Comparative Serum Proteome Analysis Indicates a Negative Correlation between a Higher Immune Level and Feed Efficiency in Pigs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Blood Collection
2.2. Protein Labeling and LC-MS/MS
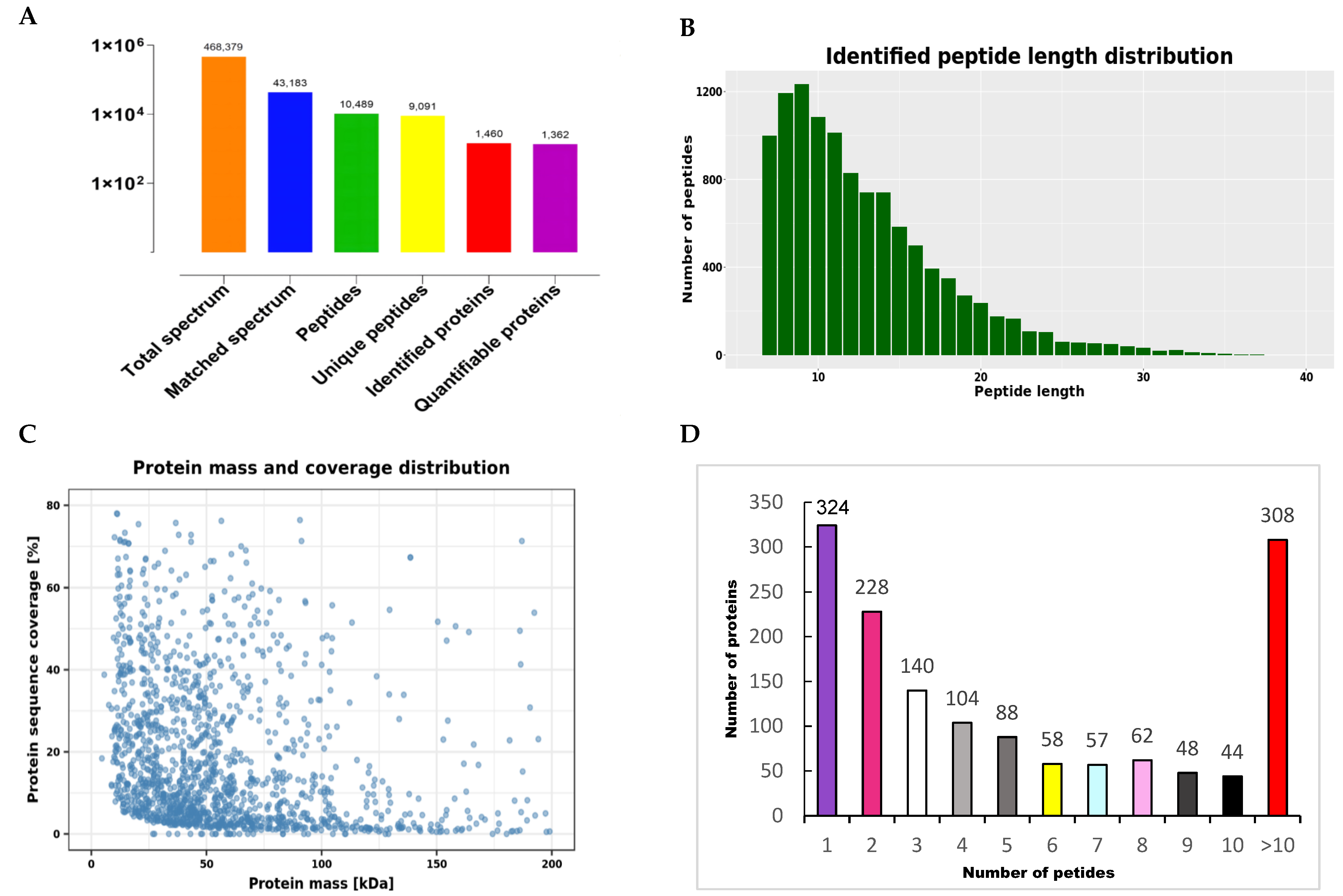
2.3. Database Search and Bioinformatics
2.4. Bioinformatics Analyses
2.5. PRM-MS Analysis
3. Results
3.1. Basic Statistics of Porcine Performance and FE
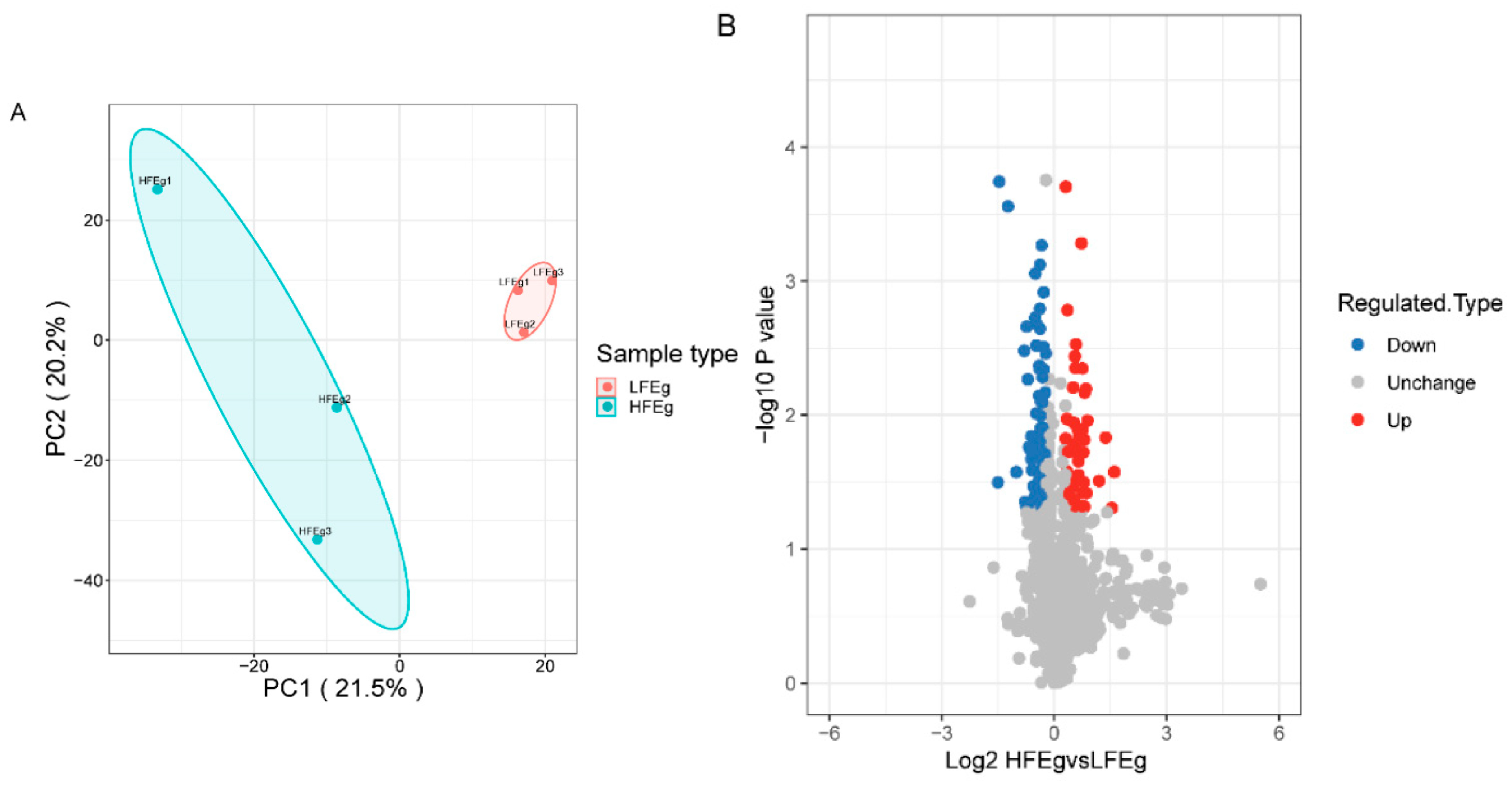
3.2. Proteomic Differences between the HFE and LFE Groups
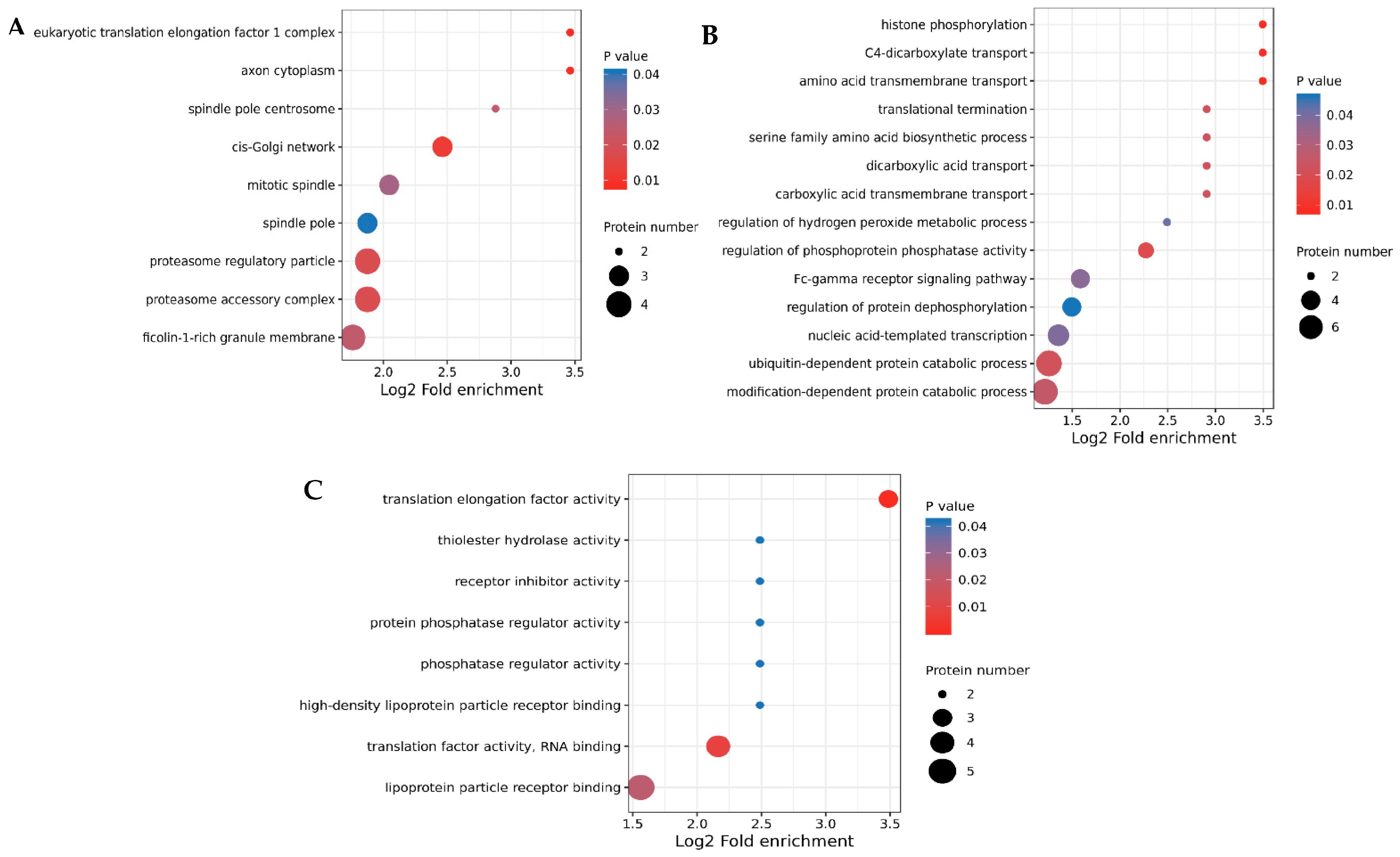
3.3. GO Annotation and KEGG Enrichment of DEPs
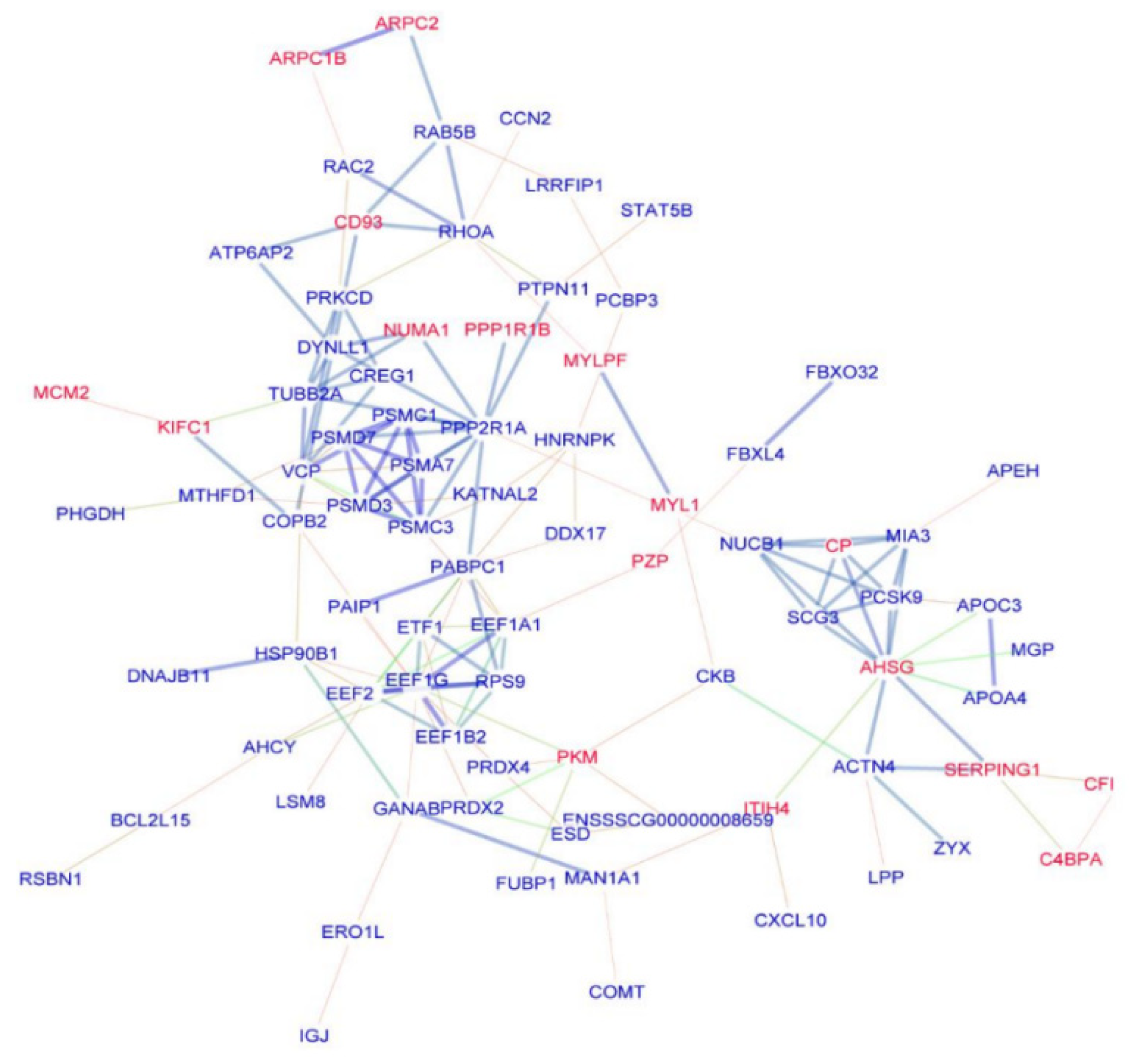
3.4. PPIs Network Construction and Analysis
3.5. DEPs Validated by PRM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Labussière, E.; Dubois, S.; van Milgen, J. Partitioning of heat production in growing pigs as a tool to improve the determination of efficiency of energy utilization. Front. Physiol. 2013, 4, 146. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, T. Evaluating environmental impacts of selection for residual feed intake in pigs. Animal 2020, 14, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, H.; Ruesche, J.; Muller, N. Responses to weaning in two pig lines divergently selected for residual feed intake depending on diet. J. Anim. Sci. 2019, 97, 43–54. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Li, T.; Wang, W.; Gao, H.; Bai, Y.; Zhang, S. Metabolic characteristics and nutrient utilization in high-feed-efficiency pigs selected using different feed conversion ratio models. Sci. China Life Sci. 2019, 62, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.A.; Suzuki, K.; Kadowaki, H.; Shibata, T.; Oikawa, T. Genetic parameters for feed efficiency traits and their relationships with growth and carcass traits in Duroc pigs. J. Anim. Breed Genet. 