Abstract
Species of the genus Paracoccidioides cause a systemic infection in human patients. Yeast cells of Paracoccidioides spp. produce melanin in the presence of L-dihydroxyphenylalanine and during infection, which may impact the pathogen’s survival in the host. To better understand the metabolic changes that occur in melanized Paracoccidioides spp. cells, a proteomic approach was performed to compare melanized and non-melanized Paracoccidioides brasiliensis and Paracoccidioides lutzii yeast cells. Melanization was induced using L-dihydroxyphenylalanine as a precursor, and quantitative proteomics were performed using reversed-phase nano-chromatography coupled to high-resolution mass spectrometry. When comparing melanized versus non-melanized cells, 1006 and 582 differentially abundant/detected proteins were identified for P. brasiliensis and P. lutzii, respectively. Functional enrichment and comparative analysis revealed 30 important KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways in melanized P. brasiliensis and 18 in P. lutzii, while differentially abundant proteins from non-melanized cells from these species were involved in 21 and 25 enriched pathways, respectively. Melanized cells presented an abundance of additional virulence-associated proteins, such as phospholipase, proteases, superoxide dis-mutases, heat-shock proteins, adhesins, and proteins related to vesicular transport. The results suggest that L-dihydroxyphenylalanine increases the virulence of Paracoccidioides spp. through complex mechanisms involving not only melanin but other virulence factors as well.
1. Introduction
Species of the genus Paracoccidioides cause a life-threatening systemic disease in humans named para-coccidioidomycosis. Paracoccidioides brasiliensis and Paracoccidioides lutzii are the primary recognized agents of this infection, although recent taxonomic studies have proposed three additional cryptic species, formerly classified as P. brasiliensis: Paracoccidioides americana, Paracoccidioides restrepiensis, and Paracoccidioides venezuelensis [1]. This disease has two major clinical forms, the acute/subacute (or juvenile) and chronic (or adult) forms, and several clinical manifestations, with multi-organ involvement, even in patients without known immunodeficiency [2].
The species P. brasiliensis and P. lutzii are difficult to distinguish using classical microbiological techniques or by clinical presentation. However, the conidia of P. brasiliensis are globular, while P. lutzii has elongated conidia [3]. Humans infected with distinct cryptic species have similar clinical presentations [4,5,6], but serologic response to the main antigen used in the diagnosis varies [7]. Regarding virulence, P. brasiliensis appears to be more virulent in experimental models of infection, since mice infected with P. lutzii develop milder disease and P. brasiliensis persists for a prolonged time in the lungs of these animals [8].
In order to cause disease, the species of the genus Paracoccidioides have a series of virulence factors that help the fungi invade different tissues and survive the defenses orchestrated by the immune system of the host. The major putative virulence factors of the Paracoccidioides genus include the immunodominant gp43, a cell surface and secreted glycoprotein with adhesion and proteinase activities; gp70, a glycoprotein with anti-phagocytic activity; carbohydrates of the cell-wall; and melanin [9].
Melanin is a pigment with stable free radicals produced by diverse organisms, from mammals to bacteria [10]. This biopolymer is usually associated with protection mechanisms against a series of severe conditions and/or toxic compounds present in the environment, such as ultraviolet light, heavy metals and other pollutants, and extremes in temperature [11]. Melanin also protects fungi and other pathogenic microbes during infection [12]. The yeast cells of Paracoccidioides spp. produce melanin in vitro under L-dihydroxyphenylalanine (L-DOPA) supplementation and during parasitism [13]. Melanin protects P. brasiliensis against phagocytosis and antifungal drugs [14]. However, there are no data on the mechanism(s) of action associated with these protective phenotypes in melanized Paracoccidioides yeast cells.
Proteomic approaches have been applied to the genus Paracoccidioides to elucidate several aspects related to the adaptation and survival of the fungus in the host, such as macrophage interaction [15], response to hypoxia [16], and metabolism under carbon starvation [17]. Therefore, we compared the proteome of melanized and non-melanized (that is, cultured in the presence or absence of L-DOPA, respectively) P. brasiliensis and P. lutzii strains to identify possible molecular mechanisms that explain the higher survival of melanized cells under harsh conditions.
2. Materials and Methods
2.1. Fungal Strains and Culture Maintenance
The reference strains P. brasiliensis Pb18 [18] and P. lutzii Pb01 (ATCC- MYA-826) [19] were used in the experiments. Yeast cells were sub-cultured in FavaNetto Agar medium (0.3% (w/v) proteose peptone (Difco Laboratories, Sparks, MD, USA); 1% (w/v) peptone (Difco Laboratories, Sparks, MD, USA); 0.5% (w/v) beef extract (Difco Laboratories, Sparks, MD, USA); 0.5% (w/v) sodium chloride (Sigma-Aldrich, Co., St. Louis, MO, USA); 0.5% (w/v) yeast extract (Difco Laboratories, Sparks, MD, USA), 4% (w/v) glucose (Sigma-Aldrich, Co., St. Louis, MO, USA), 1.8% (w/v) agar (Difco Laboratories, Sparks, MD, USA)) and incubated at 37 °C on a weekly basis.
2.2. Melanin Production
Paracoccidioides yeast cells were harvested from seven-day-old solid FavaNetto cultures and washed three times with phosphate-buffered saline (PBS). Melanized cultures of the strains were obtained as follows: firstly, the harvested and washed cells were grown in FavaNetto liquid medium (same composition as above, but without agar) at an initial density of 1 × 106 cells/mL for six days at 37 °C, under agitation (150 rpm). After this first incubation, 1 × 108 cells were transferred to a chemically defined minimal medium (15 mM glucose, 10 mM MgSO4, 29.4 mM K2HPO4, 13 mM glycine, and 3.0 mM thiamine (all from Sigma-Aldrich Co., San Luis, MO, USA), pH 5.5] supplemented with 1 mM L-dihydroxyphenylalanine (L-DOPA) (Sigma-Aldrich, Co., San Luis, MO, USA). Control non-melanized cultures were performed in the same conditions, but without L-DOPA supplementation into the minimal medium. All cultures were incubated in a rotary shaker (Nova Ética, Vargem Grande Paulista, SP, Brazil) for six days at 150 rpm and 37 °C, collected by centrifugation (10,000× g), and washed three times in PBS. Five biological replicates were prepared for each condition.
2.3. Protein Extraction
Extracts containing total protein contents from melanized and non-melanized Pb18 and Pb01 strains were obtained by mechanical cellular disruption under liquid nitrogen. Extraction buffer (20 mM Tris-HCl pH 8.8, 2 mM CaCl2), supplemented with protease inhibitors (Complete Mini®, Roche Diagnostics, Manheim, Germany) and zirconia beads of 0.5 mm diameter (BiospecProducts, Bartlesville, OK, USA), were added to the fungal lysates. A vigorous agitation of this mixture was performed on a Mini-Beadbeater (Biospec products) for five cycles of 30 s, separated by 1 min ice bath incubations. After this procedure, the material was centrifuged (10,000× g) for 15 min at 4 °C and the Bradford method was employed to determine protein content, using bovine serum albumin (Sigma-Aldrich, Co.) for the generation of standard curves [20]. The protein profiles of the fungal extracts were evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), as described [21].
