Transcriptome Analyses of Procambarus clarkii (Girard, 1852) Under Individual Exposures to CuSO4, Pendimethalin, and Glyphosate
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals Stock Preparation, Animals and Sample Collection
2.2. Histopathological Alterations
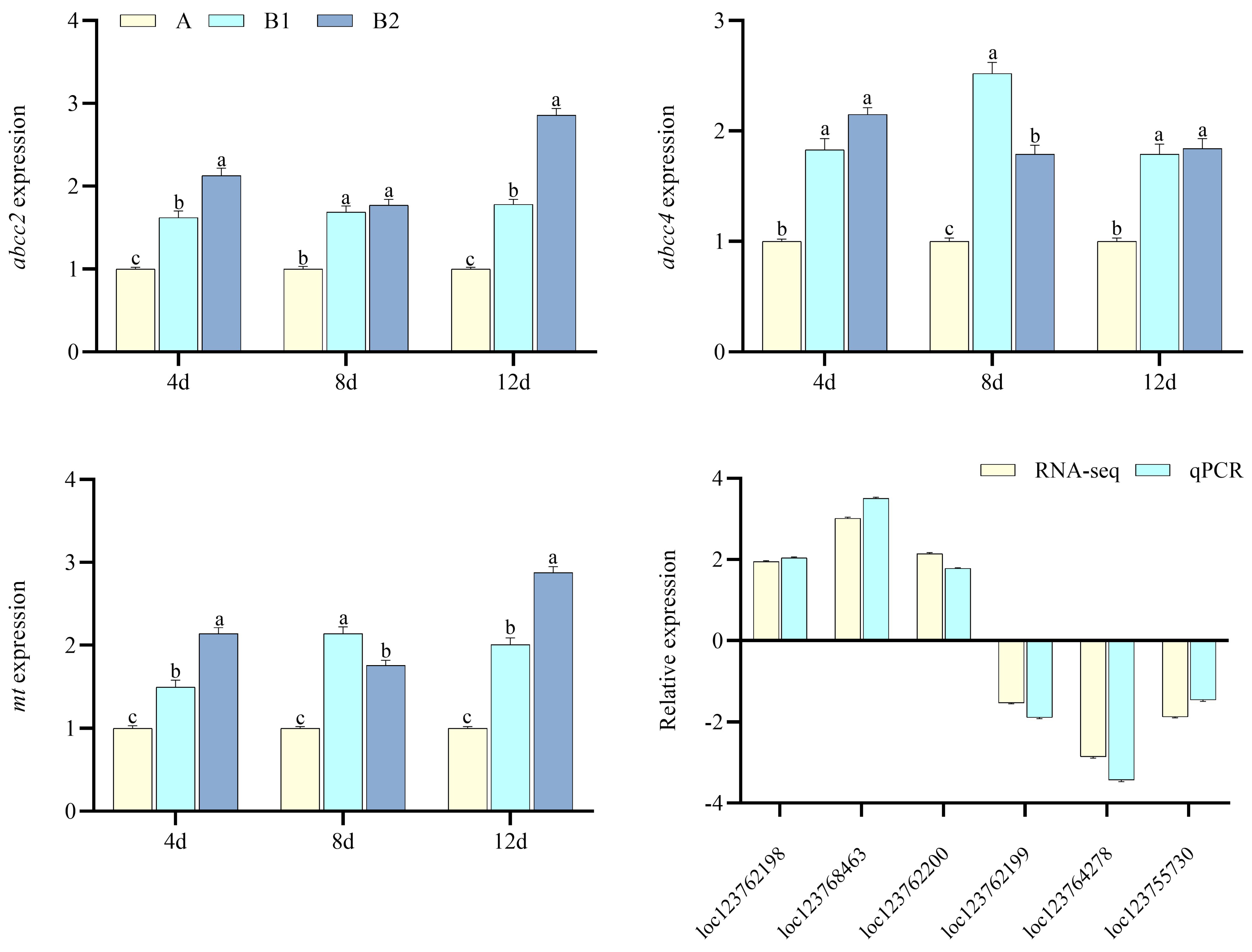
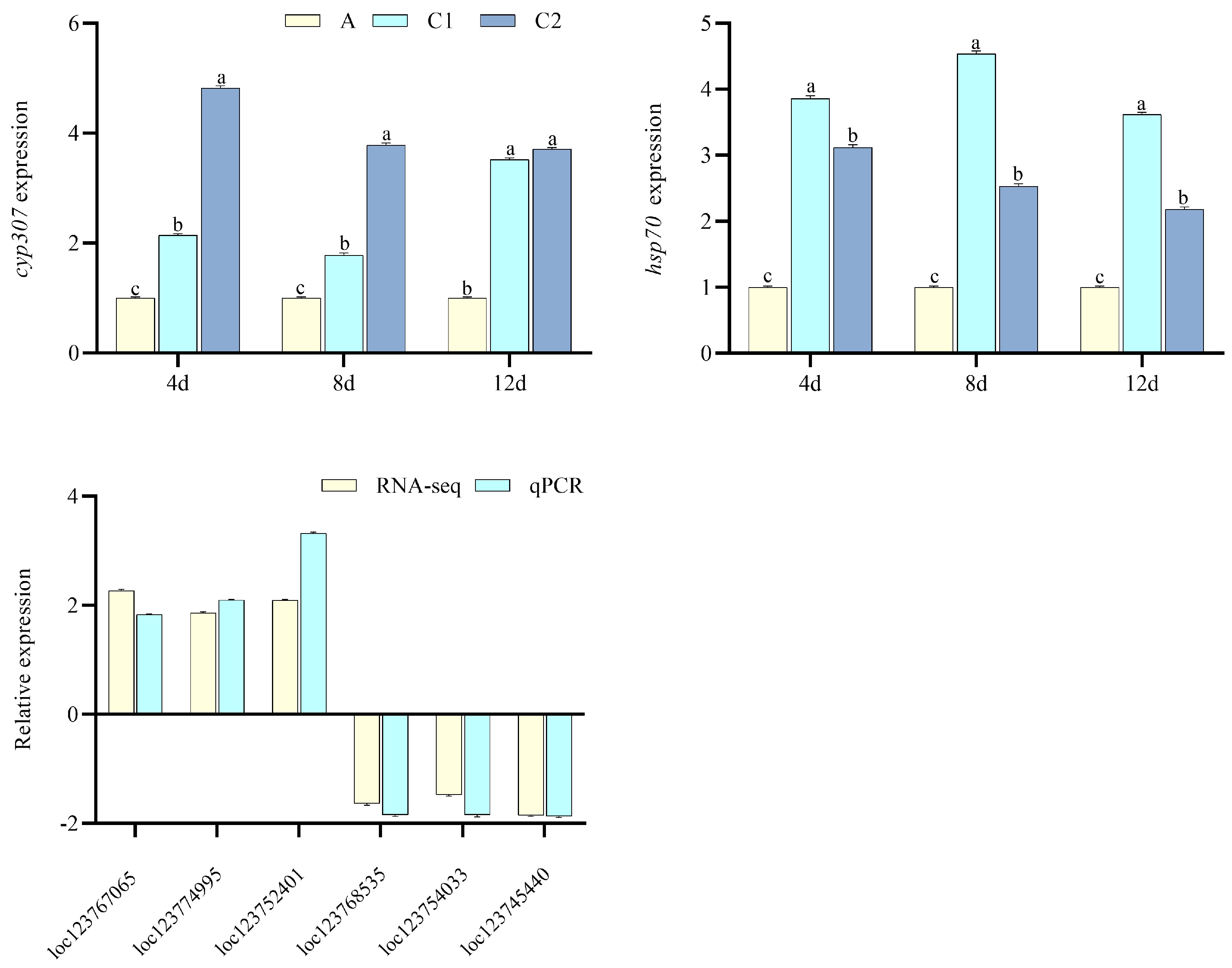
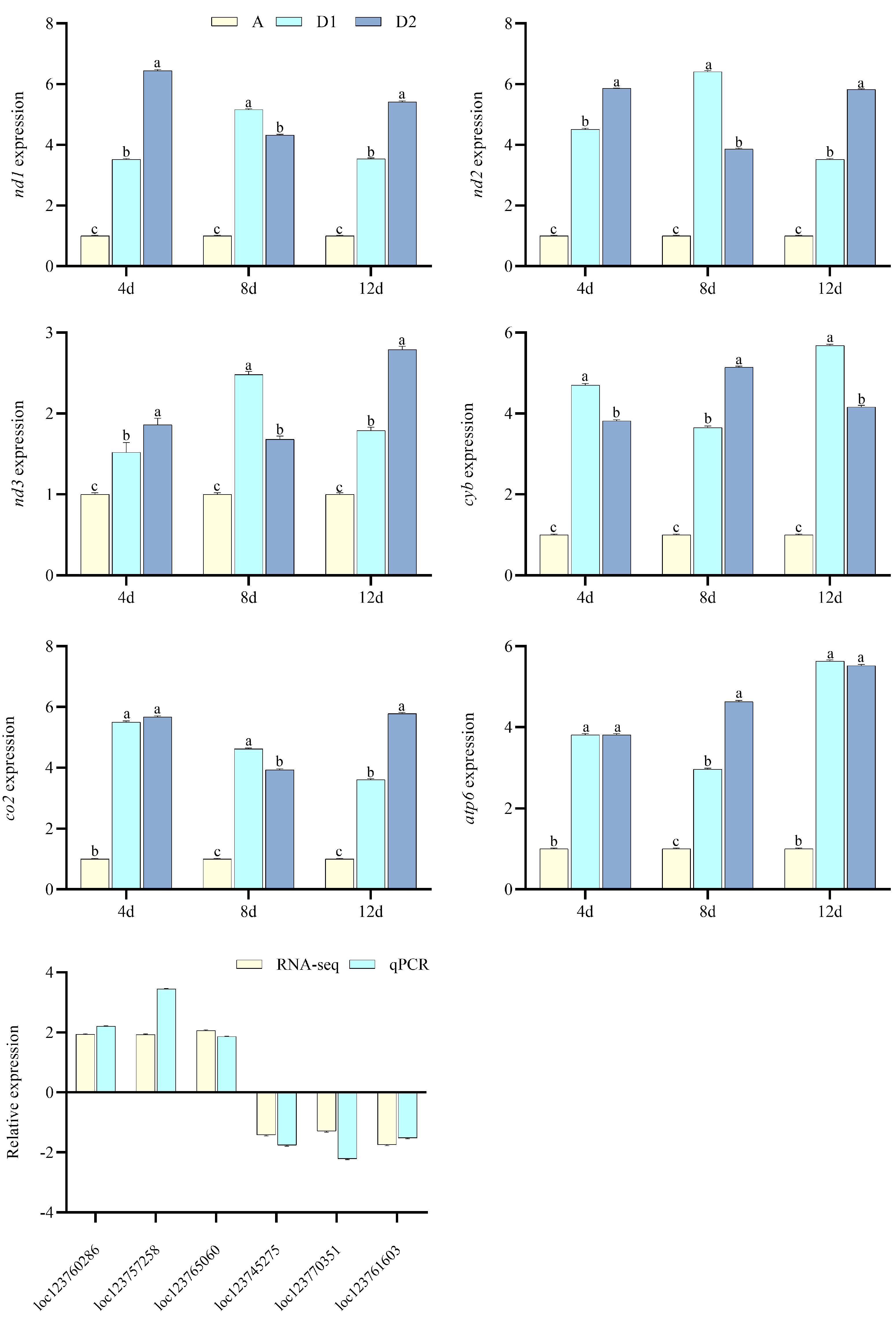
2.3. Transcriptomics and qPCR Verification
2.4. Data Statistical Analysis
3. Results
3.1. Histopathological Changes
3.2. Transcriptomics Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
Appendix A
| Gene Names | Primers | Product (bp) |
|---|---|---|
| β-actin | F: GACAAGTACAGTTGTGCGCC R: TGGCCCATACCAACCATCAC | 194 |
| abcc2 | F: CCACCTTCGCTGTGTTTGTG R: GCACTGGCTCTGTGTCTCTT | 282 |
| abcc4 | F: CAATGAGCCAATCGCTGCTC R: TCGGCTCCATATCGCTTGTC | 216 |
| mt | F: AGGAAGAGGTGAACGCCTTG R: CAGGGTGTCGCTGGATTCTT | 259 |
| loc123762198 | F: GGGAGAAGACCTTGGTGCAA R: GTGCTCTCGTGCCCAATACT | 250 |
| loc123768463 | F: GCTGCTAGACGAGCCATCAT R: GGTCATGGCCATCTTGGGAA | 269 |
| loc123762200 | F: AATCCGGCTGAAACGTCGAT R: GCCGTTTTCCACGTTTTTGTG | 210 |
| loc123762199 | F: GCAAACAGGAACGCGAGATG R: AGTGCTCTCGTGCCCAATAC | 235 |
| loc123764278 | F: ACCAGGCAACAGTCAACACA R: ACACGGCTACAGAATGCCTC | 235 |
| loc123755730 | F: AATCCACTGTGCGCCAACTA R: AGAGAGTCAAAGCCACTCGC | 162 |
| cyp307 | F: TCCCAGGACATCCGATCCTT R: CGTATCTTCCTGCCGACCTC | 251 |
| hsp70 | F: TGGCCATTCGACGTCATCAA R: AGATGGTTCCAGCGTCCTTG | 226 |
| loc123767065 | F: AGAGGAAGGACGAGCCTGAT R: GAGCTTATCGTCGAGGGTGG | 212 |
| loc123774995 | F: AAATTTGGCGGCAGTGTTCC R: TCAGTCGTCTGCACCTCAAC | 167 |
| loc123752401 | F: AGGATTAAACTGGTGGCGGG R: ATCCTCTGTCCCTCTTCGCT | 258 |
| loc123768535 | F: TGTGACTTACCGCCTTCACC R: GCCTCGTGACACTCTCAACA | 224 |
| loc123754033 | F: CTACGCATCATGGCAACGTG R: CAAGAAGGGAACTGCGACCT | 245 |
| loc123745440 | F: GAAGCGGCAGATTGAAGCAG R: CATGCCGTGAACGCGATTAG | 202 |
| nd1 | F: TCGGGTAGGAGACGTAGCAA R: GCAGTCACCAGAGTTGACGA | 237 |
| nd2 | F: TTTTCTGCTTGGTTGCCAGC R: AAATTCGCCCCTAGACCTGC | 194 |
| nd3 | F: ATCGGGTAGGAGACGTAGCA R: AGCAGTCACCAGAGTTGACG | 239 |
| cyb | F: GCTCCTGTGGTAGAGGTTGG R: AGCCCATGAAACCCAGTAGC | 299 |
| co2 | F: TGATTTGGGGCTTGAGTGGG R: TGAAAAGGAGCTGCGCCTAA | 227 |
| atp6 | F: AGGCCGCTGCTTGATATTGA R: GGTAATCGGGCCCTTCCTTT | 295 |
| loc123760286 | F: TGGTGGTAACAGCAGTAGCG R: GGCATGTAAAGGGGTCACCA | 203 |