2007, 124, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Krumah, J.D.N.; Basarab, J.A. Different measures of energetic efficiency and their phenotypic relationships with growth, feed intake, and ultrasound and carcass merit in hybrid cattle. J. Anim. Sci. 2004, 82, 2451–2459. [Google Scholar] [CrossRef]
- Kaminarides, S.; Zagari, H.; Zoidou, E. Effect of whey fat content on the properties and yields of whey cheese and serum. J. Hell Vet. Med. Soc. 2020, 71, 2149–2156. [Google Scholar] [CrossRef]
- Boddicker, N.; Gabler, N.K.; Spurlock, M.E.; Nettleton, D. Effects of ad libitum and restricted feeding on early production performance and body composition of Yorkshire pigs selected for reduced residual feed intake. Animal 2011, 5, 1344–1353. [Google Scholar] [CrossRef]
- Robert, M.; Koch, L.A.S.; Chambers, D.; Gregory, K.E. Efficiency of Feed Use in Beef Cattle. J. Anim. Sci. 1963, 22, 486–494. [Google Scholar]
- Guo, Y.M.; Zhang, Z.Y.; Ma, J.W.; Ai, H.S.; Ren, J.; Huang, L.S. A genomewide association study of feed efficiency and feeding behaviors at two fattening stages in a White Duroc × Erhualian F population. J. Anim. Sci. 2015, 93, 1481–1489. [Google Scholar] [CrossRef]
- Silva, É.F.; Lopes, M.S.; Lopes, P.S.; Gasparino, E. A genome-wide association study for feed efficiency-related traits in a crossbred pig population. Animal 2019, 13, 2447–2456. [Google Scholar] [CrossRef]
- Gondret, F.; Vincent, A.; Houée-Bigot, M.; Siegel, A.; Lagarrigue, S.; Causeur, D. A transcriptome multi-tissue analysis identifies biological pathways and genes associated with variations in feed efficiency of growing pigs. BMC Genom. 2017, 18, 244. [Google Scholar] [CrossRef] [PubMed]
- Jégou, M.; Gondret, F.; Vincent, A.; Tréfeu, C.; Gilbert, H.; Louveau, I. Whole Blood Transcriptomics is Relevant to Identify Molecular Changes in Response to Genetic Selection for Feed Efficiency and Nutritional Status in the Pig. PLoS ONE 2016, 11, e0146550. [Google Scholar] [CrossRef]
