A Molecular Tool for Rapid Detection and Traceability of Cyclospora cayetanensis in Fresh Berries and Berry Farm Soils
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extractions
2.2. Nested PCR Assay
2.3. Sensitivity of the Nested PCR Assay
2.4. Detection of C. cayetanensis in Fresh Berries and Farm Soils
2.5. Molecular Traceability of C. cayetanensis
3. Results and Discussion
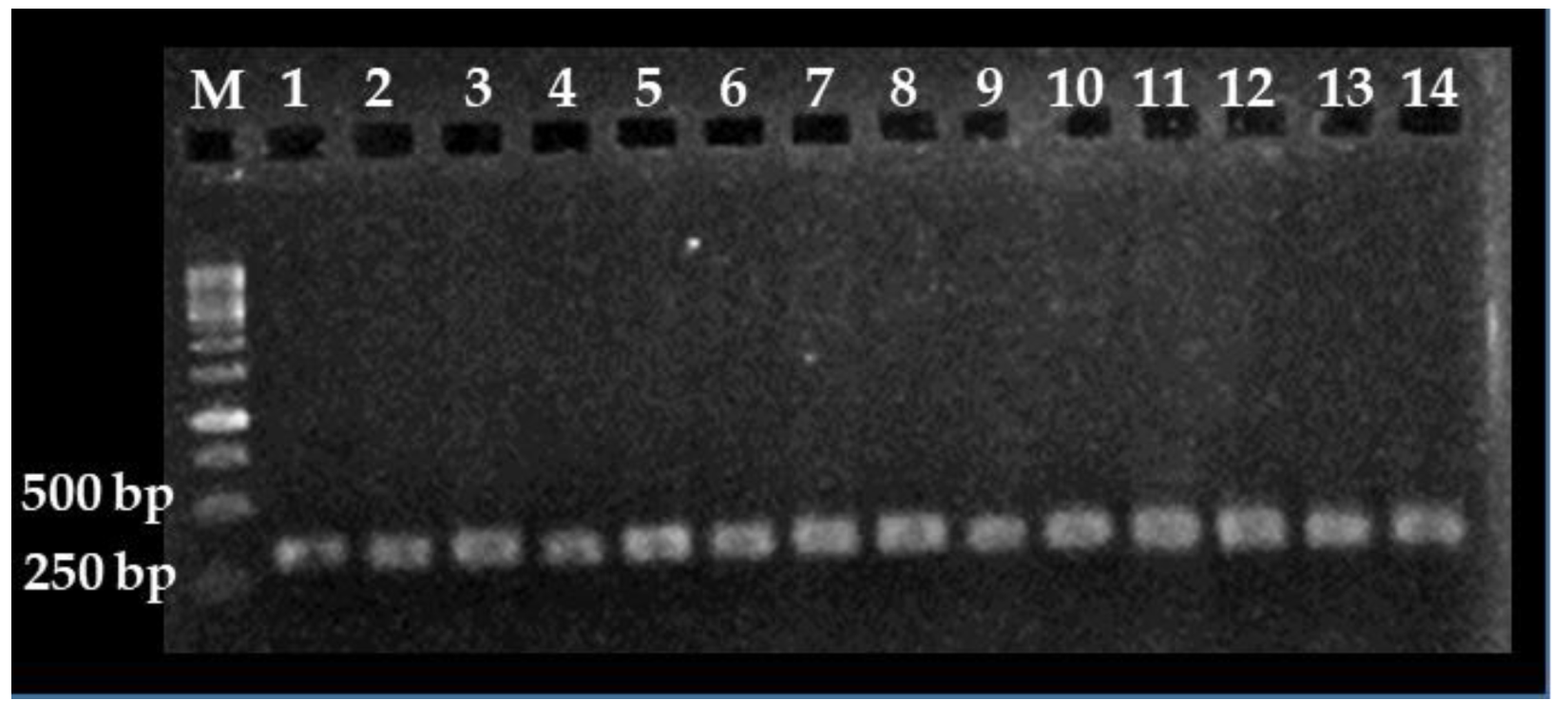
3.1. Nested PCR Standardization
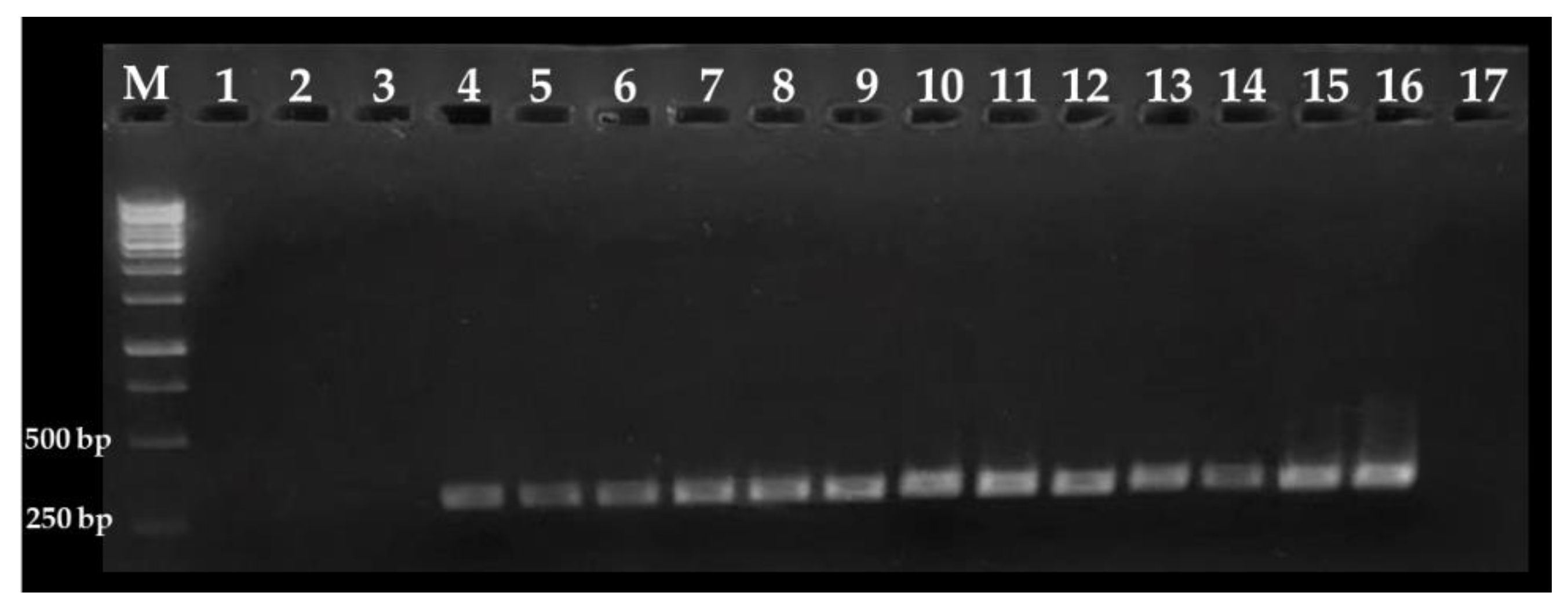
3.2. Sensitivity of the Nested PCR Assay
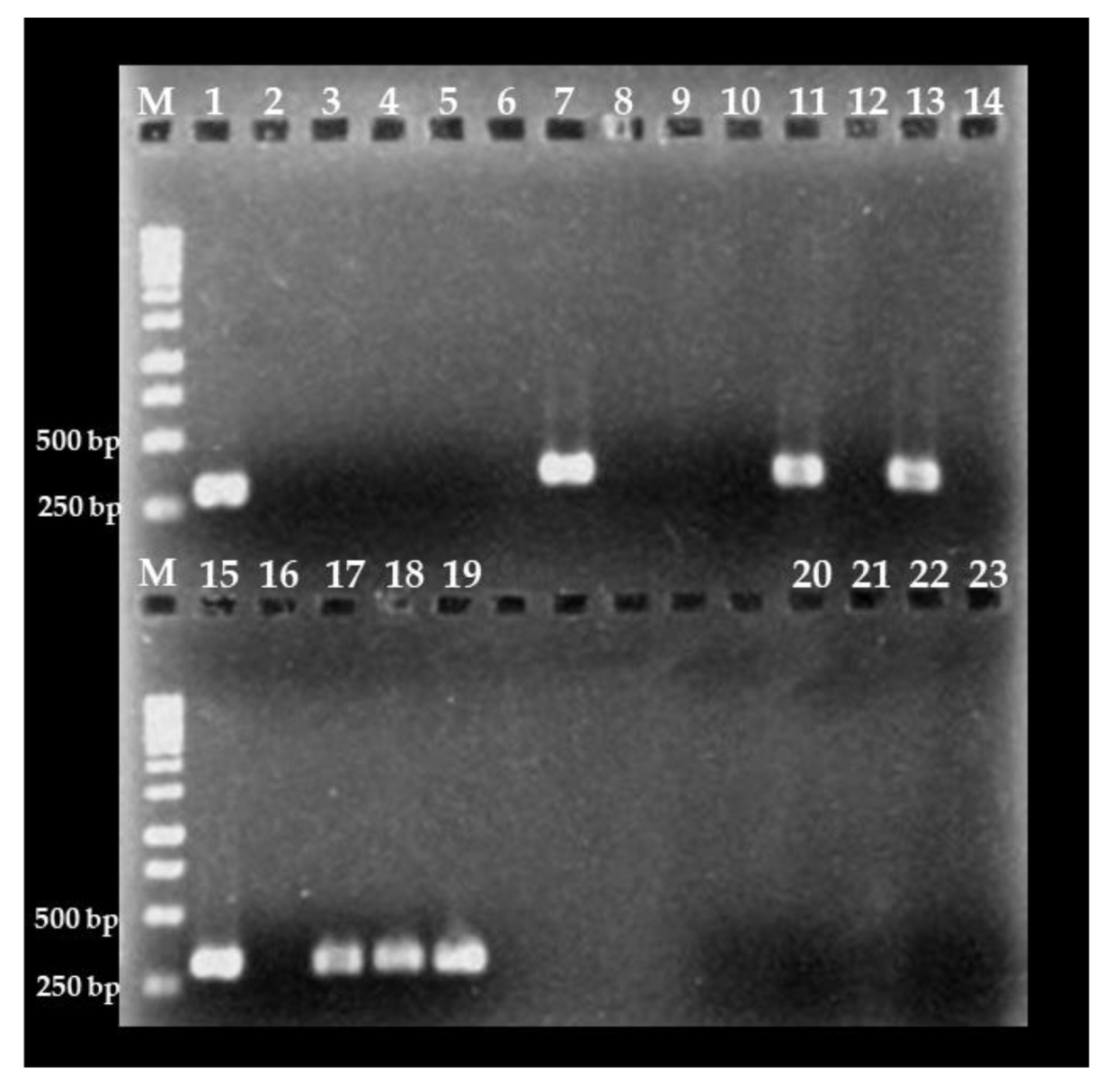
3.3. Detection of C. cayetanensis in Berries and Farm Soils
3.4. Molecular Traceability of C. cayetanensis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tefera, T.; Tysnes, K.R.; Utaaker, K.S.; Robertson, L.J. Parasite contamination of berries: Risk, occurrence, and approaches for mitigation. Food Waterborne Parasitol. 2018, 10, 23–38. [Google Scholar] [CrossRef]
- Skrovankova, S.; Sumczