rice blast fungus Pyricularia oryzae: Biosynthesis and biological function. Int. J. Mol. Sci. 2020, 21, 8698. [Google Scholar] [CrossRef]
- Haas, H.; Marzluf, G.A. NRE, the major nitrogen regulatory protein of Penicillium chrysogenum, binds specifically to elements in the intergenic promoter regions of nitrate assimilation and penicillin biosynthetic gene clusters. Curr. Genet. 1995, 28, 177–183. [Google Scholar] [CrossRef]
- Min, K.; Shin, Y.; Son, H.; Lee, J.; Kim, J.C.; Choi, G.J.; Lee, Y.W. Functional analyses of the nitrogen regulatory gene areA in Gibberella zeae. FEMS Microbiol. Lett. 2012, 334, 66–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giese, H.; Sondergaard, T.E.; Sørensen, J.L. The AreA transcription factor in Fusarium graminearum regulates the use of some nonpreferred nitrogen sources and secondary metabolite production. Fungal Biol. 2013, 117, 814–821. [Google Scholar] [CrossRef]
- Li, J.; Pan, Y.; Liu, G. Disruption of the nitrogen regulatory gene AcareA in Acremonium chrysogenum leads to reduction of cephalosporin production and repression of nitrogen metabolism. Fungal Genet. Biol. 2013, 61, 69–79. [Google Scholar] [CrossRef]
- Tudzynski, B. Nitrogen regulation of fungal secondary metabolism in fungi. Front. Microbiol. 2014, 5, 656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Sun, Z.; Shi, D.; Song, S.; Lian, L.; Shi, L.; Ren, A.; Yu, H.; Zhao, M. Dual functions of AreA, a GATA transcription factor, on influencing ganoderic acid biosynthesis in Ganoderma lucidum. Environ. Microbiol. 2019, 21, 4166–4179. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Cao, Y.; Gai, Y.; Ma, H.; Zhu, Z.; Chung, K.-R.; Li, H. Genome-Wide Identification and Functional Characterization of GATA Transcription Factor Gene Family in Alternaria alternata. J. Fungi 2021, 7, 1013. https://doi.org/10.3390/jof7121013
Chen Y, Cao Y, Gai Y, Ma H, Zhu Z, Chung K-R, Li H. Genome-Wide Identification and Functional Characterization of GATA Transcription Factor Gene Family in Alternaria alternata. Journal of Fungi. 2021; 7(12):1013. https://doi.org/10.3390/jof7121013
Chicago/Turabian StyleChen, Yanan, Yingzi Cao, Yunpeng Gai, Haijie Ma, Zengrong Zhu, Kuang-Ren Chung, and Hongye Li. 2021. "Genome-Wide Identification and Functional Characterization of GATA Transcription Factor Gene Family in Alternaria alternata" Journal of Fungi 7, no. 12: 1013. https://doi.org/10.3390/jof7121013
APA StyleChen, Y., Cao, Y., Gai, Y., Ma, H., Zhu, Z., Chung, K.-R., & Li, H. (2021). Genome-Wide Identification and Functional Characterization of GATA Transcription Factor Gene Family in Alternaria alternata. Journal of Fungi, 7(12), 1013. https://doi.org/10.3390/jof7121013





