Colonization of Wild Saccharomyces cerevisiae Strains in a New Winery
Abstract
1. Introduction
2. Material and Methods
2.1. Sampling of Winery Related Environments (WREs)
2.2. “Pied de Cuve” (PDC)
2.3. Must/Wine Samples
2.4. Illumina Sequencing
2.4.1. DNA Extraction
2.4.2. ITS Amplicon Library Preparation
2.4.3. Sequence Analysis
2.5. Yeast Isolation
2.6. Molecular Identification of Yeast Isolates
2.6.1. ITS-PCR
2.6.2. Interdelta PCR Typing
2.7. Biofilm Formation of S. cerevisiae
2.7.1. Scanning Electron Microscopy (SEM)
2.7.2. Statistical Analysis
3. Results and Discussion
3.1. Saccharomyces Status in the New Winery before the Arrival of the First Harvest
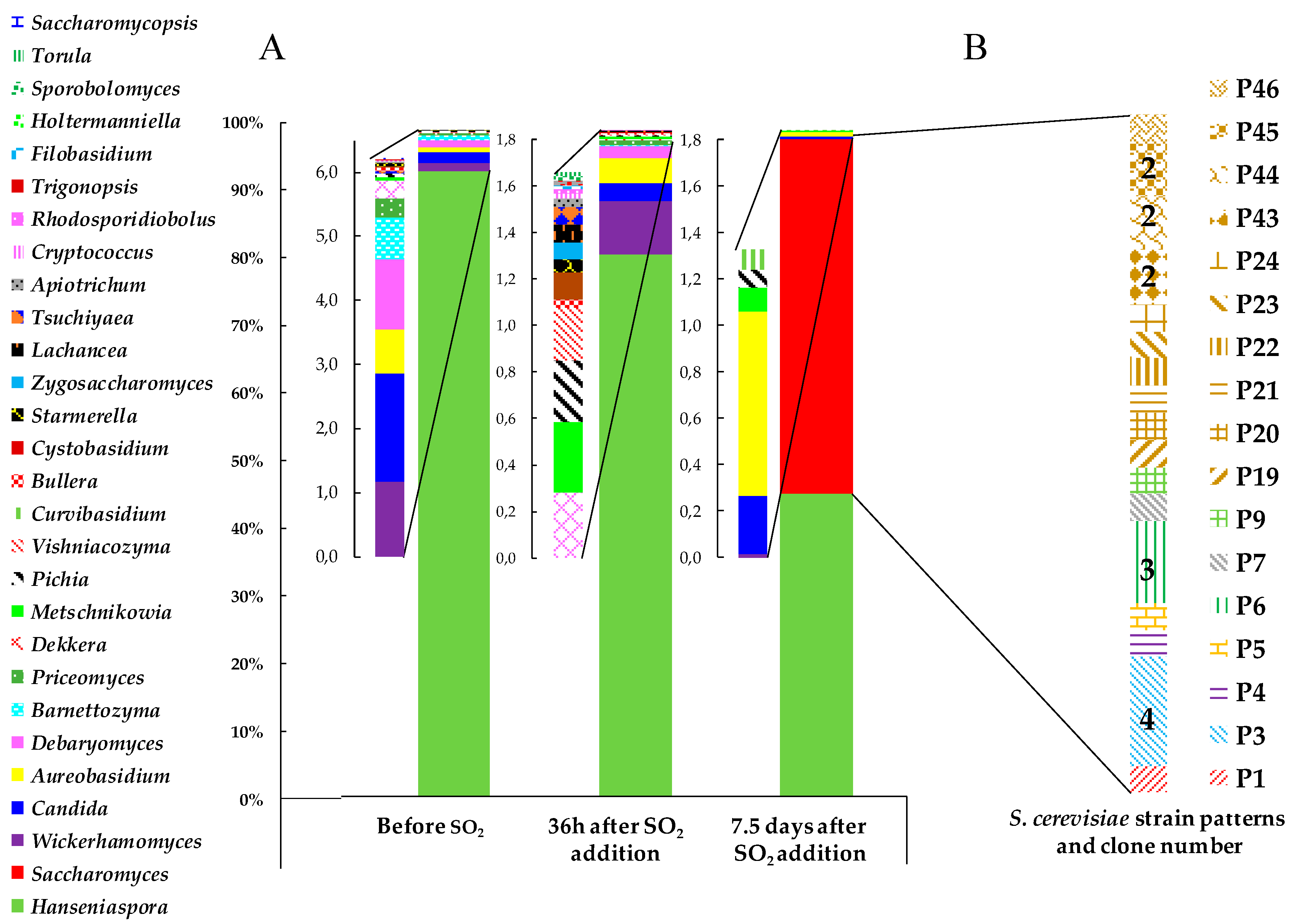
3.2. Yeast Biodiversity in ‘Pied de Cuve’ (PDC)
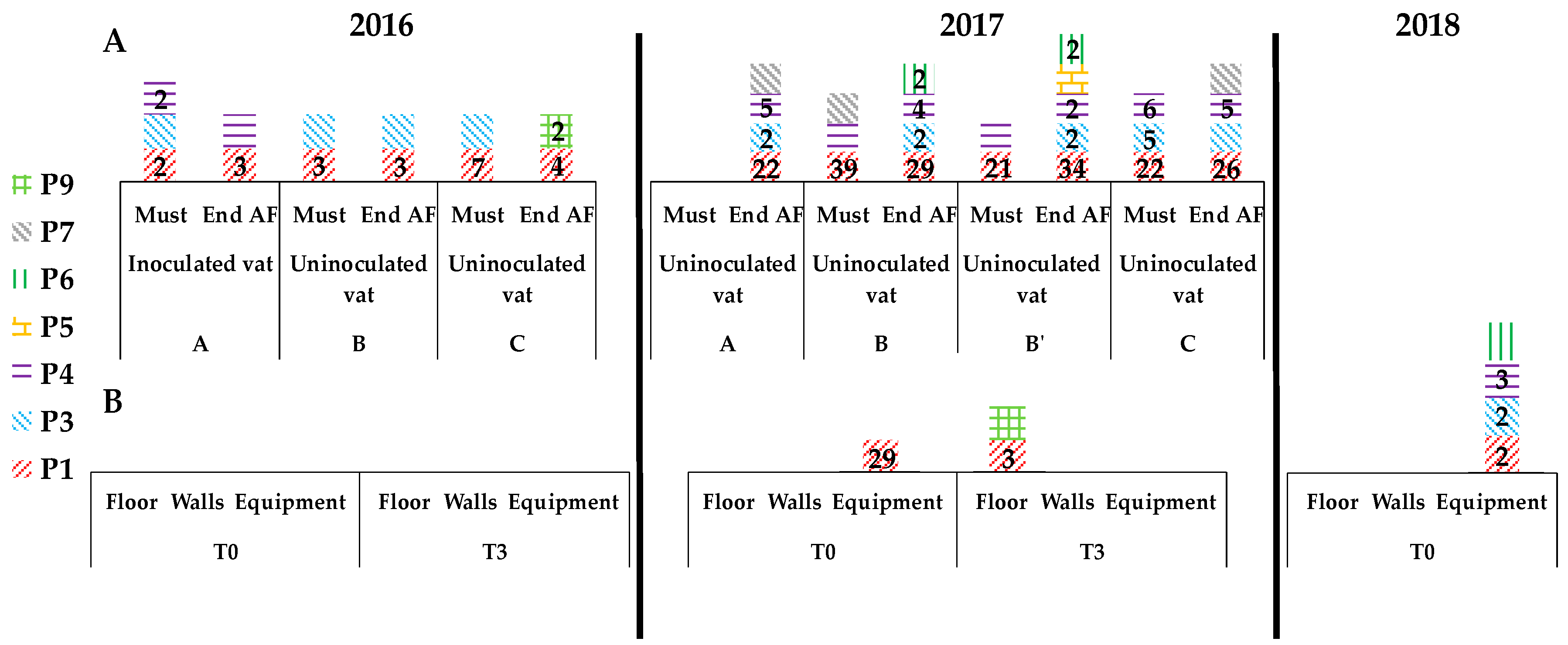
3.3. Presence of Indigenous S. cerevisiae Detected in the PDC 2016 in the New Winery during 2016, 2017 and 2018
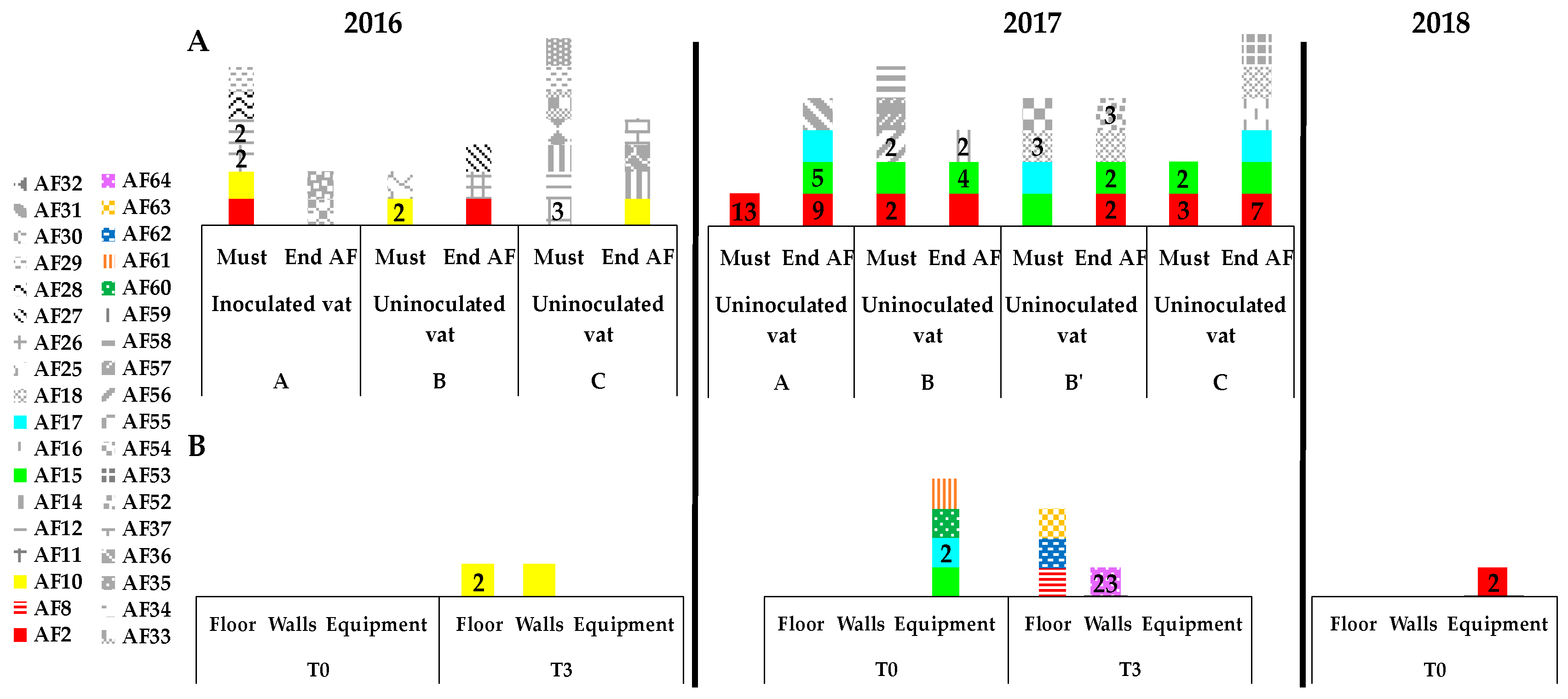
3.4. Implication of Spontaneous Fermentations on S. cerevisiae Population Found on WREs
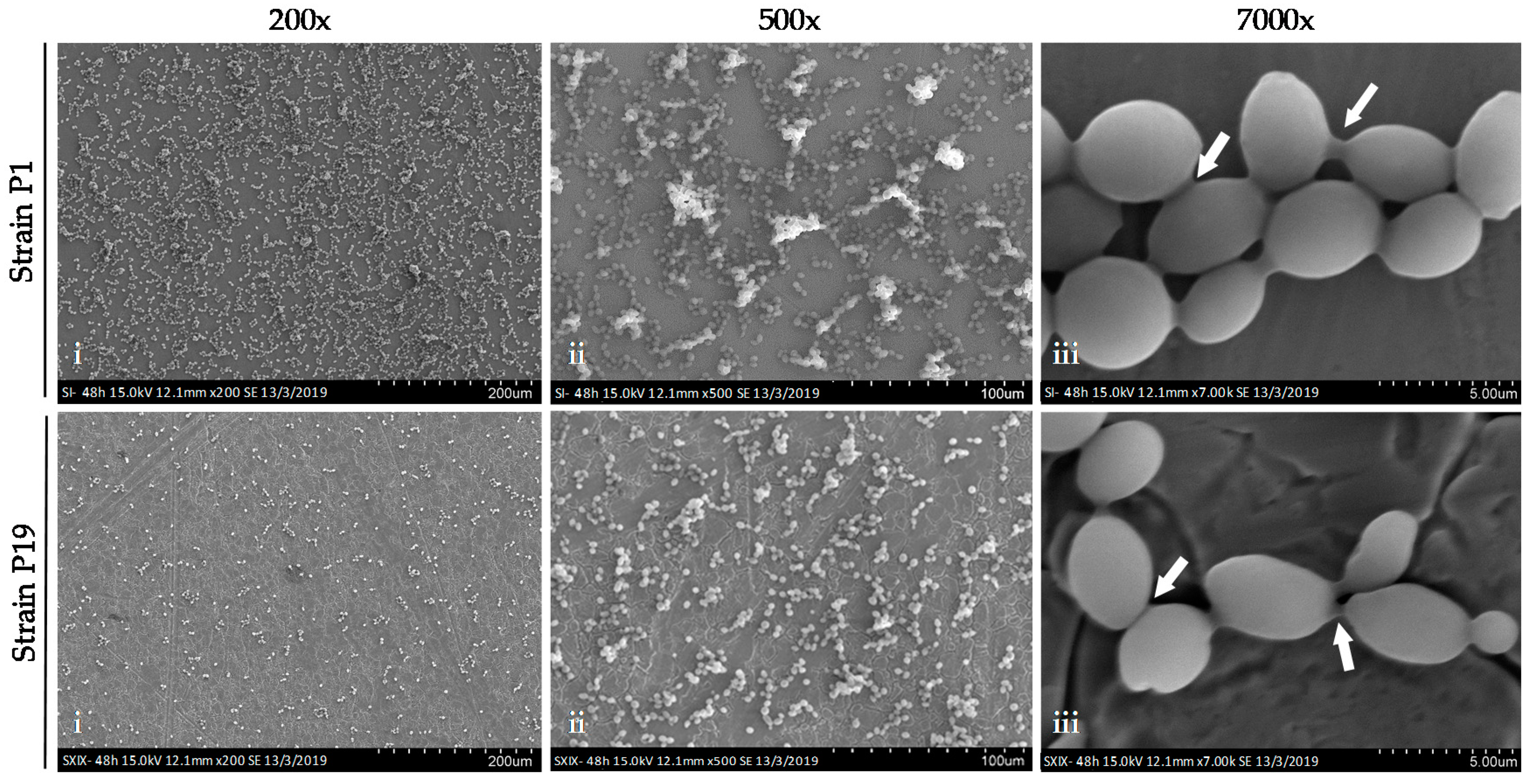
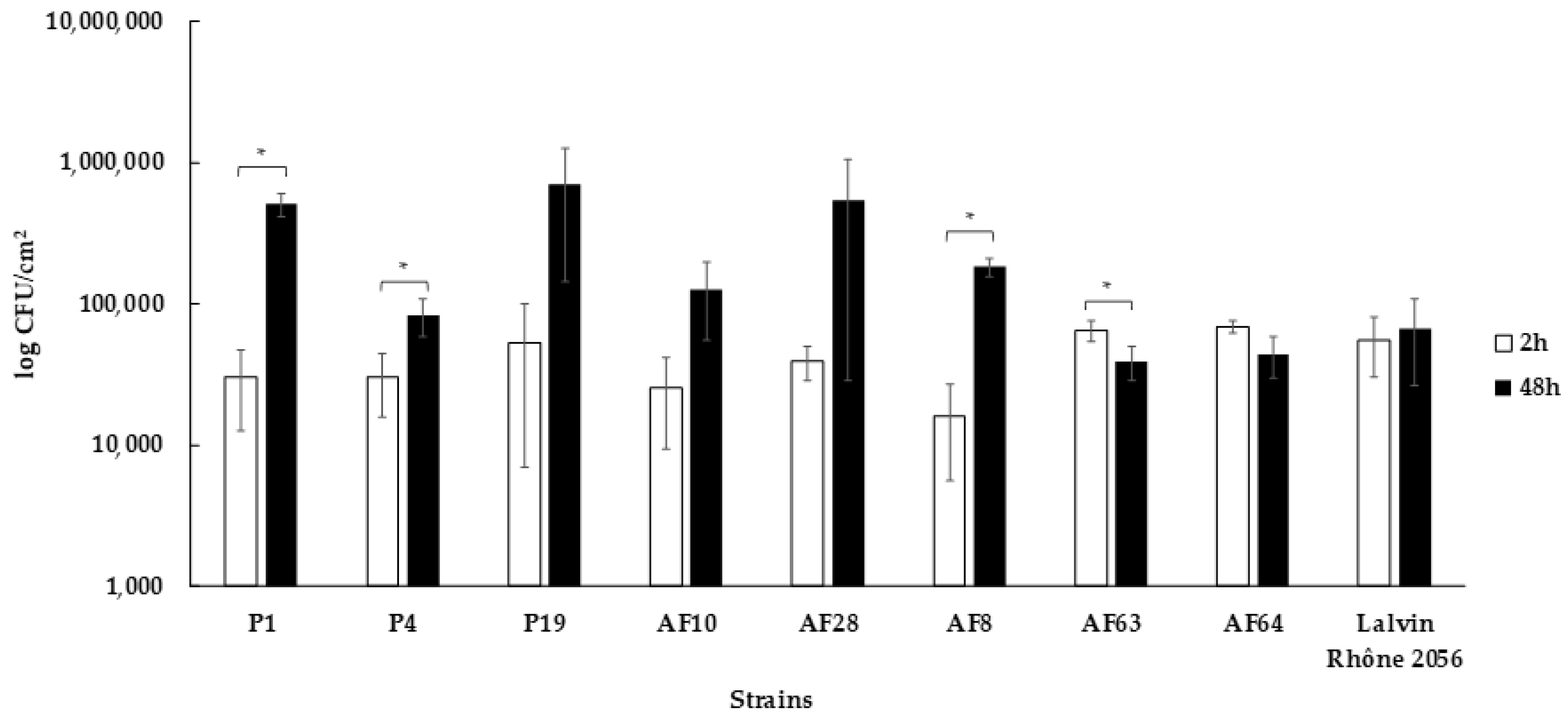
3.5. Capacity to form Biofilms on Stainless Steel
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kraus, J.K.; Reed, G.; Villettaz, J.-C. Levures sèches actives de vinification. lère partie: Fabrication et caractéristiques. OENO One 1983, 17, 93–103. [Google Scholar] [CrossRef]
- Reed, G.; Nagodawithana, T.W. Technology of yeast usage in winemaking. Am. J. Enol. Vitic. 1988, 39, 83–90. [Google Scholar]
- Schuller, D.; Casal, M. The genetic structure of fermentative vineyard-associated Saccharomyces cerevisiae populations revealed by microsatellite analysis. Antonie Leeuwenhoek 2007, 91, 137–150. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Esteban-Fernández, A.; Navascués, E.; Marquina, D.; Santos, A.; Moreno-Arribas, V. Microbial contribution to wine aroma and its intended use for wine quality improvement. Molecules 2017, 22, 189. [Google Scholar] [CrossRef]
