Novel Flu Viruses in Bats and Cattle: “Pushing the Envelope” of Influenza Infection
Abstract
1. Introduction
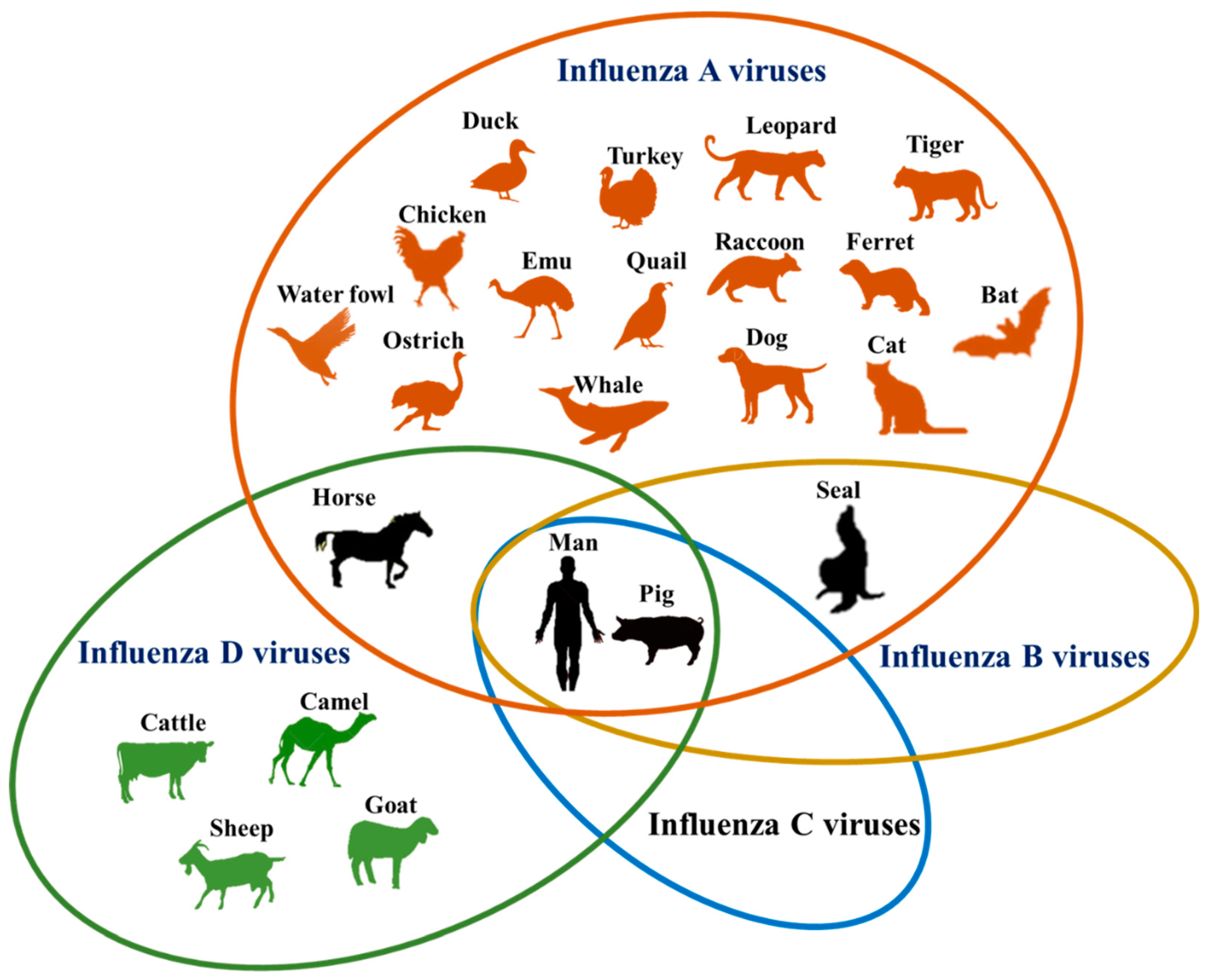
2. Influenza
3. Influenza Viruses
4. Expanding Host Range of IAVs
5. Novel Influenza Viruses in Bats
6. Role of Bats in the Ecology and Evolution of IAVs
7. Bats as “Mixing Vessels” of IAVs
8. Novel Flu Viruses in Cattle and Reclassification of Orthomyxoviridae
9. Properties of IDVs
10. Animal and Human Health Significance of IDVs
11. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Potter, C.W. A history of influenza. J. Appl. Microbiol. 2001, 91, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Morens, D.M. 1918 Influenza: The mother of all pandemics. Emerg. Infect. Dis. 2006, 12, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K. The origin and virulence of the 1918 “Spanish” influenza virus. Proc. Am. Philos. Soc. 2006, 150, 86–112. [Google Scholar] [PubMed]
- Paules, C.; Subbarao, K. Influenza. Lancet 2017, 390, 697–708. [Google Scholar] [CrossRef]
- Saunders-Hastings, P.R.; Krewski, D. Reviewing the History of Pandemic Influenza: Understanding Patterns of Emergence and Transmission. Pathogens 2016, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Crescenzo-Chaigne, B.; Barbezange, C.; van der Werf, S. Non coding extremities of the seven influenza virus type C vRNA segments: Effect on transcription and replication by the type C and type A polymerase complexes. Virol. J. 2008, 5, 132. [Google Scholar] [CrossRef] [PubMed]
- Ran, Z.; Shen, H.; Lang, Y.; Kolb, E.A.; Turan, N.; Zhu, L.; Ma, J.; Bawa, B.; Liu, Q.; Liu, H.; et al. Domestic pigs are susceptible to infection with influenza B viruses. J. Virol. 2015, 89, 4818–4826. [Google Scholar] [CrossRef] [PubMed]
- Zambon, M.C. Epidemiology and pathogenesis of influenza. J. Antimicrob. Chemother. 1999, 44 (Suppl. B), 3–9. [Google Scholar] [CrossRef]
- Webster, R.G.; Hinshaw, V.S.; Laver, W.G. Selection and analysis of antigenic variants of the neuraminidase of N2 influenza viruses with monoclonal antibodies. Virology 1982, 117, 93–104. [Google Scholar] [CrossRef]
- Garten, R.J.; Davis, C.T.; Russell, C.A.; Shu, B.; Lindstrom, S.; Balish, A.; Sessions, W.M.; Xu, X.; Skepner, E.; Deyde, V.; et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009, 325, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Kawaoka, Y.; Krauss, S.; Webster, R.G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 1989, 63, 4603–4608. [Google Scholar] [PubMed]
- Li, C.; Hatta, M.; Watanabe, S.; Neumann, G.; Kawaoka, Y. Compatibility among polymerase subunit proteins is a restricting factor in reassortment between equine H7N7 and human H3N2 influenza viruses. J. Virol. 2008, 82, 11880–11888. [Google Scholar] [CrossRef] [PubMed]
- Gerber, M.; Isel, C.; Moules, V.; Marquet, R. Selective packaging of the influenza A genome and consequences for genetic reassortment. Trends Microbiol. 2014, 22, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Goto, H.; Muramoto, Y.; Noda, T.; Kawaoka, Y. The genome-packaging signal of the influenza A virus genome comprises a genome incorporation signal and a genome-bundling signal. J. Virol. 2013, 87, 11316–11322. [Google Scholar] [CrossRef] [PubMed]
