Description of Antimicrobial-Resistant Escherichia coli and Their Dissemination Mechanisms on Dairy Farms
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture Collection and Selection of Isolates
2.2. Antimicrobial Resistance Phenotypes
2.3. Whole Genome Sequencing
2.4. Clonal Lineages and Clones
3. Results
3.1. Phenotypic Resistance and the Related AMR Genes or AMR-Associated Mutations
3.1.1. Description of AMR Phenotypes
3.1.2. Description of AMR Genotypes
3.1.3. Comparison between AMR Phenotypes and Genotypes
3.1.4. Comparison between MIC and the Presence of AMR Genes or AMR-Associated Mutations
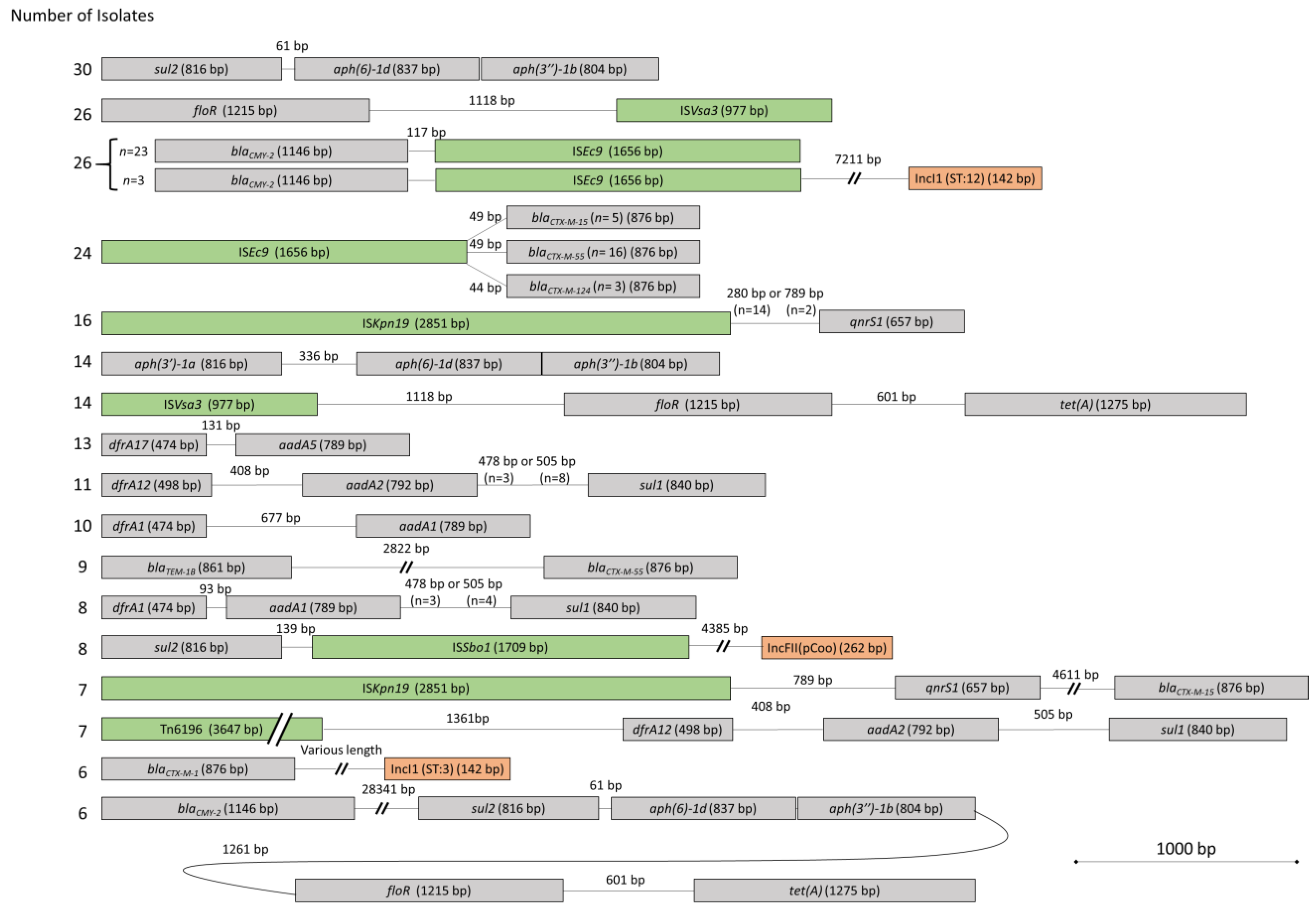
3.2. Mobile Genetic Elements and Proximity of AMR Genes
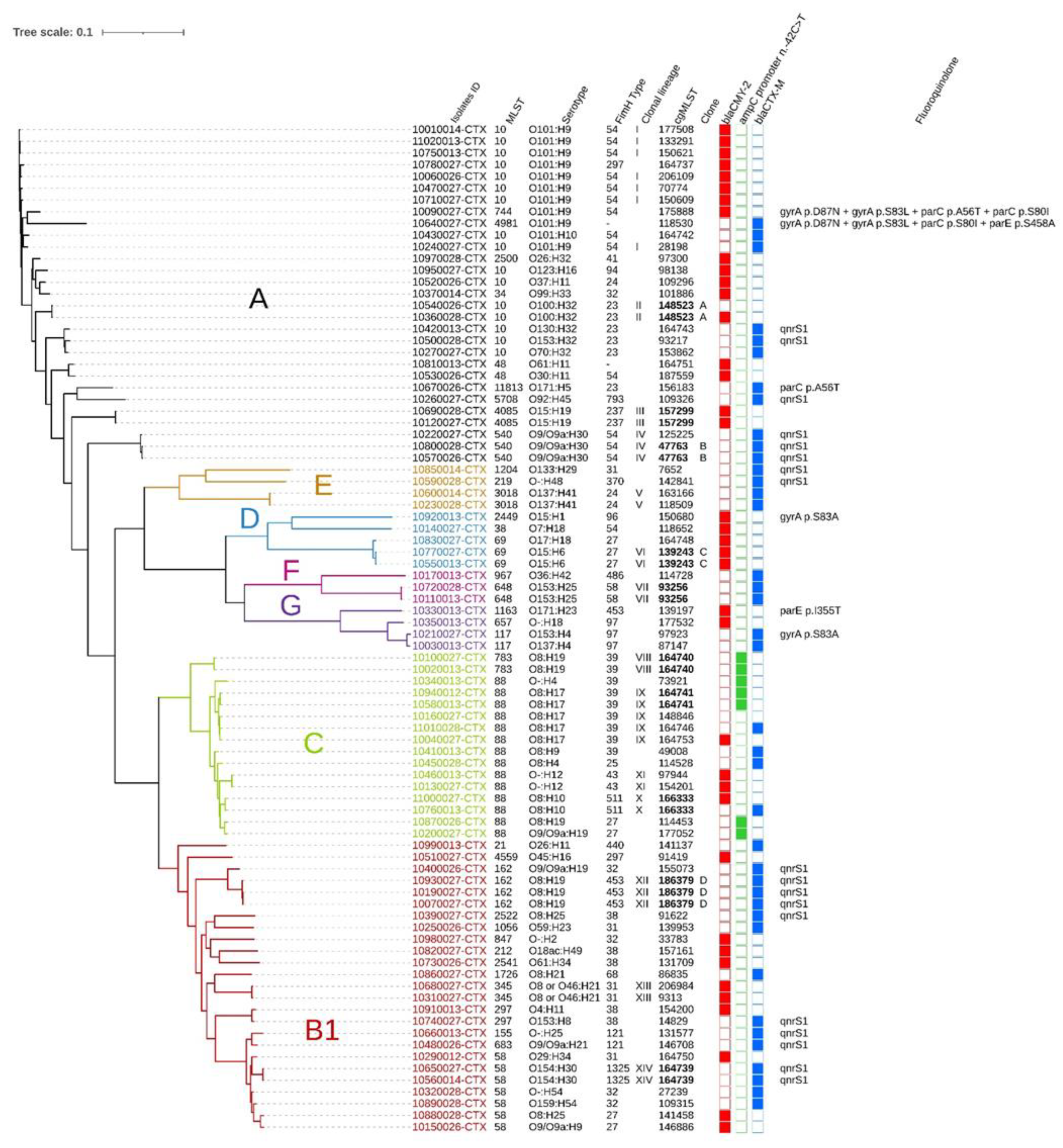
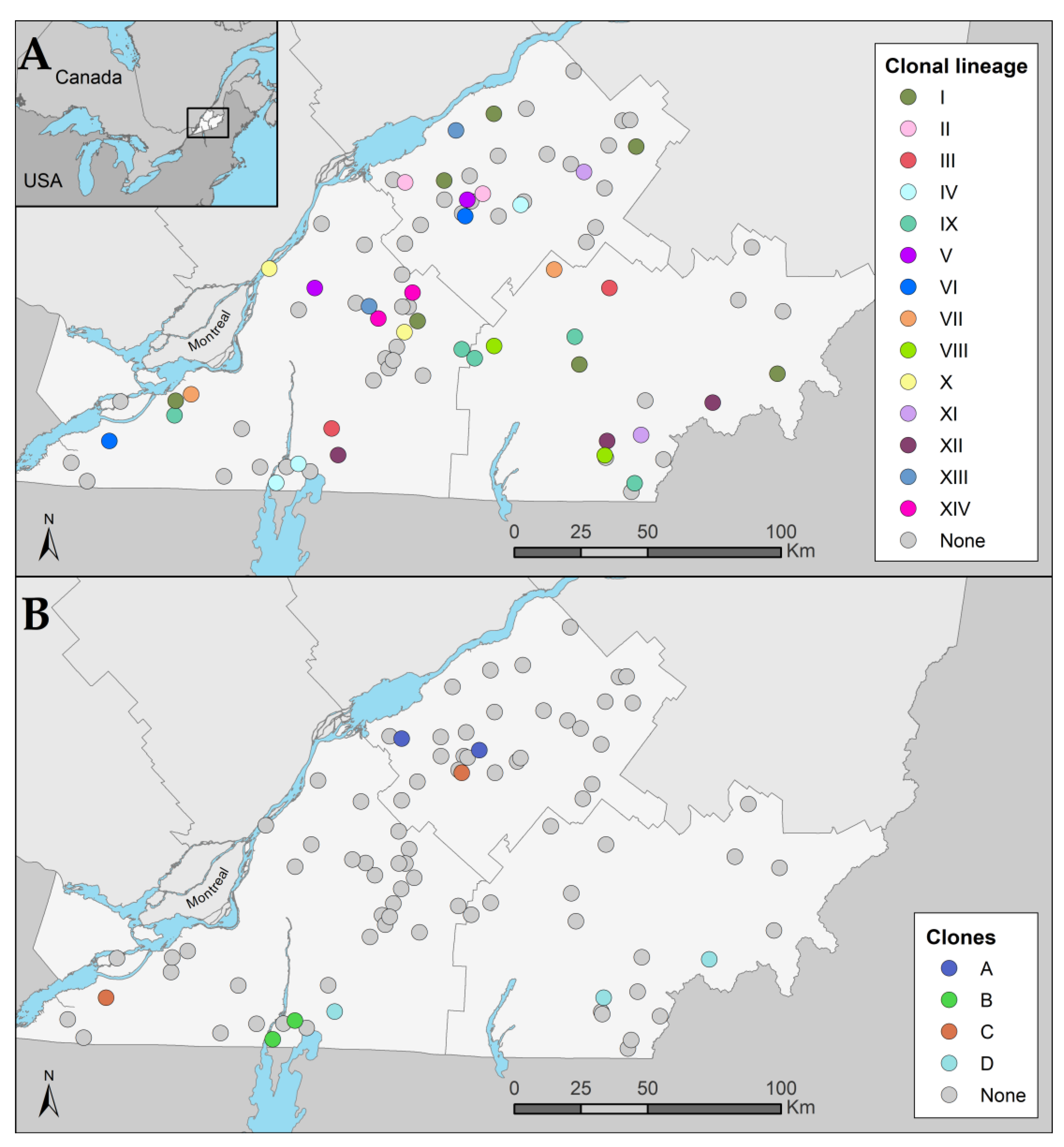
3.3. Clonal Dissemination between Farms
3.3.1. Identification of Clonal Lineages, Clones, and Their Associated Characteristics
3.3.2. Phylogroup and β-Lactam Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Antibiotic Resistance—Facts Sheet. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 8 February 2023).
- WHO. Critically Important Antimicrobials for Human Medicine, 6th ed.; WHO: Geneva, Switzerland, 2019.
