Simple Summary
Parainfluenza virus 5 (PIV5), a member of the Orthorubulavirus genus within the Paramyxoviridae family, infects diverse mammalian species, including humans. In this study, our primary objective was to isolate porcine epidemic diarrhea virus (PEDV) from intestinal tissue samples of infected piglets. Unexpectedly, we successfully isolated the PIV5 strain (SC2024) from diarrhea piglets. Genetic analysis revealed that SC2024 clustered within the same branch as most previously reported swine-derived PIV5 strains in China, and shared some genetic characteristics with strains isolated from diverse hosts, including tigers, pangolins, lesser pandas, and ticks. Collectively, these findings enhance our comprehension of PIV5 epidemiology in swine populations and highlight its cross-species transmission potential.
Abstract
Parainfluenza virus 5 (PIV5) is recognized as a pathogen capable of infecting diverse animal species and humans, posing a potential threat to public health. In this study, the PIV5 strain SC2024 was isolated from the PEDV-positive intestinal tissue of piglets. Using next-generation sequencing (NGS), we determined the full genome length of the virus to be 15,246 nucleotides (nt). Phylogenetic analysis showed that SC2024 grouped into the same branch (Lineage 2.2) as most of the previously reported swine-origin PIV5 strains in China. Additionally, it exhibited genetic similarity with isolates from various hosts, such as tigers, pangolins, lesser pandas, and ticks. Overall, these findings expand our comprehension of PIV5 epidemiology in swine populations, and underscore the importance of further research and surveillance.
1. Introduction
Parainfluenza virus 5 (PIV5), now reclassified as Orthorubulavirus mammalis, is a negative-sense, single-stranded RNA virus belonging to the genus Orthorubulavirus of the family Paramyxoviridae [1,2,3,4]. The PIV5 genome comprises 15,246 nucleotides in length and encodes eight viral proteins: nucleoprotein (NP), V protein, phosphoprotein (P), matrix protein (M), fusion protein (F), small hydrophobic protein (SH), hemagglutinin–neuraminidase protein (HN), and large protein (L) [5,6]. Notably, the V and P proteins are generated from the V/P gene; these two proteins share an overlapping genomic region and are generated through a specific RNA editing mechanism [7,8,9].
Originally, PIV5 was identified in primary monkey kidney cells in 1954 [10]. Since then, it has been widely detected across diverse host species, including humans, dogs, pigs, cats, calves, equines, geese, pangolins, and lesser pandas, with varying clinical manifestations reported [2,11,12,13,14,15,16,17,18]. For instance, in calves, PIV5 is associated with severe respiratory illness and neurological disorders, contributing to high morbidity rates [11,19]. In dogs, PIV5 is recognized as a common pathogen, resulting in moderate respiratory illness or neurological dysfunction; however, severe clinical signs may develop when co-infected with other respiratory viruses or bacteria [20,21,22,23]. Of note, accumulating evidence has indicated that it has significant potential for cross-species transmission [2,14,24,25,26]. Supporting this, the swine PIV5 strain GX2020, the equine PIV5 strain XJ033, and the human PIV5 reference strains were clustered within the same clade and exhibited a close genetic relationship with the human-derived strain AGS [13,15]. Cross-species transmission has also been documented in coyotes, ferrets, and rodents [27,28].
In pigs, the first PIV5 isolate designated, SER, was identified in the lung tissue of a sow co-infected with porcine reproductive and respiratory syndrome virus (PRRSV) [29]. Subsequently, more PIV5 strains were isolated from pigs with respiratory and diarrheal symptoms in China and South Korea [30,31,32,33]. In this study, the primary objective was to isolate pandemic PEDV strains from the intestinal tissues of PEDV-infected piglets; however, we unexpectedly isolated a PIV5 strain. Of note, a recent study demonstrated that PIV5 had been associated with diarrhea in pigs [32]. Therefore, our isolation of PIV5 from diarrheic piglets might provide further evidence suggesting a potential link between PIV5 and porcine diarrhea. To better understand PIV5 epidemiology in swine, we performed genetic variation and evolutionary analyses on this isolate. Collectively, this study enhances our understanding of PIV5′s epidemiological status in swine populations.
2. Materials and Methods
2.1. Cells and Clinical Samples
Vero cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Grand Island, New York, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, Grand Island, New York, USA) at 37 °C in a humidified 5% CO2 incubator, as previously described [34]. Three intestinal tissues were collected from piglets exhibiting clinical diarrhea on a pig farm in Yibin, China.
2.2. Virus Isolation and Identification
Intestinal tissues were first screened for major enteric viruses, including PEDV, transmissible gastroenteritis virus (TGEV), porcine rotavirus (PoRV), and porcine deltacoronavirus (PDCoV) using reverse-transcription PCR (RT-PCR). Briefly, intestinal tissues were homogenized in DMEM under sterile conditions. Total RNA was extracted from tissue homogenates using a TIANamp Virus DNA/RNA Kit (Tiangen, Beijing, China), followed by cDNA synthesis with the PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Beijing, China), according to the manufacturers’ instructions. Viral nucleic acids were detected via PCR using virus-specific primers (Table S1) under the following conditions: 95 °C for 3 min; 30 cycles of 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s; and a final extension of 72 °C for 7 min.