2.4. Sample Processing for Shotgun Proteomics
For each sample, the volume equivalent to 100 μg was dried on a centrifugal vacuum concentrator (SpeedVac, Thermo Fischer Scientific, Waltham, MA, USA) and then submitted to trypsin digestion. Samples were initially suspended in 20 μL of 0.4 M ammonium bicarbonate, 8 M urea, followed by the addition of 5 μL of 0.1 M dithiothreitol and incubation at 37 °C for 30 min. Then, 5 μL of 0.4 M iodoacetamide were added and incubated for 15 min at room temperature in the dark. Samples were diluted by the addition of 130 μL of Milli-Q water followed by trypsin (Promega, Madison, WI, USA) addition at 1/50 (m/m) of enzyme to substrate and sequential incubation for 16 h at 37 °C and 45 min at 56 °C; the reaction was stopped with 20 μL of 10% (v/v) formic acid. Samples were desalted with in-lab generated columns packed with Poros R2 resin (Life Technologies). Columns were initially activated with 100% acetonitrile (CH3CN), followed by equilibration with 1% (v/v) trifluoroacetic acid (TFA). Samples were applied to the columns and subjected to five washes with 0.1% TFA solution. The elution was carried out with four washes of 0.1% TFA in 70% CH3CN. Samples were dried on a centrifugal vacuum concentrator (SpeedVac) and stored at −20 °C until use. Prior to mass spectrometry (MS), each sample was suspended in 20 μL of 1% formic acid, and its peptide concentration was estimated by absorbance measurement at 280 nm (1.000 absorbance unit ≅ 1.000 μg/μL) on a NanoDrop 2000 spectrophotometer (Thermo Fischer Scientific); all samples were normalized to 0.675 μg/μL.
2.5. Mass Spectrometry Analysis
The desalted tryptic digests from the five biological replicates from the strains P. brasiliensis Pb18 and P. lutzii Pb01 were each analyzed in three technical replicates by reversed-phase nano-chromatography coupled to high-resolution nano-electrospray ionization mass spectrometry. Chromatography was performed using a Dionex Ultimate 3000 RSLCnano system (Thermo Fischer Scientific). Samples (4 µL per run) were initially applied, at 2 μL/min of 0.1% (v/v) formic acid in water, to a 2-cm-long trap column (100 μm inner diameter), packed with Magic C18-AQ 200 Å 5 μm matrix (MichromBioresources, Auburn, CA, USA). Next, peptides were submitted to chromatographic separation on a 26-cm-long column (75 μm inner diameter), packed with ReproSil-Pur C18-AQ 120 Å 1.9 μm matrix (Dr. Maisch GmbH, Germany) directly into a laser-pulled capillary with a 4 μm tip. Fractionation was performed at 200 nL/min having 0.1% (v/v) formic acid in water and 0.1% (v/v) formic acid in acetonitrile as mobile phases A and B, respectively. Elution was carried out on a gradient from 2% to 40% B for 162 min; concentration was increased to 80% B in 4 min and maintained for 2 min more. The eluted peptides were introduced directly into a Q Exactive Plus Orbitrap instrument. Ionization was achieved by applying 1.9 kV to the source, setting the capillary temperature to 250 °C, and alternate current (radiofrequency) level of the S-lenses at 60 V. The complete MS1 scans (300 to 1500 m/z) were acquired in the profile mode with one micro-scan at 70,000 resolution and an automatic gain control (AGC) target value of 1 × 106 with a maximum injection time of 100 ms. The 12 most intense precursor ions within both isolation window and offset of 2.0 and 0.5 m/z, respectively, were selected for HCD (higher energy collision dissociation) fragmentation with a collision energy normalized to 30%. The MS2 spectra (200 to 2000 m/z) were acquired in centroid mode with one micro-scan at 17,500 resolution and an AGC target value of 5×104 with a maximum injection time of 50 ms. Dynamic exclusion was set to 60 s, whereas peaks with unassigned charge or those with z = 1 were rejected.
2.6. Protein Identification and Quantitation Following Mass Spectrometry
Data were analyzed using the PatternLab for Proteomics 4.0 pipeline [22]. Separate reference databases were downloaded for each species from the UniProt proteomes repository, comprising 8811 entries for Pb01 (https://www.uniprot.org/proteomes/UP000002059) and 8399 entries for Pb18 (https://www.uniprot.org/proteomes/UP000001628). The “Generate Search Database” tool was used to add to each database common contaminants and decoy (reverse) sequences for all entries. MS data from each species sample runs were searched against their specific database using the Comet search tool [23], which is integrated into PatternLab. The parameters were set as follows: precursor mass error tolerance of 40 ppm, trypsin as the enzyme, semi-tryptic cleavage, maximum number of cleavage loss of 2, cysteine carbamido-methylation (+57.02146 Da) as fixed modification, and deamination of asparagine and/or glutamine (+0.98400 Da) as variable modifications. Values for fragment bin tolerance, fragment bin offset, and theoretical fragment ions were 0.02, 0, and default peak shape (flanking peaks), respectively. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [24] and received the dataset identifier PXD021711. Spectral validation was performed using SEPro algorithm (Search Engine Processor) [25]; false discovery rate (FDR) was calculated from the number of decoy sequences identified, so that a maximum limit of 1% FDR, at peptide and protein levels, was established and only identifications with a mass error tolerance of 5 ppm were accepted. Unique peptide spectral count analysis was used for the quantitative comparisons between melanized and non-melanized cells of each species in the TFold module of PatternLab for Proteomics; normalization was done by total spectral count. Only proteins that were detected in at least three (out of five) biological replicates, for each condition, were submitted to relative quantitation with cut-off values for Benjamini–Hochberg q-value, F-stringency, and L-stringency of 0.01, 0.03, and 0.60, respectively; proteins that satisfied both the fold-change and statistical (p-value) criteria were further submitted to protein interaction and metabolic pathway analyses.
2.7. Bioinformatic Analyses
The Uniprot resource for protein sequence and annotation (https://www.uniprot.org/) was used to describe functions of proteins with quantitative differences on both conditions of each Paracoccidioides species. Enrichment analyses of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (https://www.genome.jp/kegg/pathway.html) were used to functionally categorize the regulated proteins and interactions between proteins were studied on the STRING v.11 server [26], available at https://string-db.org/. Proteins associated with virulence, stress response, and pathogenesis of paracoccidioidomycosis were retrieved from indexed publications [9,27,28,29,30,31,32,33] and searched into the proteomic data generated in this study. Heat maps were constructed using the GraphPad Prism 8.4.2 software.
3. Results
3.1. Fungal Melanization and Protein Extract Control
Both Pb18 and Pb01 strains, P. brasiliensis and P. lutzii, respectively, produced melanized cells after six days of incubation at 37 °C, exclusively in the presence of L-DOPA (Figure 1). The quality of the protein extracts obtained from melanized and non-melanized cells was evaluated by SDS-PAGE under reducing conditions (Figure S1). Proteins within a broad range of molecular mass were observed on both species; for some bands, a variation of the staining intensity was observed, indicating a possible differential abundance of proteins.
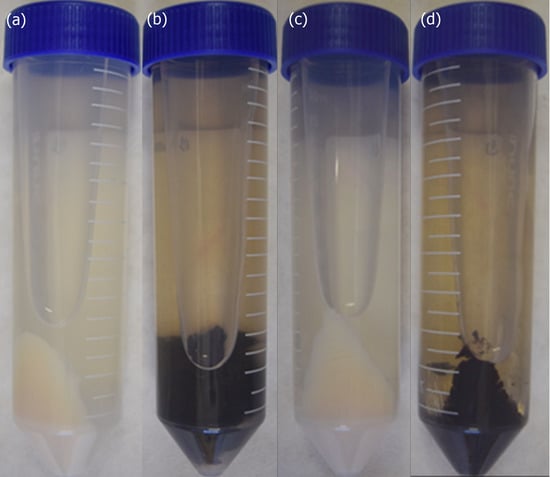
Figure 1.
L-dihydroxyphenylalanine (L-DOPA) induces melanization in Paracoccidioides species. Pellets of Paracoccidioides brasiliensis Pb18 yeast cells after six days of growth in minimal medium alone (a) or supplemented with L-DOPA (b). Pellets of Paracoccidioides lutzii Pb01 yeast cells after six days of growth in minimal medium (c) or supplemented with L-DOPA (d).