- Awosile, B.; Eisnor, J.; Saab, M.E.; Heider, L.; McClure, J.T. Occurrence of extended-spectrum β-lactamase and AmpC-producing Escherichia coli in retail meat products from the Maritime Provinces, Canada. Can. J. Microbiol. 2021, 67, 537–547. [Google Scholar] [CrossRef]
- Afema, J.A.; Ahmed, S.; Besser, T.E.; Jones, L.P.; Sischo, W.M.; Davis, M.A. Molecular Epidemiology of Dairy Cattle-Associated Escherichia coli Carrying bla(CTX-M) Genes in Washington State. Appl. Environ. Microbiol. 2018, 84, e02430-17. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, D.A.; Li, C.; Mukherjee, S.; Hsu, C.H.; Bodeis Jones, S.; Gaines, S.A.; Kabera, C.; Loneragan, G.H.; Torrence, M.; Harhay, D.M.; et al. Whole-Genome Sequence Analysis of CTX-M Containing Escherichia coli Isolates from Retail Meats and Cattle in the United States. Microb. Drug Resist. 2018, 24, 939–948. [Google Scholar] [CrossRef]
- Jahanbakhsh, S.; Smith, M.G.; Kohan-Ghadr, H.R.; Letellier, A.; Abraham, S.; Trott, D.J.; Fairbrother, J.M. Dynamics of extended-spectrum cephalosporin resistance in pathogenic Escherichia coli isolated from diseased pigs in Quebec, Canada. Int. J. Antimicrob. Agents 2016, 48, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.L.; Caffrey, N.P.; Nóbrega, D.B.; Cork, S.C.; Ronksley, P.E.; Barkema, H.W.; Polachek, A.J.; Ganshorn, H.; Sharma, N.; Kellner, J.D.; et al. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: A systematic review and meta-analysis. Lancet Planet. Health 2017, 1, e316–e327. [Google Scholar] [CrossRef]
- Jahanbakhsh, S.; Letellier, A.; Fairbrother, J.M. Circulating of CMY-2 β-lactamase gene in weaned pigs and their environment in a commercial farm and the effect of feed supplementation with a clay mineral. J. Appl. Microbiol. 2016, 121, 136–148. [Google Scholar] [CrossRef]
- Vounba, P.; Arsenault, J.; Bada-Alambédji, R.; Fairbrother, J.M. Pathogenic potential and the role of clones and plasmids in beta-lactamase-producing E. coli from chicken faeces in Vietnam. BMC Vet. Res. 2019, 15, 106. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration (FDA). National Antimicrobial Resistance Monitoring Surveillance, NARMS Now: Integrated Data. Available online: https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/narms-now-integrated-data (accessed on 8 February 2023).