- Fournier, E.; Moules, V.; Essere, B.; Paillart, J.C.; Sirbat, J.D.; Cavalier, A.; Rolland, J.P.; Thomas, D.; Lina, B.; Isel, C.; et al. Interaction network linking the human H3N2 influenza A virus genomic RNA segments. Vaccine 2012, 30, 7359–7367. [Google Scholar] [CrossRef] [PubMed]
- Fournier, E.; Moules, V.; Essere, B.; Paillart, J.C.; Sirbat, J.D.; Isel, C.; Cavalier, A.; Rolland, J.P.; Thomas, D.; Lina, B.; et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res. 2012, 40, 2197–2209. [Google Scholar] [CrossRef] [PubMed]
- Gavazzi, C.; Isel, C.; Fournier, E.; Moules, V.; Cavalier, A.; Thomas, D.; Lina, B.; Marquet, R. An in vitro network of intermolecular interactions between viral RNA segments of an avian H5N2 influenza A virus: Comparison with a human H3N2 virus. Nucleic Acids Res. 2013, 41, 1241–1254. [Google Scholar] [CrossRef] [PubMed]
- Gavazzi, C.; Yver, M.; Isel, C.; Smyth, R.P.; Rosa-Calatrava, M.; Lina, B.; Moules, V.; Marquet, R. A functional sequence-specific interaction between influenza A virus genomic RNA segments. Proc. Natl. Acad. Sci. USA 2013, 110, 16604–16609. [Google Scholar] [CrossRef] [PubMed]
- Cauldwell, A.V.; Long, J.S.; Moncorge, O.; Barclay, W.S. Viral determinants of influenza A virus host range. J. Gen. Virol. 2014, 95, 1193–1210. [Google Scholar] [CrossRef] [PubMed]
- Jeong, O.M.; Kim, M.C.; Kim, M.J.; Kang, H.M.; Kim, H.R.; Kim, Y.J.; Joh, S.J.; Kwon, J.H.; Lee, Y.J. Experimental infection of chickens, ducks and quails with the highly pathogenic H5N1 avian influenza virus. J. Vet. Sci. 2009, 10, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Nelli, R.K.; Dunham, S.P.; Kuchipudi, S.V.; White, G.A.; Baquero-Perez, B.; Chang, P.; Ghaemmaghami, A.; Brookes, S.M.; Brown, I.H.; Chang, K.C. Mammalian innate resistance to highly pathogenic avian influenza H5N1 virus infection is mediated through reduced proinflammation and infectious virus release. J. Virol. 2012, 86, 9201–9210. [Google Scholar] [CrossRef] [PubMed]
- Parrish, C.R.; Murcia, P.R.; Holmes, E.C. Influenza virus reservoirs and intermediate hosts: Dogs, horses, and new possibilities for influenza virus exposure of humans. J. Virol. 2015, 89, 2990–2994. [Google Scholar] [CrossRef] [PubMed]
- Fiorentini, L.; Taddei, R.; Moreno, A.; Gelmetti, D.; Barbieri, I.; De Marco, M.A.; Tosi, G.; Cordioli, P.; Massi, P. Influenza A pandemic (H1N1) 2009 virus outbreak in a cat colony in Italy. Zoonoses Public Health 2011, 58, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Hatta, M.; Zhong, G.; Gao, Y.; Nakajima, N.; Fan, S.; Chiba, S.; Deering, K.M.; Ito, M.; Imai, M.; Kiso, M.; et al. Characterization of a Feline Influenza A(H7N2) Virus. Emerg. Infect. Dis. 2018, 24, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Kim, H.; Na, W.; Hong, M.; Park, S.J.; Moon, H.; Kang, B.; Lyoo, K.S.; Yeom, M.; Jeong, D.G.; et al. Canine susceptibility to human influenza viruses (A/pdm 09H1N1, A/H3N2 and B). J. Gen. Virol. 2015, 96, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Kuchipudi, S.V.; Tellabati, M.; Sebastian, S.; Londt, B.Z.; Jansen, C.; Vervelde, L.; Brookes, S.M.; Brown, I.H.; Dunham, S.P.; Chang, K.C. Highly pathogenic avian influenza virus infection in chickens but not ducks is associated with elevated host immune and pro-inflammatory responses. Vet. Res. 2014, 45, 118. [Google Scholar] [CrossRef] [PubMed]