| loc123757258 | F: TGCGTTCCAAATCTCTGGCT R: ATAAGGTTGACTGGGGCTGC | 191 |
| loc123765060 | F: GGCTGAACGTCAACCTCAGA R: CCAGCCATGACAACAGGGAT | 280 |
| loc123745275 | F: TTCTCGCCCAGCTCAACAAT R: GGCTGCAGATAACGTCCCTT | 262 |
| loc123770351 | F: TGGCAAGACTGCGATTGCTA R: GGAAGAATTCCCCCATGGCA | 254 |
| loc123761603 | F: CCGTCGTCCACACTATCACC R: CTGTGCGGTAAACCCATCCT | 186 |
References
- Wang, Z.; Yang, L.; Zhou, F.; Li, J.; Wu, X.; Zhong, X.; Lv, H.; Yi, S.; Gao, Q.; Yang, Z.; et al. Integrated comparative transcriptome and weighted gene co-expression network analysis provide valuable insights into the response mechanisms of crayfish (Procambarus clarkii) to copper stress. J. Hazard. Mater. 2023, 448, 130820. [Google Scholar] [CrossRef]
- Ma, X.; Zhu, F.; Jin, Q. Antibiotics and chemical disease-control agents reduce innate disease resistance in crayfish. Fish Shellfish Immunol. 2019, 86, 169–178. [Google Scholar] [CrossRef]
- Yang, L.; He, Z.; Li, X.; Jiang, Z.; Xuan, F.; Tang, B.; Bian, X. Behavior and toxicity assessment of copper nanoparticles in aquatic environment, A case study on red swamp crayfish. J. Environ. Manag. 2022, 313, 114986. [Google Scholar] [CrossRef]
- Marcussen, H.; Løjmand, H.; Dalsgaard, A.; Hai, D.M.; Holm, P.E. Copper use and accumulation in catfish culture in the Mekong Delta; Vietnam. J. Environ. Sci. Health Part A Toxic Hazard. Subst. Environ. Eng. 2014, 49, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Ivantsova, E.; Martyniuk, C.J. Environmental presence and toxicological outcomes of the herbicide pendimethalin in teleost fish. Ecotoxicology 2024, 33, 531–545. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hussain, R.; Ghaffar, A.; Afzal, G.; Saad, A.Q.; Ahmad, N.; Nazir, U.; Ahmad, H.I.; Hussain, T.; Khan, A. Clinicohematological; mutagenic; and oxidative stress induced by pendimethalin in freshwater fish bighead carp (Hypophthalmichthys nobilis). Oxid. Med. Cell Longev. 2022, 2022, 2093822. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Lopez, S.; El Ahmadie, N.; Wengrovitz, A.S.; Ganter, J.; Zhao, Y.H.; Souders, C.L., 2nd; Martyniuk, C.J. Assessing sub-lethal effects of the dinitroaniline herbicide pendimethalin in zebrafish embryos/larvae (Danio rerio). Neurotoxicol. Teratol. 2022, 89, 107051. [Google Scholar] [CrossRef]