- Government of Canada. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) Annual Report. Available online: https://www.canada.ca/en/public-health/services/surveillance/canadian-integrated-program-antimicrobial-resistance-surveillance-cipars.html (accessed on 8 February 2023).
- Zhu, D.M.; Li, Q.H.; Shen, Y.; Zhang, Q. Risk factors for quinolone-resistant Escherichia coli infection: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2020, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Fuzi, M.; Rodriguez Baño, J.; Toth, A. Global Evolution of Pathogenic Bacteria With Extensive Use of Fluoroquinolone Agents. Front. Microbiol. 2020, 11, 271. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Perfomance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI Supplement M100; CLSI: Berwyn, PA, USA, 2021. [Google Scholar]
- EUCAST. EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiologial Importance; EUCAST: Växjö, Sweden, 2017. [Google Scholar]
- Golden, A.R.; Karlowsky, J.A.; Walkty, A.; Baxter, M.R.; Denisuik, A.J.; McCracken, M.; Mulvey, M.R.; Adam, H.J.; Bay, D.; Zhanel, G.G. Comparison of phenotypic antimicrobial susceptibility testing results and WGS-derived genotypic resistance profiles for a cohort of ESBL-producing Escherichia coli collected from Canadian hospitals: CANWARD 2007-18. J. Antimicrob. Chemother. 2021, 76, 2825–2832. [Google Scholar] [CrossRef]
- Day, M.R.; Doumith, M.; Do Nascimento, V.; Nair, S.; Ashton, P.M.; Jenkins, C.; Dallman, T.J.; Stevens, F.J.; Freedman, J.; Hopkins, K.L.; et al. Comparison of phenotypic and WGS-derived antimicrobial resistance profiles of Salmonella enterica serovars Typhi and Paratyphi. J. Antimicrob. Chemother. 2018, 73, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.H.; McDermott, P.F.; Li, C.; Chen, Y.; Tadesse, D.A.; Mukherjee, S.; Bodeis-Jones, S.; Kabera, C.; Gaines, S.A.; Loneragan, G.H.; et al. WGS accurately predicts antimicrobial resistance in Escherichia coli. J. Antimicrob. Chemother. 2015, 70, 2763–2769. [Google Scholar] [CrossRef] [PubMed]
- Lardé, H.; Dufour, S.; Archambault, M.; Massé, J.; Roy, J.P.; Francoz, D. An observational cohort study on antimicrobial usage on dairy farms in Quebec, Canada. J. Dairy Sci. 2021, 104, 1864–1880. [Google Scholar] [CrossRef] [PubMed]
- Massé, J.; Lardé, H.; Fairbrother, J.M.; Roy, J.-P.; Francoz, D.; Dufour, S.; Archambault, M. Prevalence of Antimicrobial Resistance and Characteristics of Escherichia coli Isolates From Fecal and Manure Pit Samples on Dairy Farms in the Province of Québec, Canada. Front. Vet. Sci. 2021, 8, 654125. [Google Scholar] [CrossRef]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Teng, L.; Lee, S.; Ginn, A.; Markland, S.M.; Mir, R.A.; DiLorenzo, N.; Boucher, C.; Prosperi, M.; Johnson, J.; Morris, J.G., Jr.; et al. Genomic Comparison Reveals Natural Occurrence of Clinically Relevant Multidrug-Resistant Extended-Spectrum-beta-Lactamase-Producing Escherichia coli Strains. Appl. Environ. Microbiol. 2019, 85, e03030-18. [Google Scholar] [CrossRef]