For virus isolation, the tissue homogenates were centrifuged at 12,000× g for 10 min and filtered through 0.22 μm membranes to remove bacteria. Subsequently, Vero cells were inoculated with the filtered supernatant containing 10 µg/mL trypsin–EDTA(Gibco, Grand Island, New York, USA). Following a 1 h incubation, the inoculum was removed and replaced with DMEM supplemented with 10 µg/mL trypsin-EDTA [35,36]. The cells were incubated at 37 °C and examined daily for a cytopathic effect (CPE). Upon CPE observation, supernatants from infected cells were collected for viral nucleic acid extraction using the TIANamp Virus DNA/RNA Kit (Tiangen Beijing, China), followed by pathogen identification and genome sequencing via next-generation sequencing (NGS; Illumina platform, San Diego, CA, USA). Subsequently, the presence of PIV5 was further confirmed by RT-PCR using PIV5 NP gene-specific primers (Table S1).
2.3. Phylogenetic Analysis and Sequence Alignment
To examine the evolutionary relationship between the isolate PIV5 SC2024 and reference strains, the representative PIV5 sequences were obtained from GenBank database, and selected based on geographic distribution and host species. Phylogenetic analysis was performed based on the NP, F, and HN gene sequences of the isolate, along with its complete genome sequence, using MEGA 7 (Mega Limited, Auckland, New Zealand) with the neighbor-joining algorithm, 1000 bootstrap replicates, and the Kimura 2-parameter substitution model, as previously described [20,30,34,37]. For genetic characterization, the nucleotide sequences and deduced amino acid sequences of the NP, F, and HN genes were aligned and analyzed using Geneious Prime software (version 2022.2, Biomatters, Auckland, New Zealand) with the Clustal Omega program.
3. Results
3.1. Isolation and Identification of PIV5
To identify viral infections in intestinal tissues, RT-PCR was performed using virus-specific primers for PEDV, TGEV, PoRV, and PDCoV (Table S1). As shown in Figure 1A, only PEDV tested positive among all tissue samples. Subsequently, the tissue homogenates were inoculated onto Vero cells. And the CPE was observed after three passages in one of the tissue samples (Figure 1B). Notably, RT-PCR failed to detect the PEDV genome. Then, NGS was performed to identify the CPE causative pathogen using an Illumina platform. As shown in Figure 1C, NGS generated 183,941 reads with a 100% coverage ratio for PIV5. To further validate the NGS results, RT-PCR confirmed that the supernatants of virus-infected cells were positive for PIV5 (Figure 1D). Then, the newly isolated virus was designated SC2024. Following sequence assembly and annotation, the complete genome of SC2024 was determined to be 15,246 nt in length, containing a 5′ UTR, a 3′ UTR, and six open reading frames (ORFs) that code for the NP, V/P, M, F, NH, and L proteins. Then, the complete genome sequence was submitted in GenBank under accession number PV395591.1.
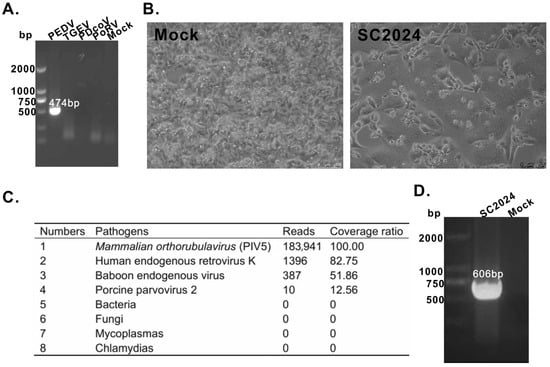
Figure 1.
Isolation and identification of PIV5. (A) PCR verification of virus infection in intestinal tissues. (B) CPE of PIV5 on Vero cells. (C) Identification of PIV5 via NGS. (D) RT-PCR verification of PIV5.
3.2. Phylogenetic Analysis
To elucidate the genetic relationships between PIV5 SC2024 and the reference strains, phylogenetic trees were constructed using the neighbor joining method via MEGA 7.0 software, based on the NP, F, and HN genes, and complete genomic sequences. As shown in Figure 2, all phylogenetic trees revealed that PIV5 strains divided into two distinct lineages (Lineage 1 and Lineage 2), with lineage 2 further diverging into two sub-lineages (Lineage 2.1 and Lineage 2.2). Significantly, strain SC2024 clustered within the tertiary sub-lineage 2.2 alongside most Chinese swine-derived strains, which also included strains isolated from pangolins, tigers, lesser pandas, and ticks.