- Puryear, W.B.; Keogh, M.; Hill, N.; Moxley, J.; Josephson, E.; Davis, K.R.; Bandoro, C.; Lidgard, D.; Bogomolni, A.; Levin, M.; et al. Prevalence of influenza A virus in live-captured North Atlantic gray seals: A possible wild reservoir. Emerg. Microbes Infect. 2016, 5, e81. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Li, Y.; Rivailler, P.; Conrardy, C.; Castillo, D.A.; Chen, L.M.; Recuenco, S.; Ellison, J.A.; Davis, C.T.; York, I.A.; et al. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. USA 2012, 109, 4269–4274. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.; Recuenco, S.; Gomez, J.; et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013, 9, e1003657. [Google Scholar] [CrossRef] [PubMed]
- Smith, I.; Wang, L.F. Bats and their virome: An important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 2013, 3, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Memish, Z.A.; Mishra, N.; Olival, K.J.; Fagbo, S.F.; Kapoor, V.; Epstein, J.H.; Alhakeem, R.; Durosinloun, A.; Al Asmari, M.; Islam, A.; et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013, 19, 1819–1823. [Google Scholar] [CrossRef] [PubMed]
- Brook, C.E.; Dobson, A.P. Bats as “special” reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015, 23, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.L.; Firth, C.; Conte, J.M.; Williams, S.H.; Zambrana-Torrelio, C.M.; Anthony, S.J.; Ellison, J.A.; Gilbert, A.T.; Kuzmin, I.V.; Niezgoda, M.; et al. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. USA 2013, 110, 8194–8199. [Google Scholar] [CrossRef] [PubMed]
- Moreira, E.A.; Locher, S.; Kolesnikova, L.; Bolte, H.; Aydillo, T.; Garcia-Sastre, A.; Schwemmle, M.; Zimmer, G. Synthetically derived bat influenza A-like viruses reveal a cell type- but not species-specific tropism. Proc. Natl. Acad. Sci. USA 2016. [Google Scholar] [CrossRef] [PubMed]
- Juozapaitis, M.; Aguiar Moreira, E.; Mena, I.; Giese, S.; Riegger, D.; Pohlmann, A.; Hoper, D.; Zimmer, G.; Beer, M.; Garcia-Sastre, A.; et al. An infectious bat-derived chimeric influenza virus harbouring the entry machinery of an influenza A virus. Nat. Commun. 2014, 5, 4448. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Ma, J.; Liu, Q.; Bawa, B.; Wang, W.; Shabman, R.S.; Duff, M.; Lee, J.; Lang, Y.; Cao, N.; et al. Characterization of uncultivable bat influenza virus using a replicative synthetic virus. PLoS Pathog. 2014, 10, e1004420. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, J.; Ma, J.; Lang, Y.; Nietfeld, J.; Li, Y.; Duff, M.; Li, Y.; Yang, Y.; Liu, H.; et al. Pathogenicity of modified bat influenza virus with different M genes and its reassortment potential with swine influenza A virus. J. Gen. Virol. 2017, 98, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Noronha, J.M.; Liu, M.; Squires, R.B.; Pickett, B.E.; Hale, B.G.; Air, G.M.; Galloway, S.E.; Takimoto, T.; Schmolke, M.; Hunt, V.; et al. Influenza virus sequence feature variant type analysis: Evidence of a role for NS1 in influenza virus host range restriction. J. Virol. 2012, 86, 5857–5866. [Google Scholar] [CrossRef] [PubMed]