3.2. Proteomic Analysis of Melanized and Non-Melanized Cells
The proteomic approach identified 2586 protein entries for the P. brasiliensis Pb18 strain and 1962 for the P. lutzii Pb01 strain. In the Pb18 proteome, 471 proteins were exclusively detected in non-melanized cells and 72 in melanized cells. Similarly, 275 proteins were only detected in the proteome of non-melanized Pb01 cells and 71 in melanized cells proteome (Figure S2). Among the 2043 proteins detected in extracts from both melanized and non-melanized Pb18 cells, 463 presented a statistically-supported (p- and q-values < 0.01) differential abundance between the two conditions tested; 245 were more abundant in non-melanized and 218 in melanized cells (Table S1-“Conditions&Volcano Plot” and “Blue” sheets). Regarding P. lutzii, 1,=616 proteins were detected in proteomes from the two studied conditions; 236 of these presented a statistically-supported (p- and q-values < 0.01) difference in abundance: 108 were more abundant in melanized cells and 128 in non-melanized cells (Table S2—“Conditions&Volcano Plot” and “Blue” sheets).
3.3. Enriched Pathways
The proteins differentially abundant and exclusively detected, for both culture conditions of each Paracoccidioides species, were clustered and analyzed using the STRING database. Both Pb18 and Pb01 proteins yielded networks (Figure S3) with more interactions than expected, presenting protein–protein interaction (PPI) enrichment values of less than 10−16 and 2.77 × 10−9, respectively. Pathways related to carbon, nucleotide, amino acid metabolisms, and other biological processes were enriched in the species studied, as presented in Table 1.

Table 1.
Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways observed for proteins with differential abundance in proteomes of melanized and non-melanized cells of Paracoccidioides brasiliensis and Paracoccidioides lutzii.
3.3.1. Carbon Metabolism
The KEGG pathways enrichment analysis revealed 49 proteins associated with carbon metabolism in Pb18 and 24 in Pb01 (Figure S4). Twelve proteins were common to the two species. Among the Pb18 proteins, 37 were more abundant in melanized cells, nine were more abundant in non-melanized cells, and three were exclusively detected in non-melanized cells. All proteins associated with the oxidative phosphorylation were more abundant in melanized cells, as well as the late enzymes of the glycolytic pathway, isocitrate lyase, one of the key enzymes of the glyoxylate cycle, and the enzymes transketolase and trans-aldolase of the non-oxidative phase of the pentose phosphate pathway (Figure 2).
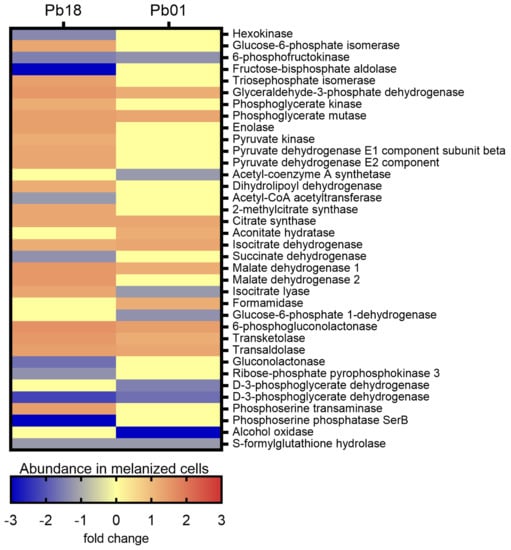
Figure 2.
Comparative heatmap of the abundance in melanized cells of proteins from carbon metabolism pathways enriched in P. brasiliensis Pb18 and P. lutzii Pb01.
Most enzymes related to methane metabolism and the citrate cycle were more abundant in non-melanized cells. In Pb01, transketolase and trans-aldolase were also enriched, as well as some enzymes from glycolysis and citrate cycle. However, the key enzyme of glycolysis, hexokinase (PADG_03813), was less abundant in melanized cells of Pb18 strain, as well as phosphofructokinase (PADG_00192 and PAAG_01583), in melanized cells of both strains studied (Figure 2). Although the oxidative phosphorylation was not enriched in Pb01, the beta and delta subunits of ATP synthase (PAAG_08037 and PAAG_05605) were more abundant in melanized cells (fold changes of 1.3 and 1.7, respectively), similarly to P. brasiliensis. In contrast, isocitrate lyase was less abundant in melanized cells of P. lutzii. The enzymes involved in β-oxidation of fatty acids enoyl-CoA hydratase and acyl-CoA dehydrogenase were found to be more abundant in melanized Pb18. However, this pathway was not enriched in the Pb01 proteome. In fact, the only enzyme related to fatty acid metabolism with differential abundance in this strain was 3-ketoacyl-CoA thiolase (PAAG_02664), with a fold change of −1.3.
3.3.2. Nucleotide Metabolism
The KEGG pathways enrichment analysis showed 37 proteins associated with nucleotide metabolism in Pb18 and 18 in Pb01 (Figure S5). Fifteen proteins were common to the two strains studied. In general, most enzymes from both purine and pyrimidine synthesis were more abundant (14 proteins in the Pb18 proteome and six in the Pb01 proteome) or exclusively detected (15 proteins in the Pb18 proteome and eight in the Pb01 proteome) in non-melanized cells of both species. The abundance profile of differentially abundant proteins related to the nucleotide metabolism was similar for the two Paracoccidioides strains analyzed, except for uracil phosphoribosyl-transferase (PADG_01100 and PAAG_06643), which was found to be more abundant in Pb18 melanized cells but less abundant in melanized cells of Pb01 (Figure 3).
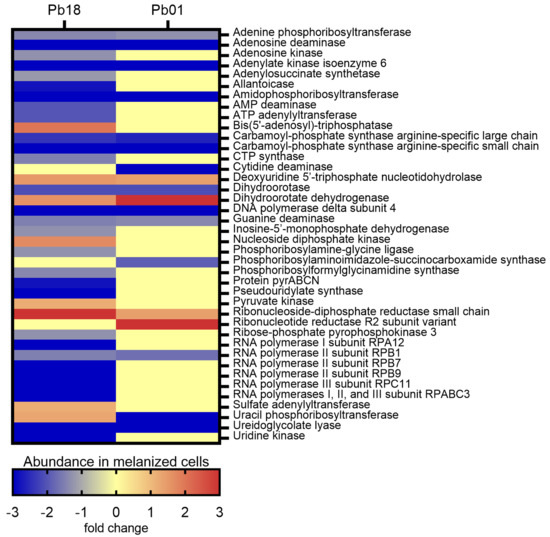
Figure 3.
Comparative heatmap of the abundance in melanized cells of proteins from nucleotide metabolism pathways enriched in P. brasiliensis Pb18 and P. lutzii Pb01.
3.3.3. Amino Acid Metabolism
The KEGG pathways enrichment analysis showed 65 exclusive proteins or proteins with differential abundance associated with amino acid metabolism in Pb18 and 51 in Pb01 (Figure S6). Twenty-two proteins were common to the two organisms. In Pb18, most pathways related to amino acid metabolism were more abundant in non-melanized cells, except for six enzymes of the tyrosine metabolism, another six from the valine, leucine, and isoleucine degradation, as well as two enzymes from the glycine, serine, and threonine metabolism and one from the phenylalanine, tyrosine, and tryptophan biosynthesis. A similar profile was observed in the Pb01 proteomes, with a predominance of enzymes more abundant in non-melanized cells. Melanized cells presented five abundant proteins related to tyrosine metabolism, two related to valine, leucine, and isoleucine degradation, two enzymes related to glycine, serine, and threonine metabolism, and one related to valine, leucine, and isoleucine biosynthesis. Supporting that degradation of amino acids generates precursors for the citrate cycle, we observed that the enzyme 3-oxoacid CoA transferase (PADG_04939), related to the degradation of leucine to acetyl CoA, was more abundant in the melanized Pb18 proteome. Enzymes related to the tyrosine degradation to fumarate presented differential abundance among melanized and non-melanized cells of both strains. They include fumarylaceto-acetase (PADG_08465 and PAAG_ 08163), homogentisate 1,2-dioxygenase (PADG_08466 and PAAG_08164), 4-hydroxyphenylpyruvate dioxygenase (PADG_08468 and PAAG_08166), phenylpyruvate tautomerase (PADG_03671), and two isoforms of aspartate aminotransferase (PADG_03686 and PADG_01404). Surprisingly, a third aspartate aminotransferase isoform (PAAG_02603) was less abundant in melanized Pb01 yeast cells (Figure 4).