- Berbegal, C.; Spano, G.; Fragasso, M.; Grieco, F.; Russo, P.; Capozzi, V. Starter cultures as biocontrol strategy to prevent Brettanomyces bruxellensis proliferation in wine. Appl. Microbiol. Biotechnol. 2018, 102, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Fleet, G.H. Yeast interactions and wine flavour. Int. J. Food Microbiol. 2003, 86, 11–22. [Google Scholar] [CrossRef]
- Fleet, G.H. Wine yeasts for the future. FEMS Yeast Res. 2008, 8, 979–995. [Google Scholar] [CrossRef] [PubMed]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef]
- Lu, Y.; Huang, D.; Lee, P.-R.; Liu, S.-Q. Assessment of volatile and non-volatile compounds in durian wines fermented with four commercial non-Saccharomyces yeasts. J. Sci. Food Agric. 2016, 96, 1511–1521. [Google Scholar] [CrossRef]
- Petruzzi, L.; Capozzi, V.; Berbegal, C.; Corbo, M.R.; Bevilacqua, A.; Spano, G.; Sinigaglia, M. Microbial resources and enological significance: Opportunities and benefits. Front. Microbiol. 2017, 8, 995. [Google Scholar] [CrossRef]
- Medina, K.; Boido, E.; Dellacassa, E.; Carrau, F. Growth of non-Saccharomyces yeasts affects nutrient availability for Saccharomyces cerevisiae during wine fermentation. Int. J. Food Microbiol. 2012, 157, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.S. Nature and origin of wine quality. In Wine Tasting: A Professional Handbook, 2nd ed.; Jackson, R.S., Ed.; Academic Press: Burlington, MA, USA, 2009; pp. 387–426. [Google Scholar]
- Ubeda Iranzo, J.F.; González Magaña, F.; González Viñas, M.A. Evaluation of the formation of volatiles and sensory characteristics in the industrial production of white wines using different commercial strains of the genus Saccharomyces. Food Control 2000, 11, 143–147. [Google Scholar] [CrossRef]
- Clavijo, A.; Calderón, I.L.; Paneque, P. Yeast assessment during alcoholic fermentation inoculated with a natural “pied de cuve” or a commercial yeast strain. World J. Microbiol. Biotechnol. 2011, 27, 1569–1577. [Google Scholar] [CrossRef]
- Li, E.; Liu, C.; Liu, Y. Evaluation of yeast diversity during wine fermentations with direct inoculation and pied de cuve method at an industrial scale. J. Microbiol. Biotechnol. 2012, 22, 960–966. [Google Scholar] [CrossRef]