- Jarmasz, J.S.; Stirton, H.; Davie, J.R.; Del Bigio, M.R. DNA methylation and histone post-translational modification stability in post-mortem brain tissue. Clin. Epigenetics 2019, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Branciamore, S.; Gogoshin, G.; Rodin, A.S. Dependency Between Protein-Protein Interactions and Protein Variability and Evolutionary Rates in Vertebrates: Observed Relationships and Stochastic Modeling. J. Mol. Evol. 2019, 87, 184–198. [Google Scholar] [CrossRef] [PubMed]
- Kong, B.W.; Lassiter, K.; Piekarski-Welsher, A.; Dridi, S. Proteomics of Breast Muscle Tissue Associated with the Phenotypic Expression of Feed Efficiency within a Pedigree Male Broiler Line: I. Highlight on Mitochondria. PLoS ONE 2016, 11, e0155679. [Google Scholar] [CrossRef]
- Fu, L.; Xu, Y.; Hou, Y.; Qi, X.; Zhou, L.; Liu, H.; Luan, Y.; Jing, L. Proteomic analysis indicates that mitochondrial energy metabolism in skeletal muscle tissue is negatively correlated with feed efficiency in pigs. Sci. Rep. 2017, 7, 45291. [Google Scholar] [CrossRef]
- Yan, X.; Wu, Y.; Zhong, F.; Jiang, Q.; Zhou, T.; Guo, Y.; Yang, X.; Liang, J.; Joshua Liao, D.; Lan, G. iTRAQ and PRM-based quantitative proteomics in T2DM-susceptible and -tolerant models of Bama mini-pig. Gene 2018, 675, 119–127. [Google Scholar] [CrossRef]
- Nkukwana, T.T. Global poultry production: Current impact and future outlook on the South African poultry industry. S. Afr. J. Anim. Sci. 2018, 48, 869–884. [Google Scholar] [CrossRef]
- De, A.; Ali, M.A.; Chutia, T.; Onteru, S.K. Comparative serum proteome analysis reveals potential early pregnancy-specific protein biomarkers in pigs. Reprod. Fert. Develop. 2019, 31, 613–631. [Google Scholar] [CrossRef]