- Merola, C.; Fabrello, J.; Matozzo, V.; Faggio, C.; Iannetta, A.; Tinelli, A.; Crescenzo, G.; Amorena, M.; Perugini, M. Dinitroaniline herbicide pendimethalin affects development and induces biochemical and histological alterations in zebrafish early-life stages. Sci. Total Environ. 2022, 828, 154414. [Google Scholar] [CrossRef]
- Kim, M.; Cho, M.; Kim, S.H.; Lee, Y.; Jo, M.R.; Moon, Y.S.; Im, M.H. Monitoring and risk assessment of pesticide residues in fishery products using GC-MS/MS in South Korea. Toxics 2024, 12, 299. [Google Scholar] [CrossRef]
- Hong, T.; Park, H.; An, G.; Park, J.; Song, G.; Lim, W. Fluchloralin induces developmental toxicity in heart; liver; and nervous system during early zebrafish embryogenesis. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2023, 271, 109679. [Google Scholar] [CrossRef]
- Lacy, B.; Rivera, M.; Flores, L.; Rahman, M.S. Combined effects of high temperature and pesticide mixture exposure on free-swimming behaviors and hepatic cytochrome P450 1A expression in goldfish; Carassius auratus. J. Toxicol. Environ. Health Part A 2023, 86, 144–165. [Google Scholar] [CrossRef]
- Lacy, B.; Rahman, M.S. Interactive effects of high temperature and pesticide exposure on oxidative status; apoptosis; and renin expression in kidney of goldfish, Molecular and cellular mechanisms of widespread kidney damage and renin attenuation. J. Appl. Toxicol. 2022, 42, 1787–1806. [Google Scholar] [CrossRef]
- Lacy, B.; Rahman, M.S.; Rahman, M.S. Potential mechanisms of Na+/K+-ATPase attenuation by heat and pesticides co-exposure in goldfish, role of cellular apoptosis; oxidative/nitrative stress; and antioxidants in gills. Environ. Sci. Pollut. Res. Int. 2022, 29, 57376–57394. [Google Scholar] [CrossRef]
- Yan, B.; Lei, L.; Chen, X.; Men, J.; Sun, Y.; Guo, Y.; Yang, L.; Wang, Q.; Han, J.; Zhou, B. Glyphosate and glufosinate-ammonium in aquaculture ponds and aquatic products, Occurrence and health risk assessment. Environ. Pollut. 2022, 296, 118742. [Google Scholar] [CrossRef] [PubMed]
