Unconventional Constituents and Shared Molecular Architecture of the Melanized Cell Wall of C. neoformans and Spore Wall of S. cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. S. cerevisiae Spore Growth Conditions
2.2. Spore Cell Wall Sample Preparation
2.3. C. neoformans Cell Growth Conditions
2.4. Melanin Ghost Sample Preparation
2.5. Solid-State NMR Spectroscopy
3. Results
3.1. The 13C CPMAS ssNMR Spectra of S. cerevisiae Spore Walls and C. neoformans Melanin “Ghosts” Display Several Similarities that Suggest Common Structural Elements
3.2. S. cerevisiae Spore Cell Walls and C. neoformans Melanin Ghosts Are both Comprised of Chitinous Polysaccharides, Tyrosine-Derived Constituents, and Neutral Lipids
3.3. 2D 13C-13C ssNMR Implicates Triglycerides as a Component of S. cerevisiae Spore Cell Walls
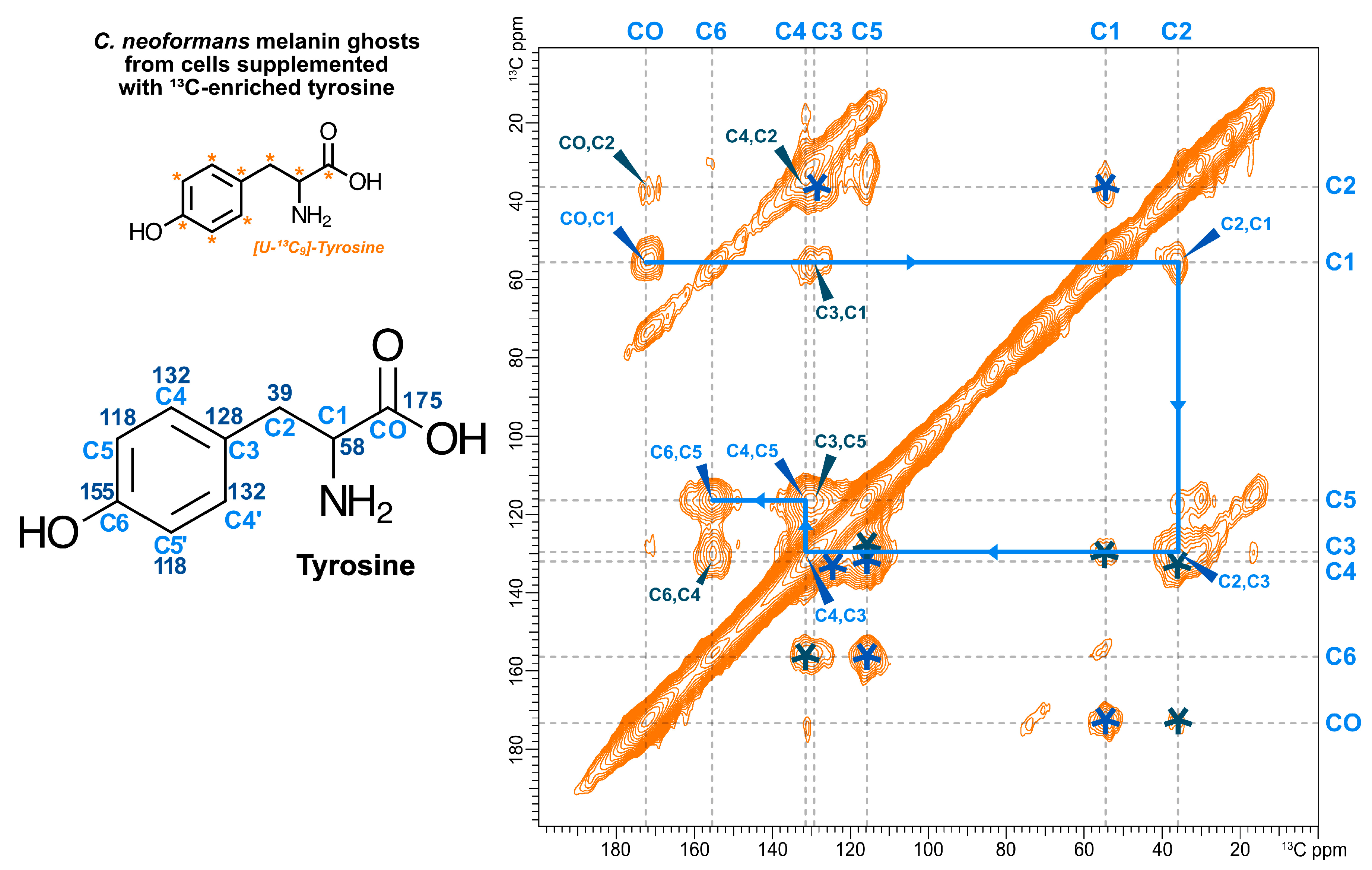
3.4. Exogenously Provided 13C-Enriched Tyrosine is Incorporated into the C. neoformans Melanized Cell Wall as Non-Pigment Constituent
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Free, S.J. Fungal Cell Wall Organization and Biosynthesis. In Advances in Genetics; Friedmann, T., Dunlap, J.C., Goodwin, S.F., Eds.; Elsevier: Waltham, MA, USA, 2013; Volume 81, ISBN 9780124076778. [Google Scholar]
- Brunke, S.; Mogavero, S.; Kasper, L.; Hube, B. Virulence factors in fungal pathogens of man. Curr. Opin. Microbiol. 2016, 32, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.P. The cell wall: A carbohydrate armour for the fungal cell. Mol. Microbiol. 2007, 66, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Herrera, J.; Ortiz-Castellanos, L. Cell wall glucans of fungi. A review. Cell Surf. 2019, 5, 100022. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.P.; Beauvais, A. Functional duality of the cell wall. Curr. Opin. Microbiol. 2014, 20, 111–117. [Google Scholar] [CrossRef]
- Mukaremera, L.; Lee, K.K.; Wagener, J.; Wiesner, D.L.; Gow, N.A.R.; Nielsen, K. Titan cell production in Cryptococcus neoformans reshapes the cell wall and capsule composition during infection. Cell Surf. 2018, 1, 15–24. [Google Scholar] [CrossRef]
- Hernández-Chávez, M.J.; Pérez-García, L.A.; Niño-Vega, G.A.; Mora-Montes, H.M. Fungal strategies to evade the host immune recognition. J. Fungi 2017, 3, 51. [Google Scholar] [CrossRef]
- Hopke, A.; Brown, A.J.P.; Hall, R.A.; Wheeler, R.T. Dynamic fungal cell wall architecture in stress adaptation and immune evasion. Trends Microbiol. 2019, 26, 284–295. [Google Scholar] [CrossRef]
- Hasim, S.; Coleman, J.J. Targeting the fungal cell wall: Current therapies and implications for development of alternative antifungal agents. Future Med. Chem. 2019, 11, 869–883. [Google Scholar] [CrossRef]
- Rajasingham, R.; Smith, R.M.; Park, B.J.; Jarvis, J.N.; Govender, N.P.; Chiller, T.M.; Denning, D.W.; Loyse, A.; Boulware, D.R. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 2017, 17, 873–881. [Google Scholar] [CrossRef]
- Ball, V. Composite materials and films based on melanins, polydopamine, and other catecholamine-based materials. Biomimetics 2017, 2, 12. [Google Scholar] [CrossRef]