- Olumee-Shabon, Z.; Chattopadhaya, C.; Myers, M.J. Proteomics profiling of swine serum following lipopolysaccharide stimulation. Rapid communications in mass spectrometry: RCM. Rapid Commun. Mass Spectrom. 2020, 34, e8639. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, J.A.; Ziemba, E.A.; Colbert, R.; Anderson, L.B.; Sluiter, W. Altered expression of mitochondrial electron transport chain proteins and improved myocardial energetic state during late ischemic preconditioning. American journal of physiology Heart and circulatory physiology. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, H1974–H1982. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, X.; Ding, R.; Quan, J.; Ye, Y.; Gu, T.; Xu, Z. Identification of Important Proteins and Pathways Affecting Feed Efficiency in DLY Pigs by iTRAQ-Based Proteomic Analysis. Animals 2020, 10, 189. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.; Achour, B.; Al-Majdoub, Z. GASP and FASP are Complementary for LC-MS/MS Proteomic Analysis of Drug-Metabolizing Enzymes and Transporters in Pig Liver. Proteomics 2018, 18, e1800200. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lu, X.; Cheng, Z.; Tian, M. Comparative proteomic analysis of Tibetan pig spermatozoa at high and low altitudes. BMC Genom. 2019, 20, 569. [Google Scholar]
- Grubbs, J.K.; Dekkers, J.C.; Huff-Lonergan, E. Identification of potential serum biomarkers to predict feed efficiency in young pigs. J. Anim. Sci. 2016, 94, 1482–1492. [Google Scholar] [CrossRef]
- Vincent, A.; Louveau, I.; Gondret, F.; Tréfeu, C. Divergent selection for residual feed intake affects the transcriptomic and proteomic profiles of pig skeletal muscle. J. Anim. Sci. 2015, 93, 2745–2758. [Google Scholar] [CrossRef]