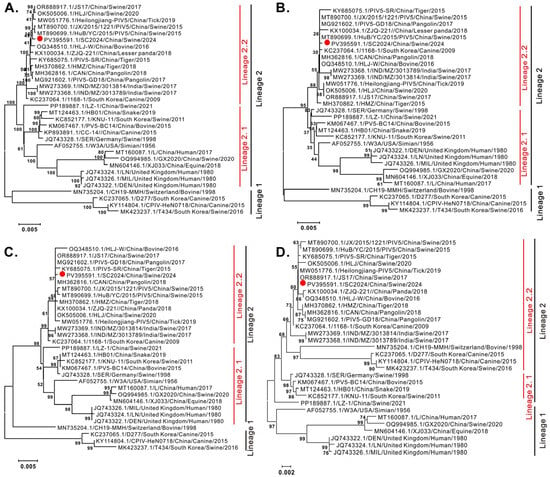
Figure 2.
Phylogenetic analysis of the PIV5 strain SC2024. Phylogenetic trees were constructed using MEGA 7 software with the neighbor joining algorithm, 1000 bootstrap replicates, and the Kimura 2-parameter substitution model. The isolate PIV5 strain SC2024 is marked with red point. (A) Phylogenetic analysis of PIV5 based on whole-genome sequences. (B) Phylogenetic analysis of PIV5 based on NP sequences. (C) Phylogenetic analysis of PIV5 based on F sequences. (D) Phylogenetic analysis of PIV5 based on HN sequences.
3.3. Sequence Identity and Alignment Analysis
Comparison of complete genomic sequences revealed that SC2024 shared 96.9–99.9% nucleotide identity with other PIV5 strains. Additionally, the NP, F, and HN genes displayed nucleotide identities of 96.8–99.9%, 95.8–99.9%, and 96.6–100% with those of other PIV5 strains, respectively. At the amino acid level, the encoded NP, F, and HN proteins showed sequence homologies of 97.6–99.8%, 94.9–99.8%, and 97.3–100% compared to their counterparts in other PIV5 strains (Table 1). Among pig-derived strains, the SC2024 demonstrated a higher degree of similarity to Chinese strains, particularly the JS17 strain. Among strains from other hosts, SC2024 showed greater similarity to strains 1168-1 (Canine), HLJ-W (Bovine), CAN (Pangolin), PIV5-GD18 (Pangolin), HMZ (Tiger), ZJQ-221 (Lesser panda), and Heilongjiang-PIV5 (Tick). Notably, the HN protein of SC2024 displayed 100% identity with that of the tick-derived strain Heilongjiang-PIV5.

Table 1.
Comparison of the nucleotide (nt) and amino acid (aa) of SC2024 with the reference PIV5 strains.
The glycoproteins F and HN, which are critical importance for viral attachment, membrane fusion, and host cell entry, were subjected to further analysis. As shown in Figure 3A, compared with the reference strains, the F protein of SC2024 harbored a novel amino acid mutation (D448G). Notably, three amino acid sites (A279, R339, A445) in the F protein were conserved across SC2024 and strains HLJ-W, CAN, PIV5-GD18, PIV5-SR, HMZ, ZJQ-221, and Heilongjiang-PIV5, which originated from diverse hosts. Correspondingly, four amino acid positions (K139, L210, T288, S447) in the HN protein of SC2024 showed complete amino acid identity with those in strains CAN, PIV5-GD18, PIV5-SR, HMZ, ZJQ-221, and Heilongjiang-PIV5 (Figure 3B). These findings suggest that the F and HN proteins may serve as key determinants in PIV5 cross-species transmission.
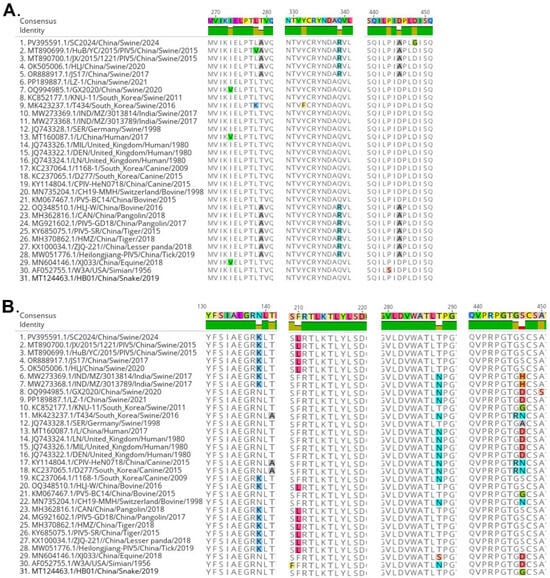
Figure 3.
Multiple-sequence alignment of F and HN protein sequences among the PIV5 strains. (A) Alignment of F protein sequences, (B) Alignment of HN protein sequences.