Figure 4.
Comparative heatmap of the abundance in melanized cells of proteins from amino acid metabolism pathways enriched in P. brasiliensis Pb18 and P. lutzii Pb01.
3.3.4. Other Metabolic Pathways
The enrichment analysis revealed another ten KEGG pathways in the Pb18 proteome: spliceosome, RNA degradation, RNA transport, protein processing in the endoplasmic reticulum, phagosome, aminoacyl-t-RNA biosynthesis, proteasome, basal transcription factors, soluble N-ethylmaleimide-sensitive-fusion attachment protein receptor (SNARE) interactions in vesicular transport, and ubiquitin-mediated proteolysis (Figure S7a). All proteins with differential abundance associated with SNARE interactions in vesicular transport and the majority (72.7%) of proteasome proteins were more abundant in melanized cells. Half the proteins associated with protein processing in the endoplasmic reticulum were also more abundant in melanized cells. The majority of proteins from the other seven pathways were more abundant in non-melanized cells. RNA degradation and RNA transport were also enriched in proteomes of Pb01, but not the other eight pathways observed in Pb18. In fact, four pathways were enriched in Pb01: RNA degradation, RNA transport, mRNA surveillance pathway, and peroxisome. Except for RNA degradation, most proteins from the other pathways were less abundant in melanized yeast cells (Figure S7b). Heat maps of each KEGG pathway enriched on both Pb18 and Pb01 strains are presented in Figure S8.
3.4. Virulence-Associated Proteins
A search was conducted for the proteins exclusively detected or with differential abundance in melanized and non-melanized cells of Pb18 and Pb01 strains. Figure 5 depicts the abundance of virulence-related proteins in melanized cells of Pb18 and Pb01 strains. Among 1006 proteins of Pb18, 34 were previously associated with fungal virulence, stress response, and/or pathogenesis of paracoccidioido-mycosis. As for Pb01, 18 proteins fulfilling the same criteria aforementioned were observed among 582 proteins.
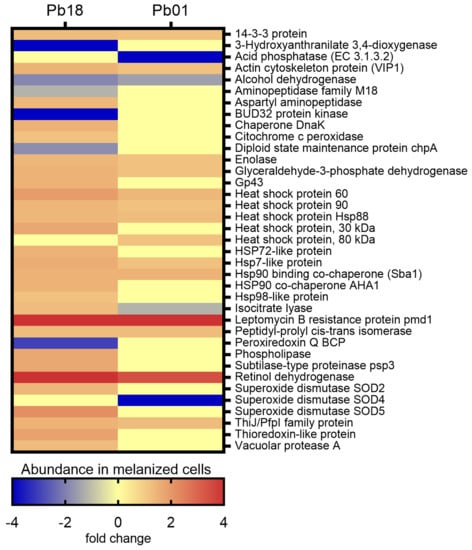
Figure 5.
Comparative heatmap of the abundance in melanized cells of virulence-related proteins in P. brasiliensis Pb18 and P. lutzii Pb01.
As depicted in Figure 5, most of the 37 virulence-related proteins observed in both Paracoccidioides species were more abundant in their melanized cells. Noteworthy, eight heat shock proteins and two co-chaperones, four proteolytic enzymes, a protein involved in multidrug resistance (Leptomycin B resistance protein pmd1), and some adhesins such as 14-3-3 protein, glyceraldehyde-3-phosphate dehydrogenase, enolase, and actin cytoskeleton protein VIP1 stand out among the abundant virulence-related proteins detected in melanized cells. The immunodominant gp43 antigen was also more abundant in melanized cells of the Pb18 yeast cells. The single protein related to virulence that was less abundant in melanized cells of both strains was alcohol dehydrogenase, which has adhesin properties.
4. Discussion
Members of the genus Paracoccidioides change their protein expression, and consequently their metabolism and virulence, as a response to physiological conditions induced in experimental settings, such as copper overload [34], copper deprivation [35], acetate availability [31], osmotic stress [36], among others. The present work demonstrates that P. brasiliensis and P. lutzii, when grown in vitro in the presence of L-DOPA, a substance potentially available for the fungus during parasitism, not only produce melanin, as already described [13,14], but also undergo metabolic shifts and present differential abundance of proteins previously related with fungal virulence.
The most studied human pathogenic fungus regarding melanization is Cryptococcus neoformans. In this species, L-DOPA only changes the expression of eight genes, most of them related to stress response [37]. On the other hand, P. brasiliensis and P. lutzii have significant changes on overall metabolism, and protein expression when L-DOPA is available during fungal growth. The differences between these two fungi might be related to the methodologies used in the studies: while in C. neoformans the early response (4 h incubation) to L-DOPA was evaluated at the transcript level, here the proteome of fully melanized cells (six days of incubation with L-DOPA) was evaluated.
The concentration of L-DOPA used in this study (1 mM) is widely used for several groups that study in vitro fungal melanization [37,38,39,40,41]. The amount of L-DOPA in infected tissues is lower than that used in experimental conditions [42], but it is continuously renewed in tissues whereas it is depleted over time in culture as it is incorporated by the fungus. However, for in vitro proteomics, a large number of cells is necessary to yield protein amounts suitable for analysis, a number also higher than that usually found in infected tissues of patients with paracoccidioido-mycosis. Therefore, the use of physiological amounts of L-DOPA in our experimental conditions using a high number of cells would be challenging, as previously demonstrated with C. neoformans [37]. Further studies are required to address whether these results correlate with in vivo conditions.
In the proteomic approach herein performed, P. brasiliensis displayed more changes than P. lutzii in response to L-DOPA. In fact, P. brasiliensis presented almost two-fold more proteins with differential abundance than P. lutzii and, besides carbon, amino acid, and nucleotide metabolisms, showed eight enriched pathways not observed in P. lutzii, which had only two exclusive enriched pathways. These differences, however, could not be associated with the known phenotypic differences between P. brasiliensis and P. lutzii. Among the enriched processes observed only in P. brasiliensis, proteasome induction also occurred in the Pb18 strain during keratinocyte infection [43], with the alpha subunit of the proteasome (PADG_02735) detected in both experimental approaches. In addition, P. brasiliensis melanized yeast cells had a higher abundance of proteins associated with vesicular transport. As previously described [44], this species is known to produce extracellular vesicles harboring proteins that could modulate the immune system, proteins associated with stress response, and enzymes involved in the acquisition of nutrients during parasitism, like lipases and the subtilase-type proteinase psp3 (PADG_07422), also more abundant in the total proteome of Pb18 melanized yeast cells described in this study. In Pb01, most peroxisomal proteins were less abundant in melanized cells. Peroxisomes are associated, among other functions, with the reduction of reactive oxygen species [45], a role that might be addressed by melanin of P. lutzii cells grown in the presence of L-DOPA.
To the best of our knowledge, this is the first comparative proteomic approach in melanized and non-melanized cells of two phylogenetically related fungal species. We have previously compared proteomes of non-melanized phylogenetically related fungal species. For example, in pathogenic species of the genus Sporothrix, enzymes related to amino acid metabolism and cell-wall remodeling are more abundant in the highly virulent Sporothrix brasiliensis species, whereas the glycolytic pathway is more active in Sporothrix schenckii and Sporothrix globosa. Moreover, the number of proteins with differential abundance is lower than that observed in this study [46]. We previously compared the proteomic profile of four distinct phylogenetic species of Paracoccidioides, including the Pb01 strain used in this study, where 301 proteins presented differential abundance [47]. The lower number of proteins may be explained by the method used in the previous study, two-dimensional gel electrophoresis and in-gel digestion, which is less sensitive than the method used in this study.