- Cordero, R.J.B.; Casadevall, A. Functions of fungal melanin beyond virulence. Fungal Biol. Rev. 2017, 31, 99–112. [Google Scholar] [CrossRef] [PubMed]
- D’Ischia, M.; Napolitano, A.; Pezzella, A.; Meredith, P.; Buehler, M.J. Melanin biopolymers: Tailoring chemical complexity for materials design. Angew. Chem. 2019. [Google Scholar] [CrossRef]
- Eisenman, H.C.; Casadevall, A. Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 2012, 93, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Polacheck, I.; Hearing, V.J.; Kwon-Chung, K.J. Biochemical studies of phenoloxidase and utilization of catecholamines in Cryptococcus neoformans. J. Bacteriol. 1982, 150, 1212–1220. [Google Scholar] [CrossRef]
- Rosas, Á.L.; Nosanchuk, J.D.; Feldmesser, M.; Cox, G.M.; McDade, H.C.; Casadevall, A. Synthesis of polymerized melanin by Cryptococcus neoformans in infected rodents. Infect. Immun. 2000, 68, 2845–2853. [Google Scholar] [CrossRef]
- Ikeda, R.; Sugita, T.; Jacobson, E.S.; Shinoda, T. Effects of melanin upon susceptibility of Cryptococcus to antifungals. Microbiol. Immunol. 2003, 47, 271–277. [Google Scholar] [CrossRef]
- Nosanchuk, J.D.; Valadon, P.; Feldmesser, M.; Casadevall, A. Melanization of Cryptococcus neoformans in Murine Infection. Mol. Cell. Biol. 1999, 19, 745–750. [Google Scholar] [CrossRef]
- Camacho, E.; Vij, R.; Chrissian, C.; Prados-Rosales, R.; Gil, D.; O’Meally, R.N.; Cordero, R.J.B.; Cole, R.N.; McCaffery, J.M.; Stark, R.E.; et al. The structural unit of melanin in the cell wall of the fungal pathogen Cryptococcus neoformans. J. Biol. Chem. 2019, 294, 10471–10489. [Google Scholar] [CrossRef]
- Camacho, E.; Chrissian, C.; Cordero, R.J.B.; Lopes, L.L.; Stark, R.E.; Casadevall, A. N-acetylglucosamine affects cryptococcus neoformans cell-wall composition and melanin architecture. Microbiology 2017, 163, 1540–1556. [Google Scholar] [CrossRef]
- Chrissian, C.; Camacho, E.; Fu, M.S.; Prados-Rosales, R.; Chatterjee, S.; Cordero, R.J.B.; Lodge, J.K.; Casadevall, A.; Stark, R.E. Melanin deposition in two Cryptococcus species depends on cell-wall composition and flexibility. J. Biol. Chem. 2020, 295, 1815–1828. [Google Scholar] [CrossRef]
- Banks, I.R.; Specht, C.A.; Donlin, M.J.; Gerik, K.J.; Levitz, S.M.; Lodge, J.K. A Chitin Synthase and Its Regulator Protein Are Critical for Chitosan Production and Growth of the Fungal Pathogen Cryptococcus neoformans. Eukaryot. Cell 2005, 4, 1902–1912. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.G.; Specht, C.A.; Donlin, M.J.; Lodge, J.K. Chitosan, the Deacetylated Form of Chitin, Is Necessary for Cell Wall Integrity in Cryptococcus neoformans. Eukaryot. Cell 2007, 6, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Prados-Rosales, R.; Itin, B.; Casadevall, A.; Stark, R.E. Solid-state NMR Reveals the Carbon-based Molecular Architecture of Melanized Cryptococcus neoformans Fungal Cells. J. Biol. Chem. 2015, 290, 13779–13790. [Google Scholar] [CrossRef] [PubMed]
- Rosas, A.L.; Nosanchuk, J.D.; Gomez, B.L.; Edens, W.A.; Henson, J.M.; Casadevall, A. Isolation and serological analyses of fungal melanins. J. Immunol. Methods 2000, 244, 69–80. [Google Scholar] [CrossRef]