4. Discussion
In this study, PIV5 strains were isolated from diverse host species [2,11,12,13,14,15,16,17,18]. This virus is associated with mild respiratory and diarrheal symptoms in pigs [32,38]. In this study, our original aim was to isolate PEDV from intestinal tissues of PEDV-affected piglets. However, we unexpectedly isolated the PIV5 strain SC2024. To better understand PIV5 epidemiology in swine, we performed genetic characterization analyses on this isolate. Our findings revealed the following: (i) phylogenetic analysis indicated that strain SC2024 was clustered within the same branch as most previously isolated swine-derived strains in China; and (ii) in terms of sequence homology and specific amino acid variation in HN and F proteins, SC2024 shared genetic similarities with the strains isolated from other host species, including tigers, pangolins, lesser pandas, and ticks.
We provide further evidence to reinforce the association between PIV5 infection and diarrhea in pigs. In swine populations, more PIV5 strains have been identified in pigs in both China and South Korea [15,30,38]. Recent serological surveys demonstrated a 75.7% PIV5 positivity rate in Chinese pig farms, indicating a high prevalence across swine facilities in China [24]. Our team successfully isolated the PIV5 strain SC2024 from the intestinal tissues of piglets co-infected with PEDV. Initially, our research aim was to isolate the prevalent strains of PEDV. However, during the virus passage process, PEDV unexpectedly disappeared, while PIV5 successfully propagated in the cells. We hypothesize that factors such as viral competition or tissue sample processing may have contributed to the failure to isolate PEDV. Similarly, a parallel study has reported the isolation of PIV5 from PEDV-affected pigs, in which PEDV also disappeared during viral passage [31]. In another study, animal trials revealed that PIV5 strain LZ-1 infection induces diarrhea in piglets, albeit with moderate pathogenicity [32]. When it comes to the pathogenicity of PIV5 in pigs, similar pathogenicity assessments of the KNU-11 strain have demonstrated either non-pathogenic or mildly pathogenic effects in pigs [38]. Given its low virulence or non-virulence nature, PIV5 represents a promising candidate for developing viral-vectored vaccines [3,39]. To date, several PIV5-based vaccine candidates have proven effective in protecting against viral infections in multiple animal models [40,41,42,43,44]. For instance, a recent report showed that PIV5-based SARS-CoV-2 vaccine could induce protective and long-lasting immunity in nonhuman primates [45,46]. However, current research on PIV5 vector vaccines primarily focuses on human and zoonotic pathogens, such as influenza viruses, coronaviruses, rabies virus (RABV), vaccinia virus (VACV), respiratory syncytial virus (RSV), HIV, or bacteria [3,39,40,41,42,43,44,45,46,47,48]. In contrast, studies targeting porcine pathogens remain significantly more limited. Thus, future experiments are required to construct a PIV5 reverse genetic system, which will pave the way for the development of vector vaccines against PRRSV and PEDV.
Our studies also provide a glimpse into the evidence of the potential cross-species transmission of PIV-5 through genetic analyses. Genome-wide phylogenetic analysis demonstrated that SC2024, together with the majority of pig-derived PIV5 strains in China, clusters within Lineage 2.2 (Figure 2A). Notably, this sub-lineage also encompasses strains originating from diverse host species, including tigers, pangolins, lesser pandas, and ticks. Similar results were observed in the phylogenetic analyses of the NP, F, and HN genes (Figure 2B–D), reinforcing the evolutionary relatedness across species. Parallel findings were reported for strain YN01 in a separate study [16]. The viral proteins F and HN facilitate viral entry and are critical targets for neutralizing antibodies in parainfluenza viruses [5,6,49]. NP is the most abundant protein in the virion, and it is believed to mediate RNA-dependent RNA polymerase activity [5,6,49]. Given their functional importance and genetic characteristics, the NP, F, and HN genes are frequently used as the basis for phylogenetic analyses across parainfluenza viruses, including Newcastle disease virus (NDV) [50] and parainfluenza virus 1–5 [33,37,51,52,53]. Supporting this cross-species potential, Ibrahim et al. also isolated a PIV5 strain from diarrheic piglets that was phylogenetically closely related to strains from lesser pandas and pigs in China. This strain had a broad host range, which was capable of infecting cell lines derived from multiple species, including pigs, humans, monkeys, cattle, dogs, cats, rabbits, hamsters, and mice [24]. Furthermore, our study also indicated that SC2024 shared some identical amino acid variation sites with the PIV5 strains of other species, including tigers, pangolins, lesser pandas, and ticks (Figure 3). Together, these results suggest that the PIV5 strain SC2024 possesses cross-species transmission potential.