Proteins related to carbon metabolism were more abundant in melanized cells of both strains studied. The citrate cycle was enriched in both strains; however, oxidative phosphorylation was only enriched in Pb18. A similar behavior was observed when different Paracoccidioides species were grown in the presence of acetate, a common carbon source within macrophages [31]. The high abundance of proteins related to the electron transport chain and oxidative phosphorylation of P. brasiliensis related to P. lutzii has been previously demonstrated [47], which could explain the results from the present study. The glyoxylate cycle appears to be differentially regulated by L-DOPA in the two studied strains since isocitrate lyase is more abundant in melanized Pb18 and less abundant in melanized Pb01. The glyoxylate cycle is associated with virulence in some human pathogenic fungi [48,49], and isocitrate lyase appears to have a role in adhesion and colonization of host tissues by P. brasiliensis [50], reinforcing the role of L-DOPA in the virulence enhancement of this species. The enzymes enoyl-CoA hydratase (PADG_01209) and acyl-CoA dehydrogenase (PADG_02244) were more abundant in melanized Pb18, indicating β-oxidation of fatty acids in this strain, which also occurs when acetate is available for fungal growth [31], and during copper deprivation [35] or experimental infection [51]. It was noteworthy that the enzymes related to ethanol production did not present differential abundance between melanized and non-melanized cells of both strains tested. Ethanol production is regulated in Paracoccidioides spp. by several factors, including growth with acetate as a carbon source [31], carbon starvation [17], response to antifungal molecules [28], and during macrophage or murine infection [51,52].
The profile of enzymes related to amino acid metabolism was also similar between Pb18 and Pb01 strains. Degradation of amino acids provides precursors for the citrate cycle (Figure S9). Amino acids are a potential nutrient source for fungal growth during parasitism. Our proteomic data show that both melanized strains have the enzymatic machinery to yield fumarate from tyrosine. Another fate of tyrosine is its conversion to L-DOPA by the enzyme tyrosinase. This enzyme was not detected in our proteomic approach, despite its theoretical role in melanin production [14]. One hypothesis to explain this result is that tyrosinase may be regulated in the early stages of fungal melanization and, therefore, cannot be detected after the fungus is fully melanized. In addition, we cannot exclude the possibility of the participation of proteins without known functions in melanin production by Paracoccidioides spp. In fact, approximately 60% of the total proteins predicted in the genomes of P. brasiliensis Pb18 and P. lutzii Pb01 do not have annotated functions. In addition, even after a functional classification of P. lutzii hypothetical proteins with gene ontology terms and protein domain annotations, more than 2500 proteins remained without a predicted function [53].
L-DOPA is described as suppressing Madurella mycetomatis growth, even at 0.1 mM [40], and to inhibit the growth of C. neoformans in concentrations above 1 mM [37]. This last concentration was not able to inhibit the growth of the two Paracoccidioides strains herein used, but some proteomic data described in this study indicate that melanized yeast cells of P. brasiliensis Pb18 have a lower transcriptional capacity than non-melanized cells. These data include the absence of several basal transcription factors in melanized cells, the lower abundance of enzymes related to the nucleotide metabolism, including five subunits of three different RNA polymerases, and the lower abundance of proteins associated with spliceosome. Interestingly, this behavior was not observed with the P. lutzii Pb01 strain, which only presented one less abundant subunit of the RNA polymerase (PAAG_05397). These results indicate that the two Paracoccidioides strains used in this study have distinct responses to L-DOPA availability. Whether these differences are strain-dependent or reflect the species-specific biological behavior of P. brasiliensis and P. lutzii remains to be elucidated.
Melanized fungal cells are more resistant to several harsh conditions faced during environmental or parasitic growth. Much of this protective effect can be explained by the biophysical properties of melanin, such as absorbance of ultraviolet light, covalent and non-covalent interaction with molecules, and scavenging of free radicals [11]. The results of the proteomic approach used in this study revealed that, besides the well-established function of melanization promotion [13,14], L-DOPA also induces the expression of several other molecules related to fungal virulence in Paracoccidioides spp. Similar behavior was observed for the Pb18 strain treated with low doses of nitric oxide [54]. We found that Pb18 presented more virulence-related proteins, with higher abundance than Pb01, which can be related to the higher virulence of this strain in experimental mice models of paracoccidioido-mycosis [8].
Eight heat shock proteins and two co-chaperones were more abundant in melanized cells. Heat shock proteins can be induced in response to biotic and abiotic factors, including high temperatures [55]. Both cultures, supplemented or not with L-DOPA, were grown in the same incubator, at the same time, discarding the hypothesis that changes in temperature could have influenced our results. Since heat shock proteins are immunologically active [56] and are thought to participate in the immune-pathogenicity of paracoccidioido-mycosis [55], this might have an implication since Paracoccidioides cells are melanized during parasitism [13].
Another relevant protein observed in the melanized Pb18 proteome was the 43-kDa glycoprotein (PADG_07615), also known as gp43. This protein is immunogenic and can bind to laminin, presenting an important role in the pathogenesis of paracoccidioido-mycosis [9]. The strain Pb18 is known to produce this antigen [57], and this work has shown that L-DOPA may enhance the production of this protein. Contrastingly, the closest P. lutzii ortholog to gp43 did not present differential abundance in our analyses, which could be associated to the protein’s low expression by this species [58]. Here again, another difference related to L-DOPA’s influence was demonstrated between the two strains studied. Moreover, the differences in the abundance of this antigen may be related to the frequent false-negative results in patients with paracoccidioido-mycosis caused by P. lutzii frequently yield when gp43 is used in serologic tests [7].
In addition, P. brasiliensis Pb18 presented two abundant superoxide dismutases in melanized cells. These enzymes are involved in the primary detoxification defense against reactive oxygen species and, in P. brasiliensis phylogenetic species PS3 (or P. restrepiensis), at least superoxide dismutase isoform 3 is required for virulence [59]. This isoform did not present differential abundance in our proteomic analysis, but the role of those two Pb18 superoxide dismutase isoforms detected in Paracoccidioides virulence has not been properly addressed so far. Interestingly, P. lutzii Pb01 presented one superoxide dismutase (isoform 4) exclusively detected in non-melanized cells, suggesting that the melanin produced by this species, when L-DOPA is available during fungal growth, may be more efficient than Pb18 melanin in countering reactive oxygen species.
5. Conclusions
Paracoccidioides spp. respond to L-DOPA availability during growth by producing melanin, changing several metabolic pathways, and expressing virulence-related proteins. Although similar, the responses of the two studied species, P. brasiliensis and P. lutzii, to L-DOPA have some differences, such as a deeper change in protein abundance and metabolic pathways in P. brasiliensis, enhanced production of the immunodominant antigen in melanized P. brasiliensis, and lower abundance of proteins associated with detoxification of reactive oxygen species, such as those involved in peroxisome and superoxide dismutase isoform 6, in melanized P. lutzii. Further studies are necessary to address if the melanin produced by these two species has, indeed, biophysical differences that may be associated with the proteomic differences regarding detoxification measures observed in the current study.