- Wang, Y.; Aisen, P.; Casadevall, A. Melanin, melanin “ghosts”, and melanin composition in Cryptococcus neoformans. Infect. Immun. 1996, 64, 2420–2424. [Google Scholar] [CrossRef]
- Chrissian, C.; Camacho, E.; Kelly, J.E.; Wang, H.; Casadevall, A.; Stark, R.E. Solid-state NMR spectroscopy identifies three classes of lipids in C. neoformans melanized cell walls and whole fungal cells. J. Biol. Chem. 2020. [Google Scholar] [CrossRef]
- Lynn, R.R.; Magee, P.T. Development of the spore wall during ascospore formation in saccharomyces cerevisiae. J. Cell Biol. 1970, 44, 688–692. [Google Scholar] [CrossRef]
- Neiman, A.M. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 2011, 189, 737–765. [Google Scholar] [CrossRef]
- Briza, P.; Winkler, G.; Kalchhauser, H.; Breitenbach, M. Dityrosine is a prominent component of the yeast ascospore wall. A proof of its structure. J. Biol. Chem. 1986, 261, 4288–4294. [Google Scholar]
- Briza, P.; Ellinger, A.; Winklerll, G.; Breitenbach, M. Characterization of a DL-Dityrosine-containing Yeast Ascospore Walls. J. Biol. Chem. 1990, 265, 15118–15123. [Google Scholar]
- Briza, P.; Ellinger, A.; Winkler, G.; Breitenbach, M. Chemical composition of the yeast ascospore wall. The second outer layer consists of chitosan. J. Biol. Chem. 1988, 263, 11569–11574. [Google Scholar] [PubMed]
- Pammer, M.; Briza, P.; Ellinger, A.; Schustert, T.; Stuckas, R.; Feldmanns, H.; Breitenbachst, M. Dit101 (csd2, cal1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast 1992, 8, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.P.C.; Kim, C.; Smith, S.O.; Neiman, A.M. A Highly Redundant Gene Network Controls Assembly of the Outer Spore Wall in S. cerevisiae. PLoS Genet. 2013, 9, e1003700. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Prados-Rosales, R.; Frases, S.; Itin, B.; Casadevall, A.; Stark, R.E. Using solid-state NMR to monitor the molecular consequences of cryptococcus neoformans melanization with different catecholamine precursors. Biochemistry 2012, 51, 6080–6088. [Google Scholar] [CrossRef] [PubMed]
- Bax, A.; Freeman, R.; Kempsell, S.P. Natural Abundance 13C-13C Coupling Observed via Double-Quantum Coherence. J. Am. Chem. Soc. 1980, 102, 4849–4851. [Google Scholar] [CrossRef]
- Lesage, A.; Auger, C.; Caldarelli, S.; Emsley, L. Determination of through-bond carbon-carbon connectivities in solid-state NMR using the INADEAUATE experiment. J. Am. Chem. Soc. 1997, 119, 7867–7868. [Google Scholar] [CrossRef]
- Bennett, A.E.; Rienstra, C.M.; Auger, M.; Lakshmi, K.V.; Griffin, R.G. Heteronuclear decoupling in rotating solids. J. Chem. Phys. 1995, 103, 6951–6958. [Google Scholar] [CrossRef]
- Takegoshi, K.; Nakamura, S.; Terao, T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001, 344, 631–637. [Google Scholar] [CrossRef]
- Morcombe, C.R.; Gaponenko, V.; Byrd, R.A.; Zilm, K.W. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J. Am. Chem. Soc. 2004, 126, 7196–7197. [Google Scholar] [CrossRef]