- Hale, B.G. Conformational plasticity of the influenza A virus NS1 protein. J. Gen. Virol. 2014, 95, 2099–2105. [Google Scholar] [CrossRef] [PubMed]
- Turkington, H.L.; Juozapaitis, M.; Tsolakos, N.; Corrales-Aguilar, E.; Schwemmle, M.; Hale, B.G. Unexpected Functional Divergence of Bat Influenza Virus NS1 Proteins. J. Virol. 2018, 92, JVI-02097. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Sun, X.; Li, Z.; Liu, Y.; Vavricka, C.J.; Qi, J.; Gao, G.F. Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus. Proc. Natl. Acad. Sci. USA 2012, 109, 18897–18902. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Shi, Y.; Lu, X.; He, J.; Gao, F.; Yan, J.; Qi, J.; Gao, G.F. Bat-derived influenza hemagglutinin H17 does not bind canonical avian or human receptors and most likely uses a unique entry mechanism. Cell Rep. 2013, 3, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Freidl, G.S.; Binger, T.; Muller, M.A.; de Bruin, E.; van Beek, J.; Corman, V.M.; Rasche, A.; Drexler, J.F.; Sylverken, A.; Oppong, S.K.; et al. Serological evidence of influenza A viruses in frugivorous bats from Africa. PLoS ONE 2015, 10, e0127035. [Google Scholar] [CrossRef] [PubMed]
- Poole, D.S.; Yu, S.; Cai, Y.; Dinis, J.M.; Muller, M.A.; Jordan, I.; Friedrich, T.C.; Kuhn, J.H. Influenza A virus polymerase is a site for adaptive changes during experimental evolution in bat cells. J. Virol. 2014, 88, 12572–12585. [Google Scholar] [CrossRef] [PubMed]
- Kuchipudi, S.V.; Nelli, R.; White, G.A.; Bain, M.; Chang, K.C.; Dunham, S. Differences in influenza virus receptors in chickens and ducks: Implications for interspecies transmission. J. Mol. Genet. Med. 2009, 3, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Lakadamyali, M.; Rust, M.J.; Zhuang, X. Endocytosis of influenza viruses. Microbes Infect. 2004, 6, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Nelli, R.K.; Kuchipudi, S.V.; White, G.A.; Perez, B.B.; Dunham, S.P.; Chang, K.-C. Comparative distribution of human and avian type sialic acid influenza receptors in the pig. BMC Vet. Res. 2010, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chothe, S.K.; Bhushan, G.; Nissly, R.H.; Yeh, Y.T.; Brown, J.; Turner, G.; Fisher, J.; Sewall, B.J.; Reeder, D.M.; Terrones, M.; et al. Avian and human influenza virus compatible sialic acid receptors in little brown bats. Sci. Rep. 2017, 7, 660. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013, 9, e1003176. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and Swine: Proposal for a new genus in the Orthomyxoviridae family. mBio 2014, 5, e00031-14. [Google Scholar] [CrossRef] [PubMed]
- Ducatez, M.F.; Pelletier, C.; Meyer, G. Influenza D virus in cattle, France, 2011–2014. Emerg. Infect. Dis. 2015, 21, 368–371. [Google Scholar] [CrossRef] [PubMed]
- Chiapponi, C.; Faccini, S.; De Mattia, A.; Baioni, L.; Barbieri, I.; Rosignoli, C.; Nigrelli, A.; Foni, E. Detection of Influenza D Virus among Swine and Cattle, Italy. Emerg. Infect. Dis. 2016, 22, 352–354. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.L.; Zhang, H.; Chen, S.N.; Zhou, X.; Lin, T.; Liu, R.; Lv, D.H.; Wen, X.H.; Wei, W.K.; Wang, D.; et al. Influenza D Virus in Animal Species in Guangdong Province, Southern China. Emerg. Infect. Dis. 2017, 23, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Endoh, M.; Kobayashi, T.; Takenaka-Uema, A.; Chambers, J.K.; Uchida, K.; Nishihara, M.; Hause, B.; Horimoto, T. Influenza D Virus Infection in Herd of Cattle, Japan. Emerg. Infect. Dis. 2016, 22, 1517–1519. [Google Scholar] [CrossRef] [PubMed]