Supplementary Materials
The following are available online at https://www.mdpi.com/2309-608X/6/4/328/s1, Figure S1: Electrophoretic profile of the protein extracts from melanized and non-melanized cells of Paracoccidioides brasiliensis Pb18 and Paracoccidioides lutzii Pb01. Figure S2: Venn diagrams presenting the relationship between the proteome of melanized and non-melanized cells of Paracoccidioides brasiliensis Pb18 and Paracoccidioides lutzii Pb01. Figure S3: Protein-protein interaction networks of the differentially abundant proteins of Paracoccidiodies brasiliensis Pb18 and Paracoccidioides lutzii Pb01. Figure S4: Enriched KEGG pathways associated with carbon metabolism in the proteomes of melanized Paracoccidioides species. (a) protein-protein interactions of 49 P. brasiliensis Pb18 proteins; (b) protein-protein interactions of 24 P. lutzii Pb01 proteins. Figure S5: Enriched KEGG pathways associated with nucleotide metabolism in the proteomes of melanized Paracoccidioides species. (a) protein-protein interactions of 37 P. brasiliensis Pb18 proteins; (b) protein-protein interactions of 18 P. lutzii Pb01 proteins. Figure S6: Enriched KEGG pathways associated with amino acid metabolism in the proteomes of melanized Paracoccidioides species. (a) protein-protein interactions of 49 P. brasiliensis Pb18 proteins; (b) protein-protein interactions of 24 P. lutzii Pb01 proteins. Figure S7: Other enriched KEGG pathways in the proteomes of melanized Paracoccidioides species. (a) protein-protein interactions of 123 P. brasiliensis Pb18 proteins from 10 different pathways; (b) protein-protein interactions of 35 P. lutzii Pb01 proteins from four different pathways. Figure S8: Comparison of the abundance in melanized cells of proteins associated with enriched pathways observed on proteomes of both Paracoccidiodies brasiliensis Pb18 and Paracoccidiodies lutzii Pb01. Figure S9: Interaction between nucleotide and amino acid metabolisms with the tricarboxylic acid and glyoxylate cycles. Amino acids in red represent potential carbon sources for the tricarboxylic acid cycle in melanized Paracoccidioides spp. cells. Table S1: Proteins with differential abundance in Paracoccidioides brasiliensis Pb18 proteome and their classification after Benjamini–Hochberg q-value, F-stringency, and L-stringency analyses. Table S2: Proteins with differential abundance in Paracoccidioides lutzii Pb01 proteome and their classification after Benjamini–Hochberg q-value, F-stringency, and L-stringency analyses.
Author Contributions
Conceptualization, R.A.-P., C.M.d.A.S. and R.M.Z.-O.; methodology, R.A.-P., M.A.A. and R.H.V.; software, R.H.V.; validation, M.A.A., M.R.d.O.T. and R.H.V.; formal analysis, R.A.-P., M.A.A., M.R.d.O.T. and R.H.V.; investigation, M.A.A., L.C.B., L.A.M.M. and R.H.V.; resources, R.A.-P., M.R.d.O.T., J.D.N., C.M.d.A.S., R.H.V. and R.M.Z.-O.; data curation, M.R.d.O.T., R.H.V.; writing—original draft preparation, R.H.V. and R.A.-P.; writing—review and editing, M.A.A., L.C.B., M.R.d.O.T., J.D.N., C.M.d.A.S., R.M.Z.-O.; visualization, R.A.-P.; supervision, R.M.Z.-O.; project administration, R.A.-P.; funding acquisition, R.A.-P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), grant number 449184/2014-5 and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brasil (Capes)-Finance Code 001. RHV is a fellow from CNPq, grant number 304523/2019-4. RMZ-O is a fellow from CNPq, grant number 302796/2017-7) and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro, grant number FAPERJ E-26/202.527/2019. JDN is supported in part by NIH AI52733.
Acknowledgments
The authors thank the Fiocruz Network of Technology Platforms for the use of core facilities (Proteômica/RJ RPT2A Espectrometria de massas) and Joelma Saldanha for sample processing assistance.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Hrycyk, M.F.; Garcia Garces, H.; de Bosco, S.M.G.; de Oliveira, S.L.; Marques, S.A.; Bagagli, E. Ecology of Paracoccidioides brasiliensis, P. lutzii and related species: Infection in armadillos, soil occurrence and mycological aspects. Med. Mycol. 2018, 56, 950–962. [Google Scholar] [CrossRef]
- Shikanai-Yasuda, M.A.; Mendes, R.P.; Colombo, A.L.; de Queiroz-Telles, F.; Kono, A.S.G.; Paniago, A.M.M.; Nathan, A.; do Valle, A.C.F.; Bagagli, E.; Benard, G.; et al. Brazilian guidelines for the clinical management of paracoccidioidomycosis. Rev. Soc. Bras. Med. Trop. 2017, 50, 715–740. [Google Scholar] [CrossRef]
- de Teixeira, M.M.; Theodoro, R.C.; de Oliveira, F.F.M.; Machado, G.C.; Hahn, R.C.; Bagagli, E.; San-Blas, G.; Soares Felipe, M.S. Paracoccidioides lutzii sp. nov.: Biological and clinical implications. Med. Mycol. 2014, 52, 19–28. [Google Scholar] [CrossRef]
- de Macedo, P.M.; de Teixeira, M.M.; Barker, B.M.; Zancopé-Oliveira, R.M.; Almeida-Paes, R.; do Francesconi Valle, A.C. Clinical features and genetic background of the sympatric species Paracoccidioides brasiliensis and Paracoccidioides americana. PLoS Negl. Trop. Dis. 2019, 13, e0007309. [Google Scholar] [CrossRef]
- Hahn, R.C.; Rodrigues, A.M.; Della Terra, P.P.; Nery, A.F.; Hoffmann-Santos, H.D.; Góis, H.M.; Fontes, C.J.F.; de Camargo, Z.P. Clinical and epidemiological features of paracoccidioidomycosis due to Paracoccidioides lutzii. PLoS Negl. Trop. Dis. 2019, 13, e0007437. [Google Scholar] [CrossRef]
- Pereira, E.F.; Gegembauer, G.; Chang, M.R.; de Camargo, Z.P.; Nunes, T.F.; Ribeiro, S.M.; de Carvalho, L.R.; Maldonado, B.M.; Mendes, R.P.; Paniago, A.M.M. Comparison of clinico-epidemiological and radiological features in paracoccidioidomycosis patients regarding serological classification using antigens from Paracoccidioides brasiliensis complex and Paracoccidioides lutzii. PLoS Negl. Trop. Dis. 2020, 14, e0008485. [Google Scholar] [CrossRef]
- De Silva, J.F.; de Oliveira, H.C.; Marcos, C.M.; Assato, P.A.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. Advances and challenges in paracoccidioidomycosis serology caused by Paracoccidioides species complex: An update. Diagn. Microbiol. Infect. Dis. 2016, 84, 87–94. [Google Scholar] [CrossRef]
- Siqueira, I.M.; Fraga, C.L.F.; Amaral, A.C.; Souza, A.C.O.; Jerônimo, M.S.; Correa, J.R.; Magalhães, K.G.; Inácio, C.A.; Ribeiro, A.M.; Burguel, P.H.; et al. Distinct patterns of yeast cell morphology and host responses induced by representative strains of Paracoccidioides brasiliensis (Pb18) and Paracoccidioides lutzii (Pb01). Med. Mycol. 2016, 54, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.A.; Grisolia, J.C.; Burger, E.; de Araujo Paula, F.B.; Dias, A.L.T.; Malaquias, L.C.C. Virulence factors of Paracoccidioides brasiliensis as therapeutic targets: A review. Antonie Van Leeuwenhoek 2020, 113, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Nosanchuk, J.D.; Casadevall, A. The contribution of melanin to microbial pathogenesis. Cell. Microbiol. 2003, 5, 203–223. [Google Scholar] [CrossRef]