- Fung, B.M.; Khitrin, A.K.; Ermolaev, K. An Improved Broadband Decoupling Sequence for Liquid Crystals and Solids. J. Magn. Reson. 2000, 142, 97–101. [Google Scholar] [CrossRef]
- Tian, S.; Garcia-Rivera, J.; Yan, B.; Casadevall, A.; Stark, R.E. Unlocking the molecular structure of fungal melanin using 13C biosynthetic labeling and solid-state NMR. Biochemistry 2003, 42, 8105–8109. [Google Scholar] [CrossRef] [PubMed]
- Polacheck, I.; Platt, Y.; Aronovitch, J. Catecholamines and virulence of Cryptococcus neoformans. Infect. Immun. 1990, 58, 2919–2922. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Frases, S.; Wang, H.; Casadevall, A.; Stark, R.E. Following fungal melanin biosynthesis with solid-state NMR: Biopolymer molecular structures and possible connections to cell-wall polysaccharides. Biochemistry 2008, 47, 4701–4710. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Prados-rosales, R.; Tan, S.; Phan, V.C.; Itin, B.; Wang, H.; Khajo, A.; Magliozzo, R.S. The melanization road more traveled by: Precursor substrate effects on melanin synthesis in cell-free and fungal cell systems. J. Biol. Chem. 2018, 293, 20157–20168. [Google Scholar] [CrossRef]
- Tanner, S. High-Resolution Solid-state Carbon-13 Nuclear Magnetic Resonance Study of Chitin. Macromolecules 1990, 23, 3576–3583. [Google Scholar] [CrossRef]
- Heux, L.; Brugnerotto, J.; Desbrières, J.; Versali, M.F.; Rinaudo, M. Solid state NMR for determination of degree of acetylation of chitin and chitosan. Biomacromolecules 2000, 1, 746–751. [Google Scholar] [CrossRef]
- Varum, K.; Anthonsen, M.; Grasdalen, H.; Smidsrod, O. 13C-N.m.r. studies of the acetylation sequences in partially N-deacetylated chitin (chitosans). Carbohydr. Res. 1991, 217, 19–27. [Google Scholar] [CrossRef]
- Varum, K.M.; Anthonsen, M.W.; Grasdalen, H.; Smidsrod, O. Determination of the degree of N-acetylation and the distri-bution of N-acetyl groups in partially N-deacetylated chitins (chitosans) by high-field n.m.r. spectroscopy. Carbohydr. Res. 1991, 211, 17–23. [Google Scholar] [CrossRef]
- Ottoy, M.H.; Varum, K.M.; Smidsrod, O. Compositional heterogeneity of heterogeneously deacetylated chitosans. Carbohydr. Polym. 1996, 29, 17–24. [Google Scholar] [CrossRef]
- Alemany, L.B. Using simple 13C NMR linewidth and relaxation measurements to make detailed chemical shift assignments in triacylglycerols and related compounds. Chem. Phys. Lipids 2002, 120, 33–44. [Google Scholar] [CrossRef]
- McKenzie, J.M.; Koch, K.R. Rapid analysis of major components and potential authentication of South African olive oils by quantitative 13C nuclear magnetic resonance spectroscopy. S. Afr. J. Sci. 2004, 100, 349–354. [Google Scholar]
- Gunstone, F.D. The 13C-NMR spectra of oils containing γ-linolenic acid. Chem. Phys. Lipids 1990, 56, 201–207. [Google Scholar] [CrossRef]
- Mannina, L.; Luchinat, C.; Emanuele, M.C.; Segre, A. Acyl positional distribution of glycerol tri-esters in vegetable oils: A 13C NMR study. Chem. Phys. Lipids 1999, 103, 47–55. [Google Scholar] [CrossRef]
- Lie Ken Jie, M.; Lam, C. 13C-Nuclear magnetic resonance spectroscopic studies of triacylglycerols of type AAA containing (Z)-and (E)-monoethylenic acyl groups. Chem. Phys. Lipids 1995, 78, 15–27. [Google Scholar] [CrossRef]