- Marçal, R.; Pacheco, M.; Guilherme, S. DNA of crayfish spermatozoa as a target of waterborne pesticides—An ex vivo approach as a tool to short-term spermiotoxicity screening. J. Hazard. Mater. 2020, 400, 123300. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Huang, Q.; Zhou, K.; Luo, X.; Long, W.; Yin, Z.; Huang, Z.; Hong, Y. Effects of glyphosate on neurotoxicity; oxidative stress and immune suppression in red swamp crayfish; Procambarus Clarkii. Aquat. Toxicol. 2024, 275, 107050. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Liu, J.; Sun, Y.; Cheng, Y. Transcriptomics; metabolomics and proteomics analyses reveal glyphosate tolerance mechanism in red swamp crayfish Procambarus clarkii. Sci. Total Environ. 2025, 958, 178068. [Google Scholar] [CrossRef]
- Huang, Y.; Li, T.; Hu, X.; Qi, D.; Li, X.; Huang, Z.; Wu, S.; Hong, Y. Dopaminergic system disruption induced by envrionmentally-realistic glyphosate leads to behavioral alteration in crayfish; Procambarus clarkii. Ecotoxicol. Environ. Saf. 2025, 301, 118509. [Google Scholar] [CrossRef]
- Lusk, M.G.; Chapman, K. Copper concentration data for water, sediments, and vegetation of urban stormwater ponds treated with copper sulfate algaecide. Data Brief 2020, 31, 105982. [Google Scholar] [CrossRef]
- Malhotra, N.; Ger, T.R.; Uapipatanakul, B.; Huang, J.C.; Chen, K.H.; Hsiao, C.D. Review of Copper and Copper Nanoparticle Toxicity in Fish. Nanomaterials 2020, 10, 1126. [Google Scholar] [CrossRef]
- Zheng, Y.; Lu, X.; Hu, J.; Sun, Y.; Zhu, H.; Xu, G. Chlorella alleviates the intestinal damage of tilapia caused by microplastics. Chemosphere 2024, 353, 141644. [Google Scholar] [CrossRef] [PubMed]
- Bernet, D.; Schmidt, H.; Meier, W.; Burkhardt-Holm, P.; Wahli, T. Histopathology in fish: Proposal for a protocol to assess aquatic pollution. J. Fish Dis. 1999, 22, 25–34. [Google Scholar] [CrossRef]
- Chen, L.; Xu, J.; Wan, W.; Xu, Z.; Hu, R.; Zhang, Y.; Zheng, J.; Gu, Z. The microbiome structure of a rice-crayfish integrated breeding model and its association with crayfish growth and water quality. Microbiol. Spectr. 2022, 10, e0220421. [Google Scholar] [CrossRef]
- Hou, J.; Styles, D.; Cao, Y.; Ye, X. The sustainability of rice-crayfish coculture systems, a mini review of evidence from Jianghan plain in China. J. Sci. Food Agric. 2021, 101, 3843–3853. [Google Scholar] [CrossRef]
- Wang, X.; Qin, Y.; Li, X.; Yan, B.; Martyniuk, C.J. Comprehensive interrogation of metabolic and bioenergetic responses of early-staged zebrafish (Danio rerio) to a commercial copper hydroxide nanopesticide. Environ. Sci. Technol. 2021, 55, 13033–13044. [Google Scholar] [CrossRef]