- Moschetti, G.; Corona, O.; Gaglio, R.; Squadrito, M.; Parrinello, A.; Settanni, L.; Barone, E.; Francesca, N. Use of fortified pied de cuve as an innovative method to start spontaneous alcoholic fermentation for red winemaking. J. Grape Wine Res. 2016, 22, 36–45. [Google Scholar] [CrossRef]
- Tello, J.; Cordero-Bueso, G.; Aporta, I.; Cabellos, J.M.; Arroyo, T. Genetic diversity in commercial wineries: Effects of the farming system and vinification management on wine yeasts. J. Appl. Microbiol. 2012, 112, 302–315. [Google Scholar] [CrossRef]
- Martiniuk, J.T.; Pacheco, B.; Russell, G.; Tong, S.; Backstrom, I.; Measday, V. Impact of commercial strain use on Saccharomyces cerevisiae population structure and dynamics in Pinot Noir vineyards and spontaneous fermentations of a Canadian winery. PLoS ONE 2016, 11. [Google Scholar] [CrossRef]
- Santamaría, P.; Garijo, P.; López, R.; Tenorio, C.; Gutiérrez, A.R. Analysis of yeast population during spontaneous alcoholic fermentation: Effect of the age of the cellar and the practice of inoculation. Int. J. Food Microbiol. 2005, 103, 49–56. [Google Scholar] [CrossRef]
- Mercado, L.; Dalcero, A.; Masuelli, R.; Combina, M. Diversity of Saccharomyces strains on grapes and winery surfaces: Analysis of their contribution to fermentative flora of Malbec wine from Mendoza (Argentina) during two consecutive years. Food Microbiol. 2007, 24, 403–412. [Google Scholar] [CrossRef]
- Santamaría, P.; López, R.; López, E.; Garijo, P.; Gutierrez, A.R. Permanence of yeast inocula in the winery ecosystem and presence in spontaneous fermentations. Eur. Food Res. Technol. 2008, 227, 1563–1567. [Google Scholar] [CrossRef]
- Grangeteau, C. Biodiversité Fongique du Raisin au Vin: Impact de l’activité anthropique. Ph.D. Thesis, Université de Bourgogne Franche-Comté, Dijon, France, 2016. [Google Scholar]
- Constantí, M.; Poblet, M.; Arola, L.; Mas, A.; Guillamón, J.M. Analysis of yeast populations during alcoholic fermentation in a newly established winery. Am. J. Enol. Vitic. 1997, 48, 339–344. [Google Scholar]