- Si, J.; Feng, L.; Gao, J.; Huang, Y.; Zhang, G.; Mo, J.; Zhu, S.; Qi, W.; Liang, J.; Lan, G. Evaluating the association between feed efficiency and the fecal microbiota of early-life Duroc pigs using 16S rRNA sequencing. AMB Express 2020, 10, 115. [Google Scholar] [CrossRef]
- Cai, W.; Casey, D.S.; Dekkers, J.C.M. Selection response and genetic parameters for residual feed intake in Yorkshire swine. J. Anim. Sci. 2008, 86, 287–298. [Google Scholar] [CrossRef]
- Kuleš, J.; Bilić, P.; Horvatić, A.; Kovačević, A.; Guillemin, N. Serum proteome profiling in canine chronic valve disease using a TMT-based quantitative proteomics approach. J. Proteomics 2020, 223, 103825. [Google Scholar] [CrossRef]
- Kavlak, A.T.; Uimari, P. Estimation of heritability of feeding behaviour traits and their correlation with production traits in Finnish Yorkshire pigs. J. Anim. Breed 2019, 136, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Saintilan, R.; Brossard, L. Phenotypic and genetic relationships between growth and feed intake curves and feed efficiency and amino acid requirements in the growing pig. Animal 2015, 9, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Saintilan, R.; Mérour, I.; Brossard, L. Genetics of residual feed intake in growing pigs: Relationships with production traits, and nitrogen and phosphorus excretion traits. J. Anim. Sci. 2013, 91, 2542–2554. [Google Scholar] [CrossRef] [PubMed]
- Horodyska, J.; Reyer, H. Transcriptome analysis of adipose tissue from pigs divergent in feed efficiency reveals alteration in gene networks related to adipose growth, lipid metabolism, extracellular matrix, and immune response. Mol. Genet. Genom. 2019, 294, 395–408. [Google Scholar] [CrossRef]
- Cokic, V.P.; Bhattacharya, B. JAK-STAT and AKT pathway-coupled genes in erythroid progenitor cells through ontogeny. J. Transl. Med. 2012, 10, 116. [Google Scholar] [CrossRef]