- Ou-Yang, K.; Feng, T.; Han, Y.; Li, J.; Ma, H. Cyhalofop-butyl and pyribenzoxim-induced oxidative stress and transcriptome changes in the muscle of crayfish (Procambarus clarkii). Sci. Total Environ. 2023, 864, 161170. [Google Scholar] [CrossRef] [PubMed]
- Morozov, A.A.; Yurchenko, V.V. Changes in the liver proteome of zebrafish (Danio rerio) exposed to glyphosate and aminomethylphosphonic acid in the presence of a humic substance. Pestic. Biochem. Physiol. 2024, 204, 106036. [Google Scholar] [CrossRef]
- Abu-Zahra, N.I.S.; Gouda, M.; Elseify, M.M.; Abass, M.E.; El-Gohary, M.S.; El-Sokary, E.T. Azolla pinnata mitigates pendimethalin induced immunotoxicity; oxidative stress and histopathological changes in Oreochromis niloticus. Sci. Rep. 2025, 15, 16226. [Google Scholar] [CrossRef]
- Zheng, T.; Jia, R.; Cao, L.; Du, J.; Gu, Z.; He, Q.; Xu, P.; Yin, G. Effects of chronic glyphosate exposure on antioxdative status; metabolism and immune response in tilapia (GIFT; Oreochromis niloticus). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2021, 239, 108878. [Google Scholar] [CrossRef]
- Bolten, J.S.; Pratsinis, A.; Alter, C.L.; Fricker, G.; Huwyler, J. Zebrafish (Danio rerio) larva as an in vivo vertebrate model to study renal function. Am. J. Physiol. Ren. Physiol. 2022, 322, F280–F294. [Google Scholar] [CrossRef]
- Xiong, G.; Hu, H.; Zhang, H.; Zhang, J.; Cao, Z.; Lu, H.; Liao, X. Cyhalofop-butyl exposure induces the severe hepatotoxicity and immunotoxicity in zebrafish embryos. Fish Shellfish Immunol. 2023, 134, 108644. [Google Scholar] [CrossRef]
- Xiong, G.; Deng, Y.; Li, J.; Cao, Z.; Liao, X.; Liu, Y.; Lu, H. Immunotoxicity and transcriptome analysis of zebrafish embryos in response to glufosinate-ammonium exposure. Chemosphere 2019, 236, 124423. [Google Scholar] [CrossRef]
- Navruz, F.Z.; Acar, Ü.; Yılmaz, S.; Kesbiç, O.S. Dose-dependent stress response of esfenvalerate insecticide on common carp (Cyprinus carpio), Evaluating blood parameters and gene expression. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2023, 272, 109711. [Google Scholar] [CrossRef]
- Ma, J.; Liu, Y.; Guo, Y.; Ma, Q.; Ji, C.; Zhao, L. Transcriptional profiling of Aflatoxin B1-induced oxidative stress and inflammatory response in macrophages. Toxins 2021, 13, 401. [Google Scholar] [CrossRef]