- Flynn, O.; Gallagher, C.; Mooney, J.; Irvine, C.; Ducatez, M.; Hause, B.; McGrath, G.; Ryan, E. Influenza D Virus in Cattle, Ireland. Emerg. Infect. Dis. 2018, 24, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Salem, E.; Cook, E.A.J.; Lbacha, H.A.; Oliva, J.; Awoume, F.; Aplogan, G.L.; Hymann, E.C.; Muloi, D.; Deem, S.L.; Alali, S.; et al. Serologic Evidence for Influenza C and D Virus among Ruminants and Camelids, Africa, 1991–2015. Emerg. Infect. Dis. 2017, 23, 1556–1559. [Google Scholar] [CrossRef] [PubMed]
- ICTV. Virus Taxonomy: 2017 Release. Available online: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/negative-sense-rna-viruses-2011/w/negrna_viruses/209/orthomyxoviridae (accessed on 5 August 2018).
- ICTV. Virus Taxonomy: 2009 Release. Available online: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/negative-sense-rna-viruses-2011/w/negrna_viruses/209/orthomyxoviridae (accessed on 5 August 2018).
- Ferguson, L.; Olivier, A.K.; Genova, S.; Epperson, W.B.; Smith, D.R.; Schneider, L.; Barton, K.; McCuan, K.; Webby, R.J.; Wan, X.F. Pathogenesis of Influenza D Virus in Cattle. J. Virol. 2016, 90, 5636–5642. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Qi, J.; Khedri, Z.; Diaz, S.; Yu, H.; Chen, X.; Varki, A.; Shi, Y.; Gao, G.F. An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism. PLoS Pathog. 2016, 12, e1005411. [Google Scholar]
- Mitra, N.; Cernicchiaro, N.; Torres, S.; Li, F.; Hause, B.M. Metagenomic characterization of the virome associated with bovine respiratory disease in feedlot cattle identified novel viruses and suggests an etiologic role for influenza D virus. J. Gen. Virol. 2016, 97, 1771–1784. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.F.; Kondov, N.O.; Deng, X.; Van Eenennaam, A.; Neibergs, H.L.; Delwart, E. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J. Virol. 2015, 89, 5340–5349. [Google Scholar] [CrossRef] [PubMed]
- Hilton, W.M. BRD in 2014: Where have we been, where are we now, and where do we want to go? Anim. Health Res. Rev. 2014, 15, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Portis, E.; Lindeman, C.; Johansen, L.; Stoltman, G. A ten-year (2000–2009) study of antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex—Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni—In the United States and Canada. J. Vet. Diagn. Investig. 2012, 24, 932–944. [Google Scholar] [CrossRef] [PubMed]
- Nickell, J.S.; White, B.J. Metaphylactic antimicrobial therapy for bovine respiratory disease in stocker and feedlot cattle. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 285–301. [Google Scholar] [CrossRef] [PubMed]
- Mosier, D. Review of BRD pathogenesis: The old and the new. Anim. Health Res. Rev. 2014, 15, 166–168. [Google Scholar] [CrossRef] [PubMed]
- Duff, G.C.; Galyean, M.L. Board-invited review: Recent advances in management of highly stressed, newly received feedlot cattle. J. Anim. Sci. 2007, 85, 823–840. [Google Scholar] [CrossRef] [PubMed]