Pigs are frequently recognized as potential intermediate hosts for viruses with zoonotic potential, such as influenza A virus (IAV) and Nipah virus (NiV), posing significant public health concerns [54,55,56,57]. In the context of PIV5, a previous study identified a human-like strain (GX2020) in pigs coinfected with PRRSV, highlighting the virus’s cross-species transmission risk [15]. Earlier research has linked six amino acid residues (22, 49, 57, 254, 378, and 460) in the HN gene to human-specific adaptations [53]. Notably, these sites in SC2024 did not exhibit mutations associated with human tropism. Furthermore, the receptor-binding domain of the PIV5 HN protein has been identified as 186QDHVS190, with cleavage sites located at E390 and Y523 [58]. Residues 37, 342, 437, and 457 are also recognized as being linked to viral entry into host cells and the generation of neutralizing antibodies [59]. In our study, these amino acids were conserved in SC2024 and other pig-derived strains within Lineage 2.2. The strain LZ-1 is considered to induce diarrhea in piglets through pathogenicity experiments [32]. However, phylogenetic tree analysis reveals that LZ-1 and SC2024 reside in distinct sub-branches (Figure 2). Amino acid sequence alignment further identified variations at positions 279, 339, and 445 in the F protein, and 139, 210, 288, and 447 in the HN protein between these two strains (Figure 3). This raises the question of whether these mutations affect viral pathogenicity, which remains unknown and requires further investigation in the future.
In summary, this study successfully isolated a novel PIV5 strain (SC2024) from PEDV-coinfected piglets, and performed a comprehensive genetic variation analysis. These findings deepen our understanding of PIV5 epidemiology in swine populations, and highlight the urgency of enhancing viral surveillance for PIV5 in pigs or other animal species.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/vetsci12070676/s1, Table S1: Primers for amplification of the genes in this study.
Author Contributions
Conceptualization, J.R. and F.Y.; methodology, investigation and validation, Y.M. and X.C.; writing—original draft preparation, Y.M.; visualization and supervision, X.C.; investigation, analysis and resources, M.M., X.G., R.S., Y.Y., Y.W., S.N., Y.Z. and W.T.; funding acquisition and writing—review and editing, J.R. and F.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the special Fund for Science and Technology Innovation Teams of Shanxi Province (grant no. 202204051001022), the grants from the National Natural Science Foundation of China (grant no. 32202786).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Conflicts of Interest
Yuling Ma was employed by the company Beijing Solarbio Science & Technology Co., Ltd.. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Choppin, P.W.; Stoeckenius, W. The Morphology of Sv5 Virus. Virology 1964, 23, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Chatziandreou, N.; Stock, N.; Young, D.; Andrejeva, J.; Hagmaier, K.; McGeoch, D.J.; Randall, R.E. Relationships and host range of human, canine, simian and porcine isolates of simian virus 5 (parainfluenza virus 5). J. Gen. Virol. 2004, 85 Pt 10, 3007–3016. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z. Parainfluenza virus 5-vectored vaccines against human and animal infectious diseases. Rev. Med. Virol. 2018, 28, e1965. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Davison, A.J.; Dempsey, D.M.; Dutilh, B.E.; Garcia, M.L.; et al. Changes to virus taxonomy and to the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2021). Arch. Virol. 2021, 166, 2633–2648. [Google Scholar] [CrossRef] [PubMed]
- Vainionpaa, R.; Hyypia, T. Biology of parainfluenza viruses. Clin. Microbiol. Rev. 1994, 7, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Henrickson, K.J. Parainfluenza viruses. Clin. Microbiol. Rev. 2003, 16, 242–264. [Google Scholar] [CrossRef] [PubMed]
- Paterson, R.G.; Leser, G.P.; Shaughnessy, M.A.; Lamb, R.A. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 1995, 208, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.Y.; Paterson, R.G.; Lamb, R.A. The RNA binding region of the paramyxovirus SV5 V and P proteins. Virology 1997, 238, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.M.; Lamb, R.A.; Paterson, R.G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell 1988, 54, 891–902. [Google Scholar] [CrossRef] [PubMed]