- Beltran, G.; Torija, M.J.; Novo, M.; Ferrer, N.; Poblet, M.; Guillamón, J.M.; Rozès, N.; Mas, A. Analysis of yeast populations during alcoholic fermentation: A six year follow- up study. Syst. Appl. Microbiol. 2002, 25, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Clavijo, A.; Calderón, I.L.; Paneque, P. Effect of the use of commercial Saccharomyces strains in a newly established winery in Ronda (Málaga, Spain). Antonie Leeuwenhoek 2011, 99, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Starmer, W.T.; Ganter, P.F.; Aberdeen, V.; Lachance, M.; Phaff, H.J. The ecological role of killer yeasts in natural communities of yeasts. Can. J. Microbiol. 1987, 33, 783–796. [Google Scholar] [CrossRef] [PubMed]
- De Ullivarri, M.F.; Mendoza, L.M.; Raya, R.R. Killer activity of Saccharomyces cerevisiae strains: Partial characterization and strategies to improve the biocontrol efficacy in winemaking. Antonie Leeuwenhoek 2014, 106, 865–878. [Google Scholar] [CrossRef] [PubMed]
- Salma, M.; Rousseaux, S.; Sequeira-Le Grand, A.; Divol, B.; Alexandre, H. Characterization of the viable but nonculturable (VBNC) state in Saccharomyces cerevisiae. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Davey, M.E.; O’Toole, G.A. Microbial biofilms: From ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef]
- O’Connell, H.A.; Kottkamp, G.S.; Eppelbaum, J.L.; Stubblefield, B.A.; Gilbert, S.E.; Gilbert, E.S. Influences of biofilm structure and antibiotic resistance mechanisms on indirect pathogenicity in a model polymicrobial biofilm. Appl. Environ. Microbiol. 2006, 72, 5013–5019. [Google Scholar] [CrossRef]
- Bastard, A.; Coelho, C.; Briandet, R.; Canette, A.; Gougeon, R.; Alexandre, H.; Guzzo, J.; Weidmann, S. Effect of biofilm formation by Oenococcus oeni on malolactic fermentation and the release of aromatic compounds in wine. Front. Microbiol. 2016, 7, 613. [Google Scholar] [CrossRef]
- Tek, E.L.; Sundstrom, J.F.; Gardner, J.M.; Oliver, S.G.; Jiranek, V. Evaluation of the ability of commercial wine yeasts to form biofilms (mats) and adhere to plastic: Implications for the microbiota of the winery environment. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- Taylor, D.L.; Walters, W.A.; Lennon, N.J.; Bochicchio, J.; Krohn, A.; Caporaso, G.J.; Pennanen, T. Accurate estimation of fungal diversity and abundance through improved lineage-specific primers optimized for Illumina Amplicon Sequencing. Appl. Environ. Microbiol. 2016, 82, 7217–7226. [Google Scholar] [CrossRef] [PubMed]