- White, S.K.; Ma, W.; McDaniel, C.J.; Gray, G.C.; Lednicky, J.A. Serologic evidence of exposure to influenza D virus among persons with occupational contact with cattle. J. Clin. Virol. 2016, 81, 31–33. [Google Scholar] [CrossRef] [PubMed]
- Longdon, B.; Brockhurst, M.A.; Russell, C.A.; Welch, J.J.; Jiggins, F.M. The evolution and genetics of virus host shifts. PLoS Pathog. 2014, 10, e1004395. [Google Scholar] [CrossRef] [PubMed]
- Davies, T.J.; Pedersen, A.B. Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc. Biol. Sci. 2008, 275, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, J.D.; Krasnov, B.R.; Poulin, R.; Nakagawa, S. A tale of two phylogenies: Comparative analyses of ecological interactions. Am. Nat. 2014, 183, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.E.; Haydon, D.T.; Antia, R. Emerging pathogens: The epidemiology and evolution of species jumps. Trends Ecol. Evol. 2005, 20, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.E.; Gowtage-Sequeria, S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005, 11, 1842–1847. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.; Scott, F.; Hudson, Z.; Howey, R.; Chase-Topping, M. Human viruses: Discovery and emergence. Philos. Trans R. Soc. 2012, 367, 2864–2871. [Google Scholar] [CrossRef] [PubMed]
- Imai, M.; Watanabe, T.; Hatta, M.; Das, S.C.; Ozawa, M.; Shinya, K.; Zhong, G.; Hanson, A.; Katsura, H.; Watanabe, S.; et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 2012, 486, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Ibrahim, M.S.; Ellakany, H.F.; Kawashita, N.; Mizuike, R.; Hiramatsu, H.; Sriwilaijaroen, N.; Takagi, T.; Suzuki, Y.; Ikuta, K. Acquisition of human-type receptor binding specificity by new H5N1 influenza virus sublineages during their emergence in birds in Egypt. PLoS Pathog. 2011, 7, e1002068. [Google Scholar] [CrossRef] [PubMed]
| Genus | Species | Genomic Segments |
|---|---|---|
| Alphainfluenzavirus | Influenza A virus | 8 |
| Betainfluenzavirus | Influenza B virus | 8 |
| Deltainfluenzavirus | Influenza D virus | 7 |
| Gammainfluenzavirus | Influenza C virus | 7 |
| Isavirus | Salmon isavirus | 8 |
| Quaranjavirus | Johnston Atoll quaranjavirus | 6 |
| Quaranfil quaranjavirus | ||
| Thogotovirus | Dhori thogotovirus | 6 |
| Thogoto thogotovirus | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuchipudi, S.V.; Nissly, R.H. Novel Flu Viruses in Bats and Cattle: “Pushing the Envelope” of Influenza Infection. Vet. Sci. 2018, 5, 71. https://doi.org/10.3390/vetsci5030071
Kuchipudi SV, Nissly RH. Novel Flu Viruses in Bats and Cattle: “Pushing the Envelope” of Influenza Infection. Veterinary Sciences. 2018; 5(3):71. https://doi.org/10.3390/vetsci5030071
Chicago/Turabian StyleKuchipudi, Suresh V., and Ruth H. Nissly. 2018. "Novel Flu Viruses in Bats and Cattle: “Pushing the Envelope” of Influenza Infection" Veterinary Sciences 5, no. 3: 71. https://doi.org/10.3390/vetsci5030071
APA StyleKuchipudi, S. V., & Nissly, R. H. (2018). Novel Flu Viruses in Bats and Cattle: “Pushing the Envelope” of Influenza Infection. Veterinary Sciences, 5(3), 71. https://doi.org/10.3390/vetsci5030071