- Eisenman, H.C.; Greer, E.M.; McGrail, C.W. The role of melanins in melanotic fungi for pathogenesis and environmental survival. Appl. Microbiol. Biotechnol. 2020, 104, 4247–4257. [Google Scholar] [CrossRef] [PubMed]
- Plonka, P.M.; Grabacka, M. Melanin synthesis in microorganisms—Biotechnological and medical aspects. Acta Biochim. Pol. 2006, 53, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Gómez, B.L.; Nosanchuk, J.D.; Díez, S.; Youngchim, S.; Aisen, P.; Cano, L.E.; Restrepo, A.; Casadevall, A.; Hamilton, A.J. Detection of melanin-like pigments in the dimorphic fungal pathogen Paracoccidioides brasiliensis in vitro and during infection. Infect. Immun. 2001, 69, 5760–5767. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, M.B.; Marques, A.F.; Nosanchuk, J.D.; Casadevall, A.; Travassos, L.R.; Taborda, C.P. Melanin in the dimorphic fungal pathogen Paracoccidioides brasiliensis: Effects on phagocytosis, intracellular resistance and drug susceptibility. Microbes Infect. 2006, 8, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Chaves, E.G.A.; Parente-Rocha, J.A.; Baeza, L.C.; Araújo, D.S.; Borges, C.L.; de Oliveira, M.A.P.; de Soares, C.M.A. Proteomic Analysis of Paracoccidioides brasiliensis During Infection of Alveolar Macrophages Primed or Not by Interferon-Gamma. Front. Microbiol. 2019, 10, 96. [Google Scholar] [CrossRef]
- De Lima, P.S.; Chung, D.; Bailão, A.M.; Cramer, R.A.; de Soares, C.M.A. Characterization of the Paracoccidioides Hypoxia Response Reveals New Insights into Pathogenesis Mechanisms of This Important Human Pathogenic Fungus. PLoS Negl. Trop. Dis. 2015, 9, e0004282. [Google Scholar] [CrossRef]
- De Lima, P.S.; Casaletti, L.; Bailão, A.M.; de Vasconcelos, A.T.R.; da Fernandes, G.R.; de Soares, C.M.A. Transcriptional and proteomic responses to carbon starvation in Paracoccidioides. PLoS Negl. Trop. Dis. 2014, 8, e2855. [Google Scholar] [CrossRef]
- Teixeira, H.C.; Calich, V.L.; Singer-Vermes, L.M.; D’Imperio-Lima, M.R.; Russo, M. Experimental paracoccidioidomycosis: Early immunosuppression occurs in susceptible mice after infection with pathogenic fungi. Braz. J. Med. Biol. Res. Rev. Bras. Pesqui. Med. E Biol. 1987, 20, 587–589. [Google Scholar]
- Teixeira, M.M.; Theodoro, R.C.; de Carvalho, M.J.A.; Fernandes, L.; Paes, H.C.; Hahn, R.C.; Mendoza, L.; Bagagli, E.; San-Blas, G.; Felipe, M.S.S. Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol. Phylogenet. Evol. 2009, 52, 273–283. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, P.C.; Lima, D.B.; Leprevost, F.V.; Santos, M.D.M.; Fischer, J.S.G.; Aquino, P.F.; Moresco, J.J.; Yates, J.R.; Barbosa, V.C. Integrated analysis of shotgun proteomic data with PatternLab for proteomics 4.0. Nat. Protoc. 2016, 11, 102–117. [Google Scholar] [CrossRef] [PubMed]
- Eng, J.K.; Jahan, T.A.; Hoopmann, M.R. Comet: An open-source MS/MS sequence database search tool. Proteomics 2013, 13, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, P.C.; Fischer, J.S.G.; Xu, T.; Cociorva, D.; Balbuena, T.S.; Valente, R.H.; Perales, J.; Yates, J.R.; Barbosa, V.C. Search engine processor: Filtering and organizing peptide spectrum matches. Proteomics 2012, 12, 944–949. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Elias Moreira, A.L.; Milhomem Cruz-Leite, V.R.; O’Hara Souza Silva, L.; Alves Parente, A.F.; Bailão, A.M.; Maria de Almeida Soares, C.; Parente-Rocha, J.A.; Ruiz, O.H.; Borges, C.L. Proteome characterization of Paracoccidioides lutzii conidia by using nanoUPLC-MSE. Fungal Biol. 2020, 124, 766–780. [Google Scholar] [CrossRef]
- Araújo, F.S.; Coelho, L.M.; do Silva, L.C.; da Silva Neto, B.R.; Parente-Rocha, J.A.; Bailão, A.M.; de Oliveira, C.M.A.; da Fernandes, G.R.; Hernández, O.; Ochoa, J.G.M.; et al. Effects of Argentilactone on the Transcriptional Profile, Cell Wall and Oxidative Stress of Paracoccidioides spp. PLoS Negl. Trop. Dis. 2016, 10, e0004309. [Google Scholar] [CrossRef]
- Borba, J.V.V.B.; Tauhata, S.B.F.; de Oliveira, C.M.A.; Ferreira Marques, M.; Bailão, A.M.; de Soares, C.M.A.; Pereira, M. Chemoproteomic identification of molecular targets of antifungal prototypes, thiosemicarbazide and a camphene derivative of thiosemicarbazide, in Paracoccidioides brasiliensis. PLoS ONE 2018, 13, e0201948. [Google Scholar] [CrossRef]
- Araújo, D.S.; Pereira, M.; Portis, I.G.; de Dos Santos Junior, A.C.M.; Fontes, W.; de Sousa, M.V.; do Assunção, L.P.; Baeza, L.C.; Bailão, A.M.; Ricart, C.A.O.; et al. Metabolic Peculiarities of Paracoccidioides brasiliensis Dimorphism as Demonstrated by iTRAQ Labeling Proteomics. Front. Microbiol. 2019, 10, 555. [Google Scholar] [CrossRef]
- Baeza, L.C.; da Mata, F.R.; Pigosso, L.L.; Pereira, M.; de Souza, G.H.M.F.; Coelho, A.S.G.; de Almeida Soares, C.M. Differential Metabolism of a Two-Carbon Substrate by Members of the Paracoccidioides Genus. Front. Microbiol. 2017, 8, 2308. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, J.F.; Farrer, R.A.; Desjardins, C.A.; Gallo, J.E.; Sykes, S.; Sakthikumar, S.; Misas, E.; Whiston, E.A.; Bagagli, E.; Soares, C.M.A.; et al. Genome Diversity, Recombination, and Virulence across the Major Lineages of Paracoccidioides. mSphere 2016, 1, e00213-16. [Google Scholar] [CrossRef] [PubMed]
- Soares, D.A.; Oliveira, M.B.; Evangelista, A.F.; Venancio, E.J.; Andrade, R.V.; Felipe, M.S.S.; Petrofeza, S. Phospholipase gene expression during Paracoccidioides brasiliensis morphological transition and infection. Mem. Inst. Oswaldo Cruz 2013, 108, 808–811. [Google Scholar] [CrossRef] [PubMed]
- Portis, I.G.; de Sousa Lima, P.; Paes, R.A.; Oliveira, L.N.; Pereira, C.A.; Parente-Rocha, J.A.; Pereira, M.; Nosanchuk, J.D.; de Almeida Soares, C.M. Copper overload in Paracoccidioides lutzii results in the accumulation of ergosterol and melanin. Microbiol. Res. 2020, 239, 126524. [Google Scholar] [CrossRef] [PubMed]
- Petito, G.; de Curcio, J.S.; Pereira, M.; Bailão, A.M.; Paccez, J.D.; Tristão, G.B.; de Morais, C.O.B.; de Souza, M.V.; de Castro Moreira Santos, A.; Fontes, W.; et al. Metabolic Adaptation of Paracoccidioides brasiliensis in Response to in vitro Copper Deprivation. Front. Microbiol. 2020, 11, 1834. [Google Scholar] [CrossRef] [PubMed]