- Nagata, N.; Matsuo, K.; Bettaieb, A.; Bakke, J. Hepatic Src homology phosphatase 2 regulates energy balance in mice. Endocrinology 2012, 153, 3158–3169. [Google Scholar] [CrossRef]
- Dodington, D.W.; Desai, H.R.; Woo, M. JAK/STAT—Emerging Players in Metabolism. Trends Endocrinol. Metab. 2018, 29, 55–65. [Google Scholar] [CrossRef]
- Moresi, V.; Adamo, S.; Berghella, L. The JAK/STAT Pathway in Skeletal Muscle Pathophysiology. Front. Physiol. 2019, 10, 500. [Google Scholar] [CrossRef]
- Linnekin, D.; Mou, S.; Deberry, C.S.; Weiler, S.R. Stem cell factor, the JAK-STAT pathway and signal transduction. Leuk. Lymphoma 1997, 27, 439–444. [Google Scholar] [CrossRef]
- Nadeau, K.; Hwa, V.; Rosenfeld, R.G. STAT5b deficiency: An unsuspected cause of growth failure, immunodeficiency, and severe pulmonary disease. J. Pediatr. 2011, 158, 701–708. [Google Scholar] [CrossRef]
- Nascimento, C.F.; Branco, R.H.; Bonilha, S.F. Residual feed intake and blood variables in young Nellore cattle. J. Anim. Sci. 2015, 93, 1318–1326. [Google Scholar] [CrossRef] [PubMed]
- Pick, E. Role of the Rho GTPase Rac in the activation of the phagocyte NADPH oxidase: Outsourcing a key task. Small GTPases 2014, 5, e27952. [Google Scholar] [CrossRef] [PubMed]
- Croker, B.A.; Tarlinton, D.M.; Cluse, L.A. The Rac2 guanosine triphosphatase regulates B lymphocyte antigen receptor responses and chemotaxis and is required for establishment of B-1a and marginal zone B lymphocytes. J. Immunol. 2002, 168, 3376–3386. [Google Scholar] [CrossRef] [PubMed]
- Walmsley, M.J.; Ooi, S.K.; Reynolds, L.F.; Smith, S.H.; Ruf, S.; Mathiot, A.; Vanes, L.; Williams, D.A.; Cancro, M.P.; Tybulewicz, V.L. Critical roles for Rac1 and Rac2 GTPases in B cell development and signaling. Science 2003, 302, 459–462. [Google Scholar] [CrossRef]