- Hull, R.N.; Minner, J.R.; Smith, J.W. New viral agents recovered from tissue cultures of monkey kidney cells. I. Origin and properties of cytopathogenic agents S.V.1, S.V.2, S.V.4, S.V.5, S.V.6, S.V.11, S.V.12 and S.V.15. Am. J. Hyg. 1956, 63, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, N.; Zhang, S.; Zhang, F.; Lian, H.; Hu, R. Parainfluenza Virus 5 as Possible Cause of Severe Respiratory Disease in Calves, China. Emerg. Infect. Dis. 2015, 21, 2242–2244. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, W.; Xiang, R.; Li, L.; Chen, J.; Zhong, R.; Xiang, H.; Chen, J. Complete Genome Sequence of Parainfluenza Virus 5 (PIV5) from a Sunda Pangolin (Manis javanica) in China. J. Wildl. Dis. 2019, 55, 947–950. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Tong, P.; Zhang, A.; Zhang, L.; Song, X.; Kuang, L. Identification and Characterization of the First Equine Parainfluenza Virus 5. Virol. Sin. 2020, 35, 245–247. [Google Scholar] [CrossRef] [PubMed]
- He, W.T.; Hou, X.; Zhao, J.; Sun, J.; He, H.; Si, W.; Wang, J.; Jiang, Z.; Yan, Z.; Xing, G.; et al. Virome characterization of game animals in China reveals a spectrum of emerging pathogens. Cell 2022, 185, 1117–1129.e8. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ren, T.; Wang, X.; Wang, H.; Chen, G.; Qin, Y.; Ouyang, K.; Chen, Y.; Huang, W.; Wei, Z. The emergence, isolation, and phylogenetic analysis of a closely related human strain of parainfluenza virus 5 from a case of porcine reproductive and respiratory syndrome in China. Virology 2024, 597, 110157. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Tao, L.; Zha, B.; Zeng, Q.; Hu, Y.; Guo, X.; Liu, P.; Gao, X.; Wu, H. First isolation and phylogenetic analysis of Parainfluenza Virus 5 from Geese. Poult. Sci. 2025, 104, 105177. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.Q.; Zhai, S.L.; Lin, T.; Liu, J.K.; Wang, H.X.; Li, B.; Zhang, H.; Zou, S.Z.; Zhou, X.; Wu, M.F.; et al. First complete genome sequence of parainfluenza virus 5 isolated from lesser panda. Arch. Virol. 2017, 162, 1413–1418. [Google Scholar] [CrossRef] [PubMed]
- Hsiung, G.D. Parainfluenza-5 virus. Infection of man and animal. Progress. Med. Virol. 1972, 14, 241–274. [Google Scholar]
- Hierweger, M.M.; Werder, S.; Seuberlich, T. Parainfluenza Virus 5 Infection in Neurological Disease and Encephalitis of Cattle. Int. J. Mol. Sci. 2020, 21, 498. [Google Scholar] [CrossRef] [PubMed]
- Oem, J.K.; Kim, S.H.; Kim, Y.H.; Lee, M.H.; Lee, K.K. Molecular characteristics of canine parainfluenza viruses type 5 (CPIV-5) isolated in Korea. Can. J. Vet. Res.-Rev. Can. Rech. Vet. 2015, 79, 64–67. [Google Scholar]
- Liu, C.; Li, X.; Zhang, J.; Yang, L.; Li, F.; Deng, J.; Tan, F.; Sun, M.; Liu, Y.; Tian, K. Isolation and genomic characterization of a canine parainfluenza virus type 5 strain in China. Arch. Virol. 2017, 162, 2337–2344. [Google Scholar] [CrossRef] [PubMed]
- Cornwell, H.J.; McCandlish, I.A.; Thompson, H.; Laird, H.M.; Wright, N.G. Isolation of parainfluenza virus SV5 from dogs with respiratory disease. Vet. Rec. 1976, 98, 301–302. [Google Scholar] [CrossRef] [PubMed]
- Viitanen, S.J.; Lappalainen, A.; Rajamaki, M.M. Co-infections with respiratory viruses in dogs with bacterial pneumonia. J. Vet. Intern. Med. 2015, 29, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, Y.M.; Zhang, W.; Werid, G.M.; Zhang, H.; Pan, Y.; Zhang, L.; Xu, Y.; Li, C.; Chen, H.; Wang, Y. Characterization of parainfluenza virus 5 from diarrheic piglet highlights its zoonotic potential. Transbound. Emerg. Dis. 2022, 69, e1510–e1525. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xu, P.; Salyards, G.W.; Harvey, S.B.; Rada, B.; Fu, Z.F.; He, B. Evaluating a parainfluenza virus 5-based vaccine in a host with pre-existing immunity against parainfluenza virus 5. PLoS ONE 2012, 7, e50144. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Ma, Y.; Jiang, Q.; Song, M.; Kang, H.; Liu, J.; Qu, L. Isolation, identification and pathogenic characteristics of tick-derived parainfluenza virus 5 in northeast China. Transbound. Emerg. Dis. 2022, 69, 3300–3316. [Google Scholar] [CrossRef] [PubMed]