- de Lagarde, M.; Vanier, G.; Arsenault, J.; Fairbrother, J.M.M. High Risk Clone: A Proposal of Criteria Adapted to the One Health Context with Application to Enterotoxigenic Escherichia coli in the Pig Population. Antibiotics 2021, 10, 244. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Perfomance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th ed.; CLSI Supplement VET01S; CLSI: Berwyn, PA, USA, 2020. [Google Scholar]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018, 4, e000192. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef]
- Roer, L.; Tchesnokova, V.; Allesøe, R.; Muradova, M.; Chattopadhyay, S.; Ahrenfeldt, J.; Thomsen, M.C.F.; Lund, O.; Hansen, F.; Hammerum, A.M.; et al. Development of a Web Tool for Escherichia coli Subtyping Based on fimH Alleles. J. Clin. Microbiol. 2017, 55, 2538–2543. [Google Scholar] [CrossRef] [PubMed]
- Edwards, P.R.; Ewing, W.H. Identification of Enterobacteriaceae; Burgess Publishing Company: Minneapolis, MN, USA, 1972; p. 362. [Google Scholar]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Stubberfield, E.; AbuOun, M.; Sayers, E.; O’Connor, H.M.; Card, R.M.; Anjum, M.F. Use of whole genome sequencing of commensal Escherichia coli in pigs for antimicrobial resistance surveillance, United Kingdom, 2018. Euro Surveill. 2019, 24, 1900136. [Google Scholar] [CrossRef]
- Mammeri, H.; Nazic, H.; Naas, T.; Poirel, L.; Léotard, S.; Nordmann, P. AmpC beta-lactamase in an Escherichia coli clinical isolate confers resistance to expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 2004, 48, 4050–4053. [Google Scholar] [CrossRef]
- Garcia-Migura, L.; Sunde, M.; Karlsmose, S.; Veldman, K.; Schroeter, A.; Guerra, B.; Granier, S.A.; Perrin-Guyomard, A.; Gicquel-Bruneau, M.; Franco, A.; et al. Establishing streptomycin epidemiological cut-off values for Salmonella and Escherichia coli. Microb. Drug Resist. 2012, 18, 88–93. [Google Scholar] [CrossRef]
- Jacoby, G.A. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 2005, 41 (Suppl. S2), S120–S126. [Google Scholar] [CrossRef]
- Arredondo-Alonso, S.; Willems, R.J.; van Schaik, W.; Schürch, A.C. On the (im)possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb. Genom. 2017, 3, e000128. [Google Scholar] [CrossRef]
- Call, D.R.; Singer, R.S.; Meng, D.; Broschat, S.L.; Orfe, L.H.; Anderson, J.M.; Herndon, D.R.; Kappmeyer, L.S.; Daniels, J.B.; Besser, T.E. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 2010, 54, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Meunier, D.; Jouy, E.; Lazizzera, C.; Doublet, B.; Kobisch, M.; Cloeckaert, A.; Madec, J.Y. Plasmid-borne florfenicol and ceftiofur resistance encoded by the floR and blaCMY-2 genes in Escherichia coli isolates from diseased cattle in France. J. Med. Microbiol. 2010, 59, 467–471. [Google Scholar] [CrossRef]
- Health Canada. Categorization of Antimicrobial Drugs Based on Importance in Human Medicine. Available online: https://www.canada.ca/en/health-canada/services/drugs-health-products/veterinary-drugs/antimicrobial-resistance/categorization-antimicrobial-drugs-based-importance-human-medicine.html (accessed on 8 February 2023).