- Yang, L.; He, T.; Xiong, F.; Chen, X.; Fan, X.; Jin, S.; Geng, Z. Identification of key genes and pathways associated with feed efficiency of native chickens based on transcriptome data via bioinformatics analysis. BMC Genom. 2020, 21, 292. [Google Scholar] [CrossRef] [PubMed]
- Barash, U.; Zohar, Y.; Wildbaum, G. Heparanase enhances myeloma progression via CXCL10 downregulation. Leukemia 2014, 28, 2178–2187. [Google Scholar] [CrossRef]
- Salzer, E.; Santos-Valente, E.; Klaver, S. B-cell deficiency and severe autoimmunity caused by deficiency of protein kinase C δ. Blood 2013, 121, 3112–3116. [Google Scholar] [CrossRef]
- Berridge, M.J.; Irvine, R.F. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature 1984, 312, 315–321. [Google Scholar] [CrossRef]
- Nishizuka, Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science 1992, 258, 607–614. [Google Scholar] [CrossRef]
- Chiba, Y.; Goto, K.; Momata, M.; Kobayashi, T.; Misawa, M. Induction of RhoA gene expression by interleukin-4 in cultured human bronchial smooth muscle cells. J. Smooth Muscle Res. 2010, 46, 217–224. [Google Scholar] [CrossRef]
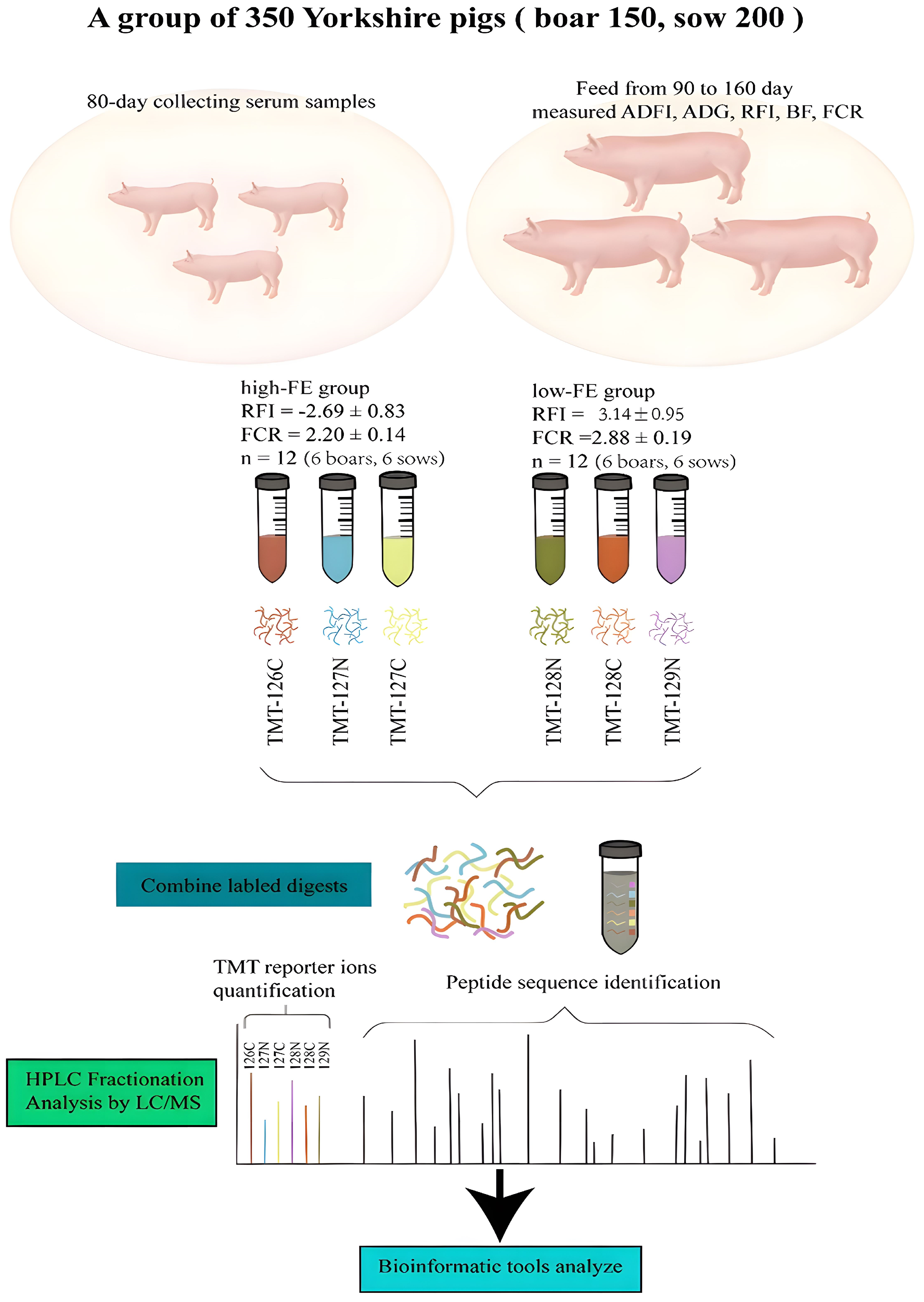

| Parameter | LFE (Mean ± SD) | HFE (Mean ±SD) | p-Value a |
|---|---|---|---|
| N | 12 | 12 | |
| RFI (kg) | 3.14 ± 0.95 | −2.69 ± 0.83 | <0.001 *** |
| FCR | 2.88 ± 0.19 | 2.20 ± 0.14 | <0.001 *** |
| ADFI (kg/d) | 2.90 ± 0.20 | 2.31 ± 0.18 | <0.001 *** |
| ADG (kg/d) | 1.01 ± 0.10 | 1.03 ± 0.07 | 0.56 |
| 100 kgBF (mm) | 13.05 ± 3.35 | 12.46 ± 2.41 | 0.62 |
| On-BW (kg) | 41.67 ± 5.46 | 40.33 ± 6.70 | 0.46 |
| Off-BW (kg) | 112.37 ± 8.66 | 110.57 ± 5.92 | 0.56 |
| Metamid-BW | 57.76 ± 4.05 | 56.59 ± 2.99 | 0.43 |
| Protein Accession | Protein Gene | Fold Change (HFE/LFE) in TMT | p-Value in TMT | Fold Change (HFE/LFE) in PRM | p-Value in PRM |
|---|---|---|---|---|---|
| F1SFI7 | AHSG | 2.52 | 1.50 × 10−2 | 2.23 | 1.13 × 10−1 |