- da Rodrigues, L.N.S.; de Brito, W.A.; Parente, A.F.A.; Weber, S.S.; Bailão, A.M.; Casaletti, L.; Borges, C.L.; de Soares, C.M.A. Osmotic stress adaptation of Paracoccidioides lutzii, Pb01, monitored by proteomics. Fungal Genet. Biol. 2016, 95, 13–23. [Google Scholar] [CrossRef]
- Eisenman, H.C.; Chow, S.-K.; Tsé, K.K.; McClelland, E.E.; Casadevall, A. The effect of L-DOPA on Cryptococcus neoformans growth and gene expression. Virulence 2011, 2, 329–336. [Google Scholar] [CrossRef]
- Nosanchuk, J.D.; Gómez, B.L.; Youngchim, S.; Díez, S.; Aisen, P.; Zancopé-Oliveira, R.M.; Restrepo, A.; Casadevall, A.; Hamilton, A.J. Histoplasma capsulatum synthesizes melanin-like pigments in vitro and during mammalian infection. Infect. Immun. 2002, 70, 5124–5131. [Google Scholar] [CrossRef]
- Figueiredo-Carvalho, M.H.G.; dos Santos, F.B.; Nosanchuk, J.D.; Zancope-Oliveira, R.M.; Almeida-Paes, R. L-Dihydroxyphenylalanine induces melanin production by members of the genus Trichosporon. FEMS Yeast Res. 2014, 14, 988–991. [Google Scholar] [CrossRef]
- Van de Sande, W.W.J.; de Kat, J.; Coppens, J.; Ahmed, A.O.A.; Fahal, A.; Verbrugh, H.; van Belkum, A. Melanin biosynthesis in Madurella mycetomatis and its effect on susceptibility to itraconazole and ketoconazole. Microbes Infect. 2007, 9, 1114–1123. [Google Scholar] [CrossRef]
- Teixeira, P.A.C.; De Castro, R.A.; Ferreira, F.R.L.; Cunha, M.M.L.; Torres, A.P.; Penha, C.V.L.Y.; Rozental, S.; Lopes-Bezerra, L.M. L-DOPA accessibility in culture medium increases melanin expression and virulence of Sporothrix schenckii yeast cells. Med. Mycol. 2010, 48, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Eisenhofer, G.; Tian, H.; Holmes, C.; Matsunaga, J.; Roffler-Tarlov, S.; Hearing, V.J. Tyrosinase: A developmentally specific major determinant of peripheral dopamine. FASEB J. 2003, 17, 1248–1255. [Google Scholar] [CrossRef] [PubMed]
- Peres da Silva, R.; Matsumoto, M.T.; Braz, J.D.; Voltan, A.R.; de Oliveira, H.C.; Soares, C.P.; Mendes Giannini, M.J.S. Differential gene expression analysis of Paracoccidioides brasiliensis during keratinocyte infection. J. Med. Microbiol. 2011, 60, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, M.C.; Nakayasu, E.S.; Matsuo, A.L.; Sobreira, T.J.P.; Longo, L.V.G.; Ganiko, L.; Almeida, I.C.; Puccia, R. Vesicle and vesicle-free extracellular proteome of Paracoccidioides brasiliensis: Comparative analysis with other pathogenic fungi. J. Proteome Res. 2012, 11, 1676–1685. [Google Scholar] [CrossRef]
- Herrero, E.; Ros, J.; Bellí, G.; Cabiscol, E. Redox control and oxidative stress in yeast cells. Biochim. Biophys. Acta 2008, 1780, 1217–1235. [Google Scholar] [CrossRef]
- Silva-Bailão, M.G.; de Lima, P.S.; Oliveira, M.M.E.; Oliveira, L.C.; Almeida-Paes, R.; Borges, C.L.; Bailão, A.M.; Coelho, A.S.G.; de Soares, C.M.A.; Zancopé-Oliveira, R.M. Comparative proteomics in the three major human pathogenic species of the genus Sporothrix. Microbes Infect. 2020. [CrossRef]
- Pigosso, L.L.; Parente, A.F.A.; Coelho, A.S.G.; Silva, L.P.; Borges, C.L.; Bailão, A.M.; de Soares, C.M.A. Comparative proteomics in the genus Paracoccidioides. Fungal Genet. Biol. 2013, 60, 87–100. [Google Scholar] [CrossRef]
- Chew, S.Y.; Ho, K.L.; Cheah, Y.K.; Ng, T.S.; Sandai, D.; Brown, A.J.P.; Than, L.T.L. Glyoxylate cycle gene ICL1 is essential for the metabolic flexibility and virulence of Candida glabrata. Sci. Rep. 2019, 9, 2843. [Google Scholar] [CrossRef]
- Ting, S.Y.; Ishola, O.A.; Ahmed, M.A.; Tabana, Y.M.; Dahham, S.; Agha, M.T.; Musa, S.F.; Muhammed, R.; Than, L.T.L.; Sandai, D. Metabolic adaptation via regulated enzyme degradation in the pathogenic yeast Candida albicans. J. Mycol. Med. 2017, 27, 98–108. [Google Scholar] [CrossRef]
- Marcos, C.M.; de Oliveira, H.C.; de Silva, J.F.; Assato, P.A.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J.S. The multifaceted roles of metabolic enzymes in the Paracoccidioides species complex. Front. Microbiol. 2014, 5, 719. [Google Scholar] [CrossRef]
- Lacerda Pigosso, L.; Baeza, L.C.; Vieira Tomazett, M.; Batista Rodrigues Faleiro, M.; Brianezi Dignani de Moura, V.M.; Melo Bailão, A.; Borges, C.L.; Alves Parente Rocha, J.; Rocha Fernandes, G.; Gauthier, G.M.; et al. Paracoccidioides brasiliensis presents metabolic reprogramming and secretes a serine proteinase during murine infection. Virulence 2017, 8, 1417–1434. [Google Scholar] [CrossRef] [PubMed]
- Parente-Rocha, J.A.; Parente, A.F.A.; Baeza, L.C.; Bonfim, S.M.R.C.; Hernandez, O.; McEwen, J.G.; Bailão, A.M.; Taborda, C.P.; Borges, C.L.; de Soares, C.M.A. Macrophage Interaction with Paracoccidioides brasiliensis Yeast Cells Modulates Fungal Metabolism and Generates a Response to Oxidative Stress. PLoS ONE 2015, 10, e0137619. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.F.F.; Novaes, E.; Pereira, M.; Soares, C.M.A.; Borges, C.L.; Salem-Izacc, S.M. In silico characterization of hypothetical proteins from Paracoccidioides lutzii. Genet. Mol. Res. 2015, 14, 17416–17425. [Google Scholar] [CrossRef] [PubMed]
- Navarro, M.V.; Chaves, A.F.A.; Castilho, D.G.; Casula, I.; Calado, J.C.P.; Conceição, P.M.; Iwai, L.K.; de Castro, B.F.; Batista, W.L. Effect of Nitrosative Stress on the S-Nitroso-Proteome of Paracoccidioides brasiliensis. Front. Microbiol. 2020, 11, 1184. [Google Scholar] [CrossRef]
- Cleare, L.G.; Zamith-Miranda, D.; Nosanchuk, J.D. Heat Shock Proteins in Histoplasma and Paracoccidioides. Clin. Vaccine Immunol. 2017, 24, 24. [Google Scholar] [CrossRef]
- Zügel, U.; Kaufmann, S.H. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin. Microbiol. Rev. 1999, 12, 19–39. [Google Scholar] [CrossRef]
- Carvalho, K.C.; Ganiko, L.; Batista, W.L.; Morais, F.V.; Marques, E.R.; Goldman, G.H.; Franco, M.F.; Puccia, R. Virulence of Paracoccidioides brasiliensis and gp43 expression in isolates bearing known PbGP43 genotype. Microbes Infect. 2005, 7, 55–65. [Google Scholar] [CrossRef]
- Leitão, N.P.; Vallejo, M.C.; Conceição, P.M.; Camargo, Z.P.; Hahn, R.; Puccia, R. Paracoccidioides lutzii Plp43 is an active glucanase with partial antigenic identity with P. brasiliensis gp43. PLoS Negl. Trop. Dis. 2014, 8, e3111. [Google Scholar] [CrossRef]
- Tamayo, D.; Muñoz, J.F.; Lopez, Á.; Urán, M.; Herrera, J.; Borges, C.L.; Restrepo, Á.; Soares, C.M.; Taborda, C.P.; Almeida, A.J.; et al. Identification and Analysis of the Role of Superoxide Dismutases Isoforms in the Pathogenesis of Paracoccidioides spp. PLoS Negl. Trop. Dis. 2016, 10, e0004481. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).