- Li, X.Z.; Mehrotra, M.; Ghimire, S.; Adewoye, L. beta-Lactam resistance and beta-lactamases in bacteria of animal origin. Vet. Microbiol. 2007, 121, 197–214. [Google Scholar] [CrossRef]
- Wittum, T.E.; Mollenkopf, D.F.; Daniels, J.B.; Parkinson, A.E.; Mathews, J.L.; Fry, P.R.; Abley, M.J.; Gebreyes, W.A. CTX-M-type extended-spectrum β-lactamases present in Escherichia coli from the feces of cattle in Ohio, United States. Foodborne Pathog. Dis. 2010, 7, 1575–1579. [Google Scholar] [CrossRef] [PubMed]
- Cormier, A.; Zhang, P.L.C.; Chalmers, G.; Weese, J.S.; Deckert, A.; Mulvey, M.; McAllister, T.; Boerlin, P. Diversity of CTX-M-positive Escherichia coli recovered from animals in Canada. Vet. Microbiol. 2019, 231, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Kurittu, P.; Khakipoor, B.; Aarnio, M.; Nykäsenoja, S.; Brouwer, M.; Myllyniemi, A.L.; Vatunen, E.; Heikinheimo, A. Plasmid-Borne and Chromosomal ESBL/AmpC Genes in Escherichia coli and Klebsiella pneumoniae in Global Food Products. Front. Microbiol. 2021, 12, 592291. [Google Scholar] [CrossRef] [PubMed]
- Lalak, A.; Wasyl, D.; Zając, M.; Skarżyńska, M.; Hoszowski, A.; Samcik, I.; Woźniakowski, G.; Szulowski, K. Mechanisms of cephalosporin resistance in indicator Escherichia coli isolated from food animals. Vet. Microbiol. 2016, 194, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, M.; Irrgang, A.; Roschanski, N.; Brenner Michael, G.; Hamprecht, A.; Rieber, H.; Käsbohrer, A.; Schwarz, S.; Rösler, U.; Kreienbrock, L.; et al. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genom. 2018, 19, 601. [Google Scholar] [CrossRef]
- Li, L.; Olsen, R.H.; Xiao, J.; Meng, H.; Peng, S.; Shi, L. Genetic context of bla (CTX-M-55) and qnrS1 genes in a foodborne Salmonella enterica serotype Saintpaul isolate from China. Front. Microbiol. 2022, 13, 899062. [Google Scholar] [CrossRef] [PubMed]
- Millar, N.; Aenishaenslin, C.; Lardé, H.; Roy, J.P.; Fourichon, C.; Francoz, D.; Paradis, M.; Dufour, S. Evidence of a decrease in sales of antimicrobials of very high importance for humans in dairy herds after a new regulation restricting their use in Quebec, Canada. Zoonoses Public Health 2022, 69, 370–381. [Google Scholar] [CrossRef] [PubMed]
- de Lagarde, M.; Fairbrother, J.M.; Archambault, M.; Dufour, S.; Francoz, D.; Massé, J.; Lardé, H.; Aenishaenslin, C.; Paradis, M.-È.; Roy, J.-P. Impact of a Regulation Restricting Critical Antimicrobial Usage on Prevalence of Antimicrobial Resistance in Escherichia coli Isolates From Fecal and Manure Pit Samples on Dairy Farms in Québec, Canada. Front. Vet. Sci. 2022, 9, 100. [Google Scholar] [CrossRef] [PubMed]
- Muloi, D.M.; Wee, B.A.; McClean, D.M.H.; Ward, M.J.; Pankhurst, L.; Phan, H.; Ivens, A.C.; Kivali, V.; Kiyong’a, A.; Ndinda, C.; et al. Population genomics of Escherichia coli in livestock-keeping households across a rapidly developing urban landscape. Nat. Microbiol. 2022, 7, 581–589. [Google Scholar] [CrossRef]
- Ludden, C.; Coll, F.; Gouliouris, T.; Restif, O.; Blane, B.; Blackwell, G.A.; Kumar, N.; Naydenova, P.; Crawley, C.; Brown, N.M.; et al. Defining nosocomial transmission of Escherichia coli and antimicrobial resistance genes: A genomic surveillance study. The Lancet. Microbe 2021, 2, e472–e480. [Google Scholar] [CrossRef]
- Bonnedahl, J.; Stedt, J.; Waldenström, J.; Svensson, L.; Drobni, M.; Olsen, B. Comparison of Extended-Spectrum β-Lactamase (ESBL) CTX-M Genotypes in Franklin Gulls from Canada and Chile. PLoS ONE 2015, 10, e0141315. [Google Scholar] [CrossRef]