- Gweon, H.S.; Oliver, A.; Taylor, J.; Booth, T.; Gibbs, M.; Read, D.S.; Griffiths, R.I.; Schonrogge, K. PIPITS: An automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol. Evol. 2015, 6, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, E2584. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Kõljalg, U.; Larsson, K.H.; Abarenkov, K.; Nilsson, R.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; et al. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005, 166, 1063–1068. [Google Scholar] [CrossRef]
- Grangeteau, C.; Gerhards, D.; von Wallbrunn, C.; Alexandre, H.; Rousseaux, S.; Guilloux-Bénatier, M. Persistence of two non-Saccharomyces yeasts (Hanseniaspora and Starmellera) in the cellar. Front. Microbiol. 2016, 7, 268. [Google Scholar] [CrossRef]
- Werner, O.; Ros, R.M.; Guerra, J. Direct amplification and NaOH extraction: Two rapid and simple methods for preparing bryophyte DNA for polymerase chain reaction (PCR). J. Bryol. 2002, 24, 127–131. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Ed.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Esteve-Zarzoso, B.; Belloch, C.; Uruburu, F.; Querol, A. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 1999, 49, 329–337. [Google Scholar] [CrossRef]
- Legras, J.-L.; Karst, F. Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterisation. FEMS Microbiol. Lett. 2003, 221, 249–255. [Google Scholar] [CrossRef]
- Rodriguez, M.E.; Orozco, H.; Cantoral, J.M.; Matallana, E.; Aranda, A. Acetyltransferase SAS2 and sirtuin SIR2, respectively, control flocculation and biofilm formation in wine yeast. FEMS Yeast Res. 2014, 14, 845–857. [Google Scholar] [CrossRef]
- Ocón, E.; Gutiérrez, A.R.; Garijo, P.; López, R.; Santamaría, P. Presence of non-Saccharomyces yeasts in cellar equipment and grape juice during harvest time. Food Microbiol. 2010, 27, 1023–1027. [Google Scholar] [CrossRef]
- Ocón, E.; Garijo, P.; Sanz, S.; Olarte, C.; Santamaría, P.; Gutiérrez, A.R. Analysis of airborne yeast in one winery over a period of one year. Food Control 2013, 30, 585–589. [Google Scholar] [CrossRef]