| F1SJW8 | SERPING1 | 1.65 | 1.30 × 10−2 | 1.26 | 1.24 × 10−1 |
| F1S133 | CFI | 2.23 | 3.10 × 10−2 | 2.17 | 1.50 × 10−1 |
| Q29594 | CKB | 0.83 | 3.50 × 10−3 | 0.39 | 4.48 × 10−3 |
| T1UNN8 | ANGPTL8 | 0.75 | 4.60 × 10−3 | 0.37 | 3.53 × 10−3 |
| G9F6X8 | P4HB | 0.78 | 2.30 × 10−2 | 0.43 | 2.60 × 10−3 |
| A0A287BAZ6 | PRDX2 | 0.78 | 8.10 × 10−3 | 0.44 | 2.93 × 10−3 |
| A0A287BKS2 | PTX3 | 0.77 | 9.60 × 10−3 | 0.37 | 4.27× 10−2 |
| I3LGB2 | PCSK9 | 0.82 | 1.80 × 10−2 | 0.49 | 2.44 × 10−4 |
| F1RHA9 | LECT2 | 0.78 | 2.20 × 10−2 | 0.50 | 1.26 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, S.; Si, J.; Zhang, H.; Qi, W.; Zhang, G.; Yan, X.; Huang, Y.; Zhao, M.; Guo, Y.; Liang, J.; et al. Comparative Serum Proteome Analysis Indicates a Negative Correlation between a Higher Immune Level and Feed Efficiency in Pigs. Vet. Sci. 2023, 10, 338. https://doi.org/10.3390/vetsci10050338
Zhu S, Si J, Zhang H, Qi W, Zhang G, Yan X, Huang Y, Zhao M, Guo Y, Liang J, et al. Comparative Serum Proteome Analysis Indicates a Negative Correlation between a Higher Immune Level and Feed Efficiency in Pigs. Veterinary Sciences. 2023; 10(5):338. https://doi.org/10.3390/vetsci10050338
Chicago/Turabian StyleZhu, Siran, Jinglei Si, Huijie Zhang, Wenjing Qi, Guangjie Zhang, Xueyu Yan, Ye Huang, Mingwei Zhao, Yafen Guo, Jing Liang, and et al. 2023. "Comparative Serum Proteome Analysis Indicates a Negative Correlation between a Higher Immune Level and Feed Efficiency in Pigs" Veterinary Sciences 10, no. 5: 338. https://doi.org/10.3390/vetsci10050338
APA StyleZhu, S., Si, J., Zhang, H., Qi, W., Zhang, G., Yan, X., Huang, Y., Zhao, M., Guo, Y., Liang, J., & Lan, G. (2023). Comparative Serum Proteome Analysis Indicates a Negative Correlation between a Higher Immune Level and Feed Efficiency in Pigs. Veterinary Sciences, 10(5), 338. https://doi.org/10.3390/vetsci10050338