- Wang, J.Y.; Zhang, L.H.; Hong, Y.H.; Cai, L.N.; Storey, K.B.; Zhang, J.Y.; Zhang, S.S.; Yu, D.N. How does mitochondrial protein-coding gene expression in Fejervarya kawamurai (Anura, Dicroglossidae) respond to extreme temperatures? Animals 2023, 13, 3015. [Google Scholar] [CrossRef]
- Lavarías, S.M.L.; Colpo, K.D.; Landro, S.M.; Ambrosio, E.S.; Rodrigues Capítulo, A.; Arrighetti, F. Deleterious effects of two pesticide formulations with different toxicological mechanisms in the hepatopancreas of a freshwater prawn. Chemosphere 2022, 286 Pt 3, 131920. [Google Scholar] [CrossRef]
- Yang, Q.; Ai, X.; Li, S.; Liu, H.; Liu, Y. Determination of pendimethalin in water; sediment; and Procambarus clarkii by high performance liquid chromatography-triple quadrupole mass spectrometry. Environ. Monit. Assess. 2019, 191, 621. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Ai, X.; Dong, J.; Yang, Y.; Zhou, S.; Liu, Y.; Xu, N. Elimination of pendimethalin in integrated rice and Procambarus clarkii breeding models and dietary risk assessments. Foods 2022, 11, 1300. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Cao, L.; Du, J.; Gao, J.; Zhang, Y.; Sun, Y.; Li, Q.; Nie, Z.; Xu, G. Effects of prometryn exposure on hepatopancreas oxidative stress and intestinal flora in Eriocheir sinensis (Crustacea, Decapoda). Antioxidants 2023, 12, 1548. [Google Scholar] [CrossRef]
- Huang, P.; Gao, J.; Du, J.; Nie, Z.; Li, Q.; Sun, Y.; Xu, G.; Cao, L. Prometryn exposure disrupts the intestinal health of Eriocheir sinensis, physiological responses and underlying mechanism. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2024, 277, 109820. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bao, Y.; Zhang, X.; Zhao, S.; Qiu, J.; Li, N.; He, J. Anaerobic biodegradation and detoxification of chloroacetamide herbicides by a novel Proteiniclasticum sediminis BAD-10T. Environ. Res. 2022, 209, 112859. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Fateh, B.; Xu, G. Effects of methomyl on the intestinal microbiome and hepatic transcriptome of tilapia; and the modifying effects of mint co-culture. Aquat. Toxicol. 2023, 263, 106675. [Google Scholar] [CrossRef]
- Liu, S.; Li, X.; Qin, S.; Zhang, H.; Zhang, T.; Zhu, J.; Lin, L.; Lian, L.; Xie, F.; Tan, H.; et al. Comprehensive study of flusulfinam in paddy water-sediment microcosms, enantioselective fate; degradation pathways; and toxicity assessment. J. Hazard. Mater. 2025, 488, 137342. [Google Scholar] [CrossRef]