- Abdo, H.; Catacchio, C.R.; Ventura, M.; D’Addabbo, P.; Alexandre, H.; Guilloux-Bénatier, M.; Rousseaux, S. The fungal life of a new winery: Establishment of the fungal consortium. Sci. Rep. 2019. (Under Review). [Google Scholar]
- Sabate, J.; Cano, J.; Esteve-Zarzoso, B.; Guillamon, J.M. Isolation and identification of yeasts associated with vineyard and winery by RFLP analysis of ribosomal genes and mitochondrial DNA. Microbiol. Res. 2002, 157, 267–274. [Google Scholar] [CrossRef]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef]
- Mortimer, R.; Polsinelli, M. On the origins of wine yeast. Res. Microbiol. 1999, 150, 199–204. [Google Scholar] [CrossRef]
- Valero, E.; Cambon, B.; Schuller, D.; Casal, M.; Dequin, S. Biodiversity of Saccharomyces yeast strains from grape berries of wine-producing areas using starter commercial yeasts. FEMS Yeast Res. 2007, 7, 317–329. [Google Scholar] [CrossRef]
- Warth, A.D. Resistance of yeast species to benzoic and sorbic acids and to sulfur dioxide. J. Food Prot. 1985, 48, 564–569. [Google Scholar] [CrossRef]
- Henick-Kling, T.; Edinger, W.; Daniel, P.; Monk, P. Selective effects of sulfur dioxide and yeast starter culture addition on indigenous yeast populations and sensory characteristics of wine. J. Appl. Microbiol. 1998, 84, 865–876. [Google Scholar] [CrossRef]
- Constantí, M.; Reguant, C.; Poblet, M.; Zamora, F.; Mas, A.; Guillamón, J.M. Molecular analysis of yeast population dynamics: Effect of sulphur dioxide and inoculum on must fermentation. Int. J. Food Microbiol. 1998, 41, 169–175. [Google Scholar] [CrossRef]
- Vezinhet, F.; Hallet, J.; Valade, M.; Poulard, A. Ecological survey of wine yeast strains by molecular methods of identification. Am. J. Enol. Vitic. 1992, 43, 83–86. [Google Scholar]