- Lie Ken Jie, M.S.F.; Mustafa, J. High-resolution nuclear magnetic resonance spectroscopy—Applications to fatty acids and triacylglycerols. Lipids 1997, 32, 1019–1034. [Google Scholar] [CrossRef]
- Alexandri, E.; Ahmed, R.; Siddiqui, H.; Choudhary, M.I.; Tsiafoulis, C.G.; Gerothanassis, I.P. High resolution NMR spectroscopy as a structural and analytical tool for unsaturated lipids in solution. Molecules 2017, 22, 1663. [Google Scholar] [CrossRef]
- Li, J.; Vosegaard, T.; Guo, Z. Applications of nuclear magnetic resonance in lipid analyses: An emerging powerful tool for lipidomics studies. Prog. Lipid Res. 2017, 68, 37–56. [Google Scholar] [CrossRef]
- Eisenman, H.C.; Mues, M.; Weber, S.E.; Frases, S.; Chaskes, S.; Gerfen, G.; Casadevall, A. Cryptococcus neoformans laccase catalyses melanin synthesis from both D- and L-DOPA. Microbiology 2007, 153, 3954–3962. [Google Scholar] [CrossRef]
- Baldrian, P. Fungal laccases-occurrence and properties. FEMS Microbiol. Rev. 2006, 30, 215–242. [Google Scholar] [CrossRef]
- Rinaudo, M.; Ã, M.R. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Świderska-Burek, U.; Polak, J.; Sulej, J.; Jarosz-Wilkołazka, A.; Paszczyński, A. Laccase properties, physiological functions, and evolution. Int. J. Mol. Sci. 2020, 21, 966. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, J.P. Utilization of low molecular weight aromatic compounds by heterobasidiomycetous yeasts: Taxonomic implications. Can. J. Microbiol. 1999, 45, 491–512. [Google Scholar] [CrossRef] [PubMed]
- Illingworth, R.F.; Rose, A.H.; Beckett, A. Changes in the lipid composition and fine structure of Saccharomyces cerevisiae during ascus formation. J. Bacteriol. 1973, 113, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Zweytick, D.; Athenstaedt, K.; Nther Daum, G. Intracellular lipid particles of eukaryotic cells. Biochim. Biophys. Acta 2000, 1469, 101–120. [Google Scholar] [CrossRef]
- Coluccio, A.; Bogengruber, E.; Conrad, M.N.; Dresser, M.E.; Briza, P.; Neiman, A.M. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 2004, 3, 1464–1475. [Google Scholar] [CrossRef]
- Ren, J.; Lin, C.P.C.; Pathak, M.C.; Temple, B.R.S.; Nile, A.H.; Mousley, C.J.; Duncan, M.C.; Eckert, D.M.; Leiker, T.J.; Ivanova, P.T.; et al. A phosphatidylinositol transfer protein integrates phosphoinositide signaling with lipid droplet metabolism to regulate a developmental program of nutrient stress-induced membrane biogenesis. Mol. Biol. Cell 2014, 25, 712–727. [Google Scholar] [CrossRef]
- Lam, C.; Santore, E.; Lavoie, E.; Needleman, L.; Fiacco, N.; Kim, C.; Neiman, A.M. A visual screen of protein localization during sporulation identifies new components of prospore membrane-associated complexes in budding yeast. Eukaryot. Cell 2014, 13, 383–391. [Google Scholar] [CrossRef]
- Hsu, T.H.; Chen, R.H.; Cheng, Y.H.; Wang, C.W.; Riezman, H. Lipid droplets are central organelles for meiosis II progression during yeast sporulation. Mol. Biol. Cell 2017, 28, 440–451. [Google Scholar] [CrossRef]