- Durchfeld, B.; Baumgartner, W.; Krakowka, S. Intranasal infection of ferrets (Mustela putorius furo) with canine parainfluenza virus. J. Vet. Med. Ser. B 1991, 38, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Davidson, W.R.; Appel, M.J.; Doster, G.L.; Baker, O.E.; Brown, J.F. Diseases and parasites of red foxes, gray foxes, and coyotes from commercial sources selling to fox-chasing enclosures. J. Wildl. Dis. 1992, 28, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Heinen, E.; Herbst, W.; Schmeer, N. Isolation of a cytopathogenic virus from a case of porcine reproductive and respiratory syndrome (PRRS) and its characterization as parainfluenza virus type 2. Arch. Virol. 1998, 143, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.N.; Lee, C. Complete genome sequence of a novel porcine parainfluenza virus 5 isolate in Korea. Arch. Virol. 2013, 158, 1765–1772. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Wang, E.; Guo, D.; Wang, X.; Su, M.; Kong, F.; Yuan, D.; Zhai, J.; Sun, D. Isolation and molecular characterization of parainfluenza virus 5 in diarrhea-affected piglets in China. J. Vet. Med. Sci./Jpn. Soc. Vet. Sci. 2018, 80, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Lin, S.; Shao, Y.; Tang, J.; Li, S.; Tan, C.; Gong, Z.; Li, H.; Wang, J.; Liu, G.; et al. Enteric pathogenicity characterization of emerging parainfluenza virus 5 in western China. Virology 2025, 604, 110409. [Google Scholar] [CrossRef] [PubMed]
- Truong, H.T.; Nguyen, V.G.; Pham, L.B.; Huynh, T.M.; Lee, J.; Hwang, S.J.; Lee, J.M.; Chung, H.C. PCR-Based Detection and Genetic Characterization of Parainfluenza Virus 5 Detected in Pigs in Korea from 2016 to 2018. Vet. Sci. 2023, 10, 414. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.L.; Tan, S.S.; Chen, X.X.; Yao, J.Y.; Niu, Z.H.; Wang, Y.; Ma, L.; Gao, X.L.; Niu, S.; Liang, L.B.; et al. Genomic Characterization and gE/gI-Deleted Strain Construction of Novel PRV Variants Isolated in Central China. Viruses 2023, 15, 1237. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Kou, Q.; Ge, X.; Zhou, L.; Guo, X.; Yang, H. Phylogenetic analysis of porcine epidemic diarrhea virus field strains prevailing recently in China. Arch. Virol. 2013, 158, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jin, Z.; Gao, Y.; Zhou, L.; Ge, X.; Guo, X.; Han, J.; Yang, H. Development of the full-length cDNA clones of two porcine epidemic diarrhea disease virus isolates with different virulence. PLoS ONE 2017, 12, e0173998. [Google Scholar] [CrossRef] [PubMed]
- Sheng, W.; Wang, K.; Gui, Y.; Qi, X.; Shen, L.; Zhang, Y.; Tang, C.; Li, X.; Tao, J.; Cao, C.; et al. Molecular characteristics and phylogenetic analysis of pigeon paramyxovirus type 1 isolates from pigeon meat farms in Shanghai (2009–2012). Sci. Rep. 2024, 14, 10741. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.N.; Park, C.K.; Kim, S.H.; Lee du, S.; Shin, J.H.; Lee, C. Characterization in vitro and in vivo of a novel porcine parainfluenza virus 5 isolate in Korea. Virus Res. 2013, 178, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zheng, L.L.; Zhao, Q.J.; Yao, Y.Z.; Zhou, F.; Wei, F.; Cai, Q.L. Parainfluenza virus 5 is a next-generation vaccine vector for human infectious pathogens. J. Med. Virol. 2023, 95, e28622. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, S.M.; Lin, Y.; Leser, G.P.; Kramer, K.A.; Haas, D.L.; Howerth, E.W.; Xu, J.; Kennett, M.J.; Durbin, R.K.; Durbin, J.E.; et al. Recombinant parainfluenza virus 5 (PIV5) expressing the influenza A virus hemagglutinin provides immunity in mice to influenza A virus challenge. Virology 2007, 362, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.M.; Johnson, J.B.; Kock, N.D.; Mizel, S.B.; Parks, G.D. Parainfluenza virus 5-based vaccine vectors expressing vaccinia virus (VACV) antigens provide long-term protection in mice from lethal intranasal VACV challenge. Virology 2011, 419, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gupta, T.; Xu, P.; Phan, S.; Pickar, A.; Yau, W.; Karls, R.K.; Quinn, F.D.; Sakamoto, K.; He, B. Efficacy of parainfluenza virus 5 (PIV5)-based tuberculosis vaccines in mice. Vaccine 2015, 33, 7217–7224. [Google Scholar] [CrossRef] [PubMed]
- Xiao, P.; Dienger-Stambaugh, K.; Chen, X.; Wei, H.; Phan, S.; Beavis, A.C.; Singh, K.; Adhikary, N.R.D.; Tiwari, P.; Villinger, F.; et al. Parainfluenza Virus 5 Priming Followed by SIV/HIV Virus-Like-Particle Boosting Induces Potent and Durable Immune Responses in Nonhuman Primates. Front. Immunol. 2021, 12, 623996. [Google Scholar] [CrossRef] [PubMed]