- Clermont, O.; Dixit, O.V.A.; Vangchhia, B.; Condamine, B.; Dion, S.; Bridier-Nahmias, A.; Denamur, E.; Gordon, D. Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol. 2019, 21, 3107–3117. [Google Scholar] [CrossRef] [PubMed]
- Skarżyńska, M.; Zaja, C.M.; Bomba, A.; Bocian, Ł.; Kozdruń, W.; Polak, M.; Wia Cek, J.; Wasyl, D. Antimicrobial Resistance Glides in the Sky-Free-Living Birds as a Reservoir of Resistant Escherichia coli With Zoonotic Potential. Front. Microbiol. 2021, 12, 656223. [Google Scholar] [CrossRef]

| Antimicrobial Class | Antimicrobial Agent | MIC (µg/mL) | Non-Susceptible (%) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | |||
| Aminoglycoside/aminocyclitol | Gentamicin | 2.5 | 54.2 | 22.0 | 0.8 | 0.8 | 19.5 | 20.3 | ||||||||||
| Neomycin | 50.0 | 0.8 | 4.2 | 9.3 | 35.6 | 49.2 | ||||||||||||
| Spectinomycin | 2.5 | 28.0 | 6.8 | 10.2 | 52.5 | 62.7 | ||||||||||||
| Streptomycin | 6.8 | 5.1 | 5.9 | 3.4 | 6.8 | 72.0 | 78.8 | |||||||||||
| β-lactam | Amoxicillin/clavu. | 1.7 | 1.7 | 40.7 | 3.4 | 50.0 | 2.5 | 55.9 | ||||||||||
| Ampicillin | 1.7 | 98.3 | 98.3 | |||||||||||||||
| Cefoxitin | 4.2 | 23.7 | 11.0 | 9.3 | 20.3 | 31.4 | 61.0 | |||||||||||
| Ceftiofur | 1.7 | 4.2 | 0.8 | 9.3 | 5.1 | 17.8 | 61.0 | 83.9 | ||||||||||
| Ceftriaxone | 6.8 | 3.4 | 5.9 | 14.4 | 20.3 | 7.6 | 11.9 | 29.7 | 89.8 | |||||||||
| Meropenem | 100.0 | 0.0 | ||||||||||||||||
| Folate pathway antagonist | Sulfisoxazole | 8.5 | 2.5 | 0.8 | 88.1 | 88.1 | ||||||||||||
| Trimethoprim/sulfa. | 16.1 | 5.1 | 1.7 | 0.8 | 0.8 | 75.4 | 75.4 | |||||||||||
| Macrolide | Azithromycin | 0.8 | 12.7 | 51.7 | 12.7 | 5.1 | 12.7 | 4.2 | 16.9 | |||||||||
| Phenicol | Chloramphenicol | 16.9 | 33.1 | 1.7 | 48.3 | 50.0 | ||||||||||||
| Florfenicola | 3.4 | 33.9 | 17.8 | 44.9 | NA | |||||||||||||
| Quinolone | Ciprofloxacin | 66.9 | 3.4 | 4.2 | 1.7 | 8.5 | 7.6 | 5.1 | 2.5 | 15.3 | ||||||||
| Danofloxacin | 74.6 | 2.5 | 5.9 | 8.5 | 8.5 | 22.9 | ||||||||||||
| Enrofloxacin | 74.6 | 3.4 | 5.1 | 9.3 | 5.1 | 2.5 | 22.0 | |||||||||||
| Nalidixic acid | 9.3 | 50.0 | 16.9 | 8.5 | 9.3 | 5.9 | 5.9 | |||||||||||
| Tetracycline | Tetracycline | 20.3 | 2.5 | 77.1 | 79.7 | |||||||||||||
| AMR Gene or Mutation | Proportion (%) | AMR Phenotype a | Antimicrobial Class |
|---|---|---|---|
| aac(3)-IIa | 2 | GEN | Aminoglycosides /Aminocyclitols |
| aac(3)-IId | 17 | GEN | |
| aac(3)-Via | 2 | GEN | |
| aadA(1,2,5,12,22,24) | 27,25,11,1,13,1 b | STR, SPT | |
| aph(3′)-Ia | 47 | NEO | |
| aph(3″)-Ib (strA) | 65 | STR | |
| aph(6)-Id (strB) | 65 | STR | |
| ampC promoter n.-42C>T | 12 | AMC, AMP, FOX | β-lactams |
| blaCARB-2 | 1 | AMP | |
| blaCMY-2 | 39 | AMC, AMP, FOX, TIO c, CRO d | |
| blaCMY-44 (blaCMY-2 like) | 2 | AMC, AMP, FOX, TIO c, CRO d | |
| blaCTX-M-(1,15,27,55,65,124) | 9,12,2,15,1,3 b | AMP, TIO c, CRO | |
| blaOXA-1 | 1 | AMC, AMP | |
| blaOXA-10 | 1 | AMP | |
| blaTEM-1A | 2 | AMP | |
| blaTEM-1B | 43 | AMP | |
| dfrA(1,5,7,8,12,14,16,17,23) | 16,8,4,1,19,20,1,11,5 b | SXT | Folate pathway antagonist |
| sul(1,2,3) | 38,72,18 b | FIS | |
| mph(A) | 21 | AZM | Macrolide |
| catA1 | 9 | CHL | Phenicol |
| cmlA1 | 4 | CHL | |
| floR | 43 | CHL, FFC | |
| gyrA (p.D87N, p.S83A, p.S83L, p.S83V) | 3,3,5,1 b | CIP, DAN c, ENR c, NAL | Quinolone |
| parC (p.A56T, p.S80I) | 2,3 b | CIP, DAN c, ENR c, NAL | |
| parE (p.I355T, p.S458A) | 1,1 b | CIP, DAN c, ENR c, NAL | |
| qnrB19 | 1 | CIP, DAN c, ENR c | |
| qnrS1 | 19 | CIP, DAN c, ENR c | |
| ARR-2 | 13 | Not tested | Rifampin |
| tet(A) | 53 | TET | Tetracycline |
| tet(B) | 36 | TET |