- Hoffmann, R.; Grabińska, K.; Guan, Z.; Sessa, W.C.; Neiman, A.M. Long-chain polyprenols promote spore wall formation in Saccharomyces cerevisiae. Genetics 2017, 207, 1371–1386. [Google Scholar] [CrossRef][Green Version]
- Kelly, J.E.; Chrissian, C.; Stark, R.E. Tailoring NMR experiments for structural characterization of amorphous biological solids: A practical guide. Solid State Nucl. Magn. Reson. 2020, 109, 1–9. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, M.; Zhang, D.; Huang, Y.; Chen, L.; Jiang, S.; Shi, K.; Li, R. Preparation, optimization, and characterization of chitosan-coated solid lipid nanoparticles for ocular drug delivery. J. Biomed. Res. 2018, 32, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Malgarim Cordenonsi, L.; Faccendini, A.; Catanzaro, M.; Bonferoni, M.C.; Rossi, S.; Malavasi, L.; Platcheck Raffin, R.; Scherman Schapoval, E.E.; Lanni, C.; Sandri, G.; et al. The role of chitosan as coating material for nanostructured lipid carriers for skin delivery of fucoxanthin. Int. J. Pharm. 2019, 567. [Google Scholar] [CrossRef] [PubMed]
- De Cristo Soares Alves, A.; Lavayen, V.; Figueiró, F.; Dallemole, D.R.; de Fraga Dias, A.; Cé, R.; Battastini, A.M.O.; Guterres, S.S.; Pohlmann, A.R.; Ball, V.; et al. Chitosan-Coated Lipid-Core Nanocapsules Functionalized with Gold-III and Bevacizumab Induced In Vitro Cytotoxicity against C6 Cell Line and In Vivo Potent Antiangiogenic Activity. Pharm. Res. 2020, 37, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Needleman, L.; Zhou, S.; Neiman, A.M. A novel assay reveals a maturation process during ascospore wall formation. J. Fungi 2017, 3, 54. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.; Berbee, M. Dating divergences in the Fungal Tree of Life: Review and new analyses. Mycologia 2006, 98, 838–849. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chrissian, C.; Lin, C.P.-C.; Camacho, E.; Casadevall, A.; Neiman, A.M.; Stark, R.E. Unconventional Constituents and Shared Molecular Architecture of the Melanized Cell Wall of C. neoformans and Spore Wall of S. cerevisiae. J. Fungi 2020, 6, 329. https://doi.org/10.3390/jof6040329
Chrissian C, Lin CP-C, Camacho E, Casadevall A, Neiman AM, Stark RE. Unconventional Constituents and Shared Molecular Architecture of the Melanized Cell Wall of C. neoformans and Spore Wall of S. cerevisiae. Journal of Fungi. 2020; 6(4):329. https://doi.org/10.3390/jof6040329
Chicago/Turabian StyleChrissian, Christine, Coney Pei-Chen Lin, Emma Camacho, Arturo Casadevall, Aaron M. Neiman, and Ruth E. Stark. 2020. "Unconventional Constituents and Shared Molecular Architecture of the Melanized Cell Wall of C. neoformans and Spore Wall of S. cerevisiae" Journal of Fungi 6, no. 4: 329. https://doi.org/10.3390/jof6040329
APA StyleChrissian, C., Lin, C. P.-C., Camacho, E., Casadevall, A., Neiman, A. M., & Stark, R. E. (2020). Unconventional Constituents and Shared Molecular Architecture of the Melanized Cell Wall of C. neoformans and Spore Wall of S. cerevisiae. Journal of Fungi, 6(4), 329. https://doi.org/10.3390/jof6040329