- Cazenave, J.; Bacchetta, C.; Repetti, M.R.; Rossi, A. Biomarker responses in fish caged in a rice field during a bifenthrin application. Environ. Res. 2024, 263 Pt 3, 120240. [Google Scholar] [CrossRef]
- Di, S.; Diao, Z.; Cang, T.; Wang, Z.; Xu, L.; Qi, P.; Zhao, H.; Liu, Z.; Wang, X. Enantioselective fate and risk assessment of chiral fungicide pydiflumetofen in rice-fish and wheat farming systems. Sci. Total Environ. 2024, 912, 169262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Y.; Shao, Y.; Wang, J.; Chen, Z.; Roß-Nickoll, M.; Schäffer, A. Conversion of rice field ecosystems from conventional to ecological farming: Effects on pesticide fate, ecotoxicity and soil properties. Environ. Manag. 2025, 75, 930–944. [Google Scholar] [CrossRef] [PubMed]
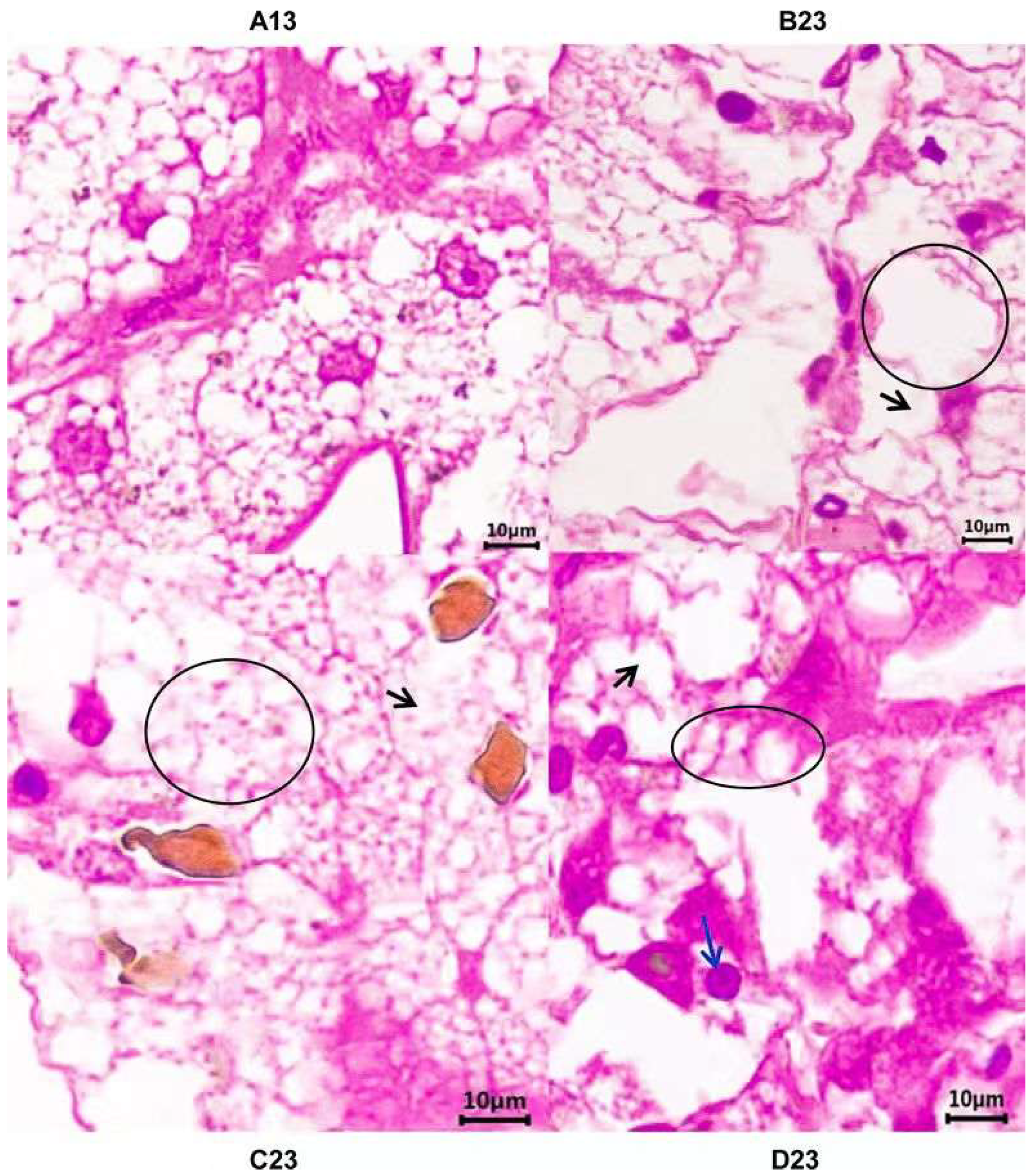
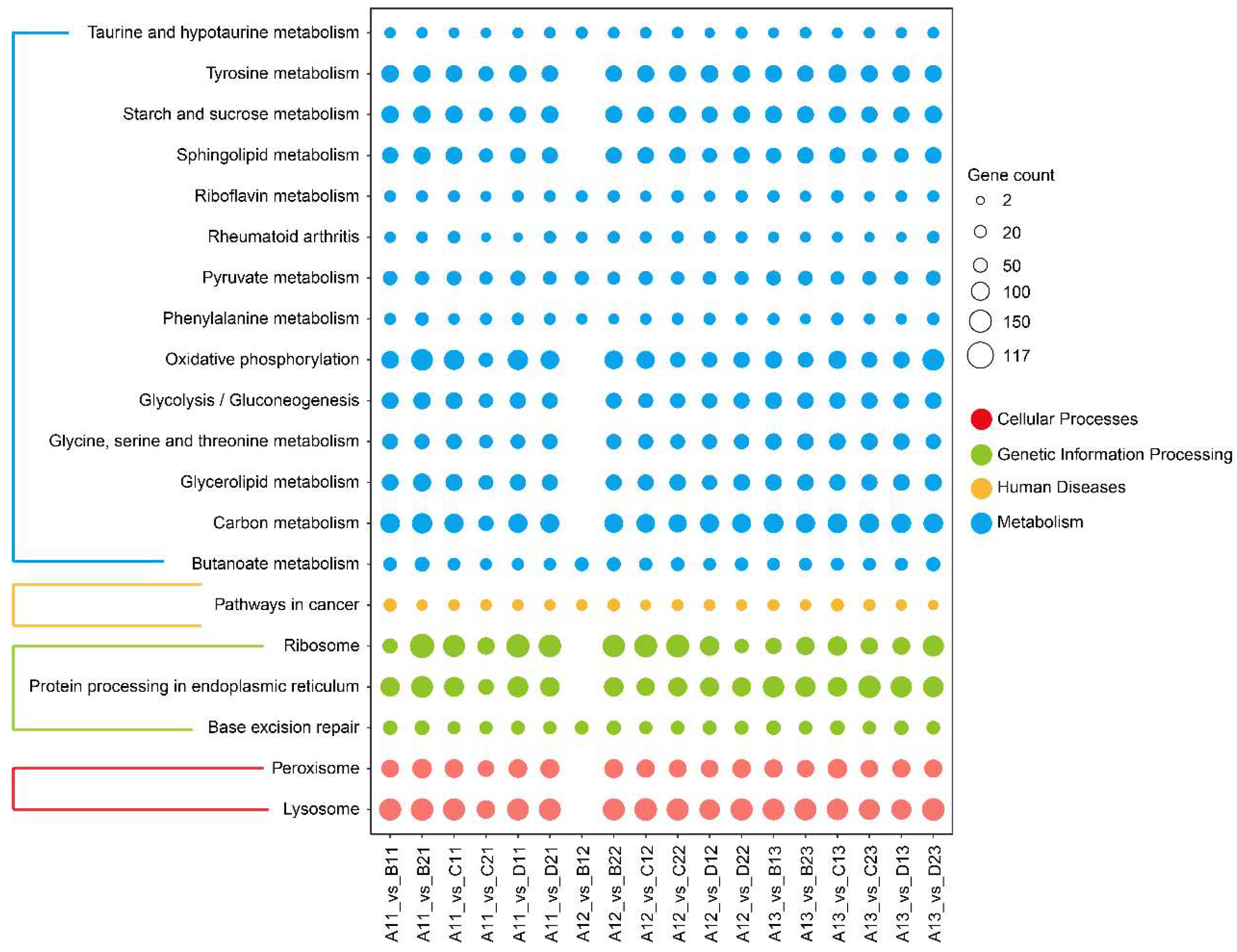

| Comparisons | Groups | DEGs—Total | DEGs—Up | DEGs—Down |
|---|---|---|---|---|
| CuSO4 vs. control | A11_vs_B11 | 2664 | 1153 | 1511 |
| A12_vs_B12 | 2464 | 1161 | 1303 | |
| A13_vs_B13 | 2512 | 1422 | 1090 | |
| A11_vs_B21 | 2585 | 1579 | 1006 | |
| A12_vs_B22 | 2561 | 1268 | 1293 | |
| A13_vs_B23 | 2372 | 1540 | 832 | |
| PND vs. control | A11_vs_C11 | 2105 | 1233 | 872 |
| A12_vs_C12 | 2583 | 1257 | 1326 | |
| A13_vs_C13 | 2711 | 1443 | 1268 | |
| A11_vs_C21 | 1837 | 1086 | 751 | |
| A12_vs_C22 | 2562 | 1237 | 1325 | |
| A13_vs_C23 | 2484 | 1426 | 1058 | |
| glyphosate vs. control | A11_vs_D11 | 2362 | 956 | 1406 |
| A12_vs_D12 | 2194 | 945 | 1249 | |
| A13_vs_D13 | 2527 | 1424 | 1103 | |
| A11_vs_D21 | 2011 | 926 | 1085 | |
| A12_vs_D22 | 2538 | 1134 | 1404 | |
| A13_vs_D23 | 2387 | 1589 | 798 |
| Inputs | Other References | Crayfish in This Study |
|---|---|---|
| CuSO4 | [3], red swamp crayfish, antioxidative enzymes decreased after exposure to copper nanoparticles for 48 h but without histological changes | irregular structure and vacuoles, pathways of ABC transporters, peroxisome, and endocytosis enriched |
| [25], 100 μg·L−1 copper hydroxide nano pesticide in zebrafish, ABC transporters pathway enriched | ||
| PND | [6], congestion, necrosis of hepatocytes, and atrophy of bighead carp hepatocytes under 0.75 mg·L−1 PND exposure | irregular structure and vacuoles; pathways of drug metabolism-cytochrome P450, cysteine and methionine metabolism, citrate cycle, fluid shear stress, and atherosclerosis enriched |