- Le Jeune, C.; Erny, C.; Demuyter, C.; Lollier, M. Evolution of the population of Saccharomyces cerevisiae from grape to wine in a spontaneous fermentation. Food Microbiol. 2006, 23, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Garijo, P.; Santamaría, P.; López, R.; Sanz, S.; Olarte, C.; Gutiérrez, A.R. The occurrence of fungi, yeasts and bacteria in the air of a Spanish winery during vintage. Int. J. Food Microbiol. 2008, 125, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Grangeteau, C.; Gerhards, D.; Rousseaux, S.; von Wallbrunn, C.; Alexandre, H.; Guilloux-Bénatier, M. Diversity of yeast strains of the genus Hanseniaspora in the winery environment: What is their involvement in grape must fermentation? Food Microbiol. 2015, 50, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Torija, M.J.; Rozès, N.; Poblet, M.; Guillamón, J.M.; Mas, A. Yeast population dynamics in spontaneous fermentations: Comparison between two different wine-producing areas over a period of three years. Antonie Leeuwenhoek 2001, 79, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Schuller, D.; Alves, H.; Dequin, S.; Casal, M. Ecological survey of Saccharomyces cerevisiae strains from vineyards in the Vinho Verde region of Portugal. FEMS Microbiol. Ecol. 2005, 51, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Nunes, J.M.; Bizerra, F.C.; Ferreira, R.C.; Colombo, A.L. Molecular identification, antifungal susceptibility profile, and biofilm formation of clinical and environmental Rhodotorula species isolates. Antimicrob. Agents Chemother. 2013, 57, 382–389. [Google Scholar] [CrossRef]
- Pu, L.; Jingfan, F.; Kai, C.; Chao-An, L.; Yunjiang, C. Phenylethanol promotes adhesion and biofilm formation of the antagonistic yeast Kloeckera apiculata for the control of blue mold on citrus. FEMS Yeast Res. 2014, 14, 536–546. [Google Scholar] [CrossRef]
- Weerasekera, M.M.; Wijesinghe, G.K.; Jayarathna, T.A.; Gunasekara, C.P.; Fernando, N.; Kottegoda, N.; Samaranayake, L.P. Culture media profoundly affect Candida albicans and Candida tropicalis growth, adhesion and biofilm development. Mem. Do Inst. Oswaldo Cruz 2016, 111, 697–702. [Google Scholar] [CrossRef]
- Kuthan, M.; Devaux, F.; Janderová, B.; Slaninová, I.; Jacq, C.; Palková, Z. Domestication of wild Saccharomyces cerevisiae is accompanied by changes in gene expression and colony morphology. Mol. Microbiol. 2003, 47, 745–754. [Google Scholar] [CrossRef]