- Spearman, P.; Jin, H.; Knopp, K.; Xiao, P.; Gingerich, M.C.; Kidd, J.; Singh, K.; Tellier, M.; Radziewicz, H.; Wu, S.; et al. Intranasal parainfluenza virus type 5 (PIV5)-vectored RSV vaccine is safe and immunogenic in healthy adults in a phase 1 clinical study. Sci. Adv. 2023, 9, eadj7611. [Google Scholar] [CrossRef] [PubMed]
- Beavis, A.C.; Xiao, P.; Gingerich, M.C.; Briggs, K.; Li, G.; Howerth, E.W.; Najera, M.; An, D.; Huang, J.C.; Mousa, J.; et al. A parainfluenza virus 5 (PIV5)-vectored intranasal SARS-CoV-2 vaccine (CVXGA1) elicits protective and long-lasting immunity in nonhuman primates. J. Virol. 2025, 99, e0199024. [Google Scholar] [CrossRef] [PubMed]
- Beavis, A.C.; Li, Z.; Briggs, K.; Gingerich, M.C.; Wrobel, E.R.; Najera, M.; An, D.; Orr-Burks, N.; Murray, J.; Patil, P.; et al. Efficacy of parainfluenza virus 5 (PIV5)-vectored intranasal COVID-19 vaccine as a single dose primer and booster against SARS-CoV-2 variants. J. Virol. 2025, 99, e0198924. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhou, M.; Gao, X.; Zhang, G.; Ren, G.; Gnanadurai, C.W.; Fu, Z.F.; He, B. A novel rabies vaccine based on a recombinant parainfluenza virus 5 expressing rabies virus glycoprotein. J. Virol. 2013, 87, 2986–2993. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, H.; Tao, M.; Han, Q.; Yu, H.; Li, J.; Lu, X.; Tong, Q.; Pu, J.; Sun, Y.; et al. Recombinant parainfluenza virus 5 expressing clade 2.3.4.4b H5 hemagglutinin protein confers broad protection against H5Ny influenza viruses. J. Virol. 2024, 98, e0112923. [Google Scholar] [CrossRef] [PubMed]
- Branche, A.R.; Falsey, A.R. Parainfluenza Virus Infection. Semin. Respir. Crit. Care Med. 2016, 37, 538–554. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Geng, Z.; Zhou, H.; Chen, P.; Qian, J.; Guo, A. Genetic Characterization, Pathogenicity, and Epidemiology Analysis of Three Sub-Genotype Pigeon Newcastle Disease Virus Strains in China. Microorganisms 2024, 12, 738. [Google Scholar] [CrossRef] [PubMed]
- Miyahara, K.; Kitada, S.; Yoshimoto, M.; Matsumura, H.; Kawano, M.; Komada, H.; Tsurudome, M.; Kusagawa, S.; Nishio, M.; Ito, Y. Molecular evolution of human paramyxoviruses. Nucleotide sequence analyses of the human parainfluenza type 1 virus NP and M protein genes and construction of phylogenetic trees for all the human paramyxoviruses. Arch. Virol. 1992, 124, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.M.; Shi, H.F.; Gao, Y.R.; Xin, J.Q.; Liu, N.H.; Xiang, W.H.; Ren, X.G.; Feng, J.K.; Zhao, L.P.; Xue, F. Isolation and genetic characterization of bovine parainfluenza virus type 3 from cattle in China. Vet. Microbiol. 2011, 149, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Charoenkul, K.; Nasamran, C.; Janetanakit, T.; Chaiyawong, S.; Bunpapong, N.; Boonyapisitsopa, S.; Tangwangvivat, R.; Amonsin, A. Molecular detection and whole genome characterization of Canine Parainfluenza type 5 in Thailand. Sci. Rep. 2021, 11, 3866. [Google Scholar] [CrossRef] [PubMed]
- Torremorell, M.; Allerson, M.; Corzo, C.; Diaz, A.; Gramer, M. Transmission of influenza A virus in pigs. Transbound. Emerg. Dis. 2012, 59 (Suppl. 1), 68–84. [Google Scholar] [CrossRef] [PubMed]
- Ma, W. Swine influenza virus: Current status and challenge. Virus Res. 2020, 288, 198118. [Google Scholar] [CrossRef] [PubMed]
- McLean, R.K.; Graham, S.P. Vaccine Development for Nipah Virus Infection in Pigs. Front. Vet. Sci. 2019, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Bruno, L.; Nappo, M.A.; Ferrari, L.; Di Lecce, R.; Guarnieri, C.; Cantoni, A.M.; Corradi, A. Nipah Virus Disease: Epidemiological, Clinical, Diagnostic and Legislative Aspects of This Unpredictable Emerging Zoonosis. Animals 2022, 13, 159. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Thompson, T.B.; Wurzburg, B.A.; Paterson, R.G.; Lamb, R.A.; Jardetzky, T.S. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 2005, 13, 803–815. [Google Scholar] [CrossRef] [PubMed]
- Baty, D.U.; Randall, R.E. Multiple amino acid substitutions in the HN protein of the paramyxovirus, SV5, are selected for in monoclonal antibody resistant mutants. Arch. Virol. 1993, 131, 217–224. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).