| Phenotype: Susceptible | Phenotype: Resistant | Sensitivity (%) | Specificity (%) | |||
|---|---|---|---|---|---|---|
| Antimicrobial | Genotype Resistant | Genotype Susceptible | Genotype Resistant | Genotype Susceptible | ||
| Gentamicin | 0 | 94 | 24 | 0 | 100 | 100 |
| Neomycin | 0 | 60 | 56 | 2 | 97 | 100 |
| Spectinomycin | 10 | 34 | 74 | 0 | 100 | 77 |
| Streptomycin | 12 | 13 | 93 | 0 | 100 | 52 |
| Amoxicillin/clavu. | 0 | 52 | 62 | 4 | 94 | 100 |
| Ampicillin | 0 | 2 | 116 | 0 | 100 | 100 |
| Cefoxitin | 0 | 46 | 61 | 11 | 85 | 100 |
| Ceftiofur | 0 | 19 | 96 | 3 | 97 | 100 |
| Ceftriaxone | 0 | 12 | 96 | 10 | 91 | 100 |
| Meropenem | 0 | 118 | 0 | 0 | NA | 100 |
| Sulfisoxazole | 0 | 14 | 104 | 0 | 100 | 100 |
| Trimethoprim/sulfa. | 4 | 25 | 89 | 0 | 100 | 86 |
| Azithromycin | 6 | 92 | 19 | 1 | 95 | 94 |
| Chloramphenicol | 1 | 58 | 57 | 2 | 97 | 98 |
| Nalidixic acid | 6 | 105 | 7 | 0 | 100 | 95 |
| Ciprofloxacin | 18 | 82 | 18 | 0 | 100 | 82 |
| Danofloxacin | 9 | 82 | 27 | 0 | 100 | 90 |
| Enrofloxacin | 10 | 82 | 26 | 0 | 100 | 89 |
| Tetracycline | 0 | 24 | 93 | 1 | 99 | 100 |
| Antimicrobial(s) | Pheno. | Geno. | Gene(s) | nb | Explanation |
|---|---|---|---|---|---|
| NEO | R | S | 2 | Unknown | |
| AMC | R | S | 4 | Unknown, only four isolates with intermediate susceptibility | |
| FOX | R | S | 11 | Unknown, most have intermediate susceptibility | |
| CRO, TIO | R | S | ampC a (-42 C->T) | 10 | Gene not reported to be associated with a phenotypic resistance to CRO or TIO |
| AZM | R | S | 1 | Unknown | |
| CHL | R | S | 2 | Unknown, only two isolates with intermediate susceptibility | |
| TET | R | S | 1 | Unknown | |
| SPT | S | R | aadA1 or aadA5 | 10 | Increase in MIC but not enough to reach clinical breakpoint |
| STR | S | R | aadA(1,5) or strA /strB | 12 | Increase in MIC but not enough to reach clinical breakpoint |
| SXT | S | R | dfrA | 4 | Increase in MIC but not enough to reach clinical breakpoint |
| AZM | S | R | mph(A) | 6 | Increase in MIC but not enough to reach clinical breakpoint |
| CHL | S | R | catA1 | 1 | Unknown, lowest coverage among catA1 (possible non-functional gene) |
| NAL, CIP, DAN, ENR | S | R | qnr, gyrA a, parC a or parE a | 18 | Increase in MIC but not enough to reach clinical breakpoint or silent mutation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massé, J.; Vanier, G.; Fairbrother, J.M.; de Lagarde, M.; Arsenault, J.; Francoz, D.; Dufour, S.; Archambault, M. Description of Antimicrobial-Resistant Escherichia coli and Their Dissemination Mechanisms on Dairy Farms. Vet. Sci. 2023, 10, 242. https://doi.org/10.3390/vetsci10040242
Massé J, Vanier G, Fairbrother JM, de Lagarde M, Arsenault J, Francoz D, Dufour S, Archambault M. Description of Antimicrobial-Resistant Escherichia coli and Their Dissemination Mechanisms on Dairy Farms. Veterinary Sciences. 2023; 10(4):242. https://doi.org/10.3390/vetsci10040242
Chicago/Turabian StyleMassé, Jonathan, Ghyslaine Vanier, John M. Fairbrother, Maud de Lagarde, Julie Arsenault, David Francoz, Simon Dufour, and Marie Archambault. 2023. "Description of Antimicrobial-Resistant Escherichia coli and Their Dissemination Mechanisms on Dairy Farms" Veterinary Sciences 10, no. 4: 242. https://doi.org/10.3390/vetsci10040242
APA StyleMassé, J., Vanier, G., Fairbrother, J. M., de Lagarde, M., Arsenault, J., Francoz, D., Dufour, S., & Archambault, M. (2023). Description of Antimicrobial-Resistant Escherichia coli and Their Dissemination Mechanisms on Dairy Farms. Veterinary Sciences, 10(4), 242. https://doi.org/10.3390/vetsci10040242