- Arroyo-López, F.N.; Salvadó, Z.; Tronchoni, J.; Guillamon, J.M.; Barno, E.; Quérol, A. Susceptibility and resistance to ethanol in Saccharomyces strains isolated from wild and fermentative environments. Yeast 2010, 27, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Nadai, C.; Treu, L.; Campanaro, S.; Giacomini, A.; Corich, V. Different mechanisms of resistance modulate sulfite tolerance in wine yeasts. Appl. Microbiol. Biotechnol. 2016, 100, 797–813. [Google Scholar] [CrossRef] [PubMed]
| Name of Fermentation Vat | Must Appellation | |
|---|---|---|
| 2016 Vintage | 2017 Vintage | |
| Pied de Cuve | - | Beaune 1er Cru—Clos de l’Ecu |
| A | A | |
| B | B and B’ | Volnay 1er Cru—Fremiets |
| C | C | Monthélie 1er Cru—Champs Fulliot |
| Yeast Genus | T0 2016 | ||
|---|---|---|---|
| Floor | Walls | Equipment | |
| Non-Saccharomyces * | 99.7 | 100.0 | 99.9 |
| Saccharomyces | 0.3 | 0.0 | 0.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdo, H.; Catacchio, C.R.; Ventura, M.; D’Addabbo, P.; Calabrese, F.M.; Laurent, J.; David-Vaizant, V.; Alexandre, H.; Guilloux-Bénatier, M.; Rousseaux, S. Colonization of Wild Saccharomyces cerevisiae Strains in a New Winery. Beverages 2020, 6, 9. https://doi.org/10.3390/beverages6010009
Abdo H, Catacchio CR, Ventura M, D’Addabbo P, Calabrese FM, Laurent J, David-Vaizant V, Alexandre H, Guilloux-Bénatier M, Rousseaux S. Colonization of Wild Saccharomyces cerevisiae Strains in a New Winery. Beverages. 2020; 6(1):9. https://doi.org/10.3390/beverages6010009
Chicago/Turabian StyleAbdo, Hany, Claudia R. Catacchio, Mario Ventura, Pietro D’Addabbo, Francesco Maria Calabrese, Julie Laurent, Vanessa David-Vaizant, Hervé Alexandre, Michèle Guilloux-Bénatier, and Sandrine Rousseaux. 2020. "Colonization of Wild Saccharomyces cerevisiae Strains in a New Winery" Beverages 6, no. 1: 9. https://doi.org/10.3390/beverages6010009
APA StyleAbdo, H., Catacchio, C. R., Ventura, M., D’Addabbo, P., Calabrese, F. M., Laurent, J., David-Vaizant, V., Alexandre, H., Guilloux-Bénatier, M., & Rousseaux, S. (2020). Colonization of Wild Saccharomyces cerevisiae Strains in a New Winery. Beverages, 6(1), 9. https://doi.org/10.3390/beverages6010009