| [8], delayed and reduced ossification of the vertebral centra, increased AchE in zebrafish by 0.5 mg·L−1 PND exposure | ||
| [11,12,13], co-exposure with high temperature, natural swimming patterns affected, apoptotic cells in the kidney and gill of goldfish found | ||
| [28], oxidative damage found in tilapia under 0.5 and 1 mg·L−1 PND exposure | ||
| glyphosate | [15], genotoxic potential in spermatozoa of crayfish by 90 μg·L−1 glyphosate exposure | narrowed hepatic sinuses, narrowed bile canaliculus, hsp70 increased, pathways of pyruvate metabolism, oxidative phosphorylation, other glycan degradation, fluid shear stress and atherosclerosis, and ribosome enriched |
| [16], neurotoxic and immunotoxic effects after 5~20 mg·L−1 glyphosate exposure in crayfish for 96 h | ||
| [17], antioxidant response, ammonia-nitrogen regulation, and energy supply of the organism enhanced in crayfish after 1.2~10.8 mg·L−1 glyphosate for 72 h | ||
| [18], 0.1~10 µg·L−1 glyphosate alerted neurotoxic and oxidative impacts in crayfish for 14 d | ||
| [27], 100 μg·L−1 in zebrafish, oxidative phosphorylation pathway enriched | ||
| [29], hsp70 increased in tilapia under 0.2~16 mg·L−1 glyphosate exposure for 28 d |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Li, J.; Liu, Z.; Wang, N.; Xu, G. Transcriptome Analyses of Procambarus clarkii (Girard, 1852) Under Individual Exposures to CuSO4, Pendimethalin, and Glyphosate. Toxics 2025, 13, 765. https://doi.org/10.3390/toxics13090765
Zheng Y, Li J, Liu Z, Wang N, Xu G. Transcriptome Analyses of Procambarus clarkii (Girard, 1852) Under Individual Exposures to CuSO4, Pendimethalin, and Glyphosate. Toxics. 2025; 13(9):765. https://doi.org/10.3390/toxics13090765
Chicago/Turabian StyleZheng, Yao, Jiajia Li, Zhuping Liu, Ning Wang, and Gangchun Xu. 2025. "Transcriptome Analyses of Procambarus clarkii (Girard, 1852) Under Individual Exposures to CuSO4, Pendimethalin, and Glyphosate" Toxics 13, no. 9: 765. https://doi.org/10.3390/toxics13090765
APA StyleZheng, Y., Li, J., Liu, Z., Wang, N., & Xu, G. (2025). Transcriptome Analyses of Procambarus clarkii (Girard, 1852) Under Individual Exposures to CuSO4, Pendimethalin, and Glyphosate. Toxics, 13(9), 765. https://doi.org/10.3390/toxics13090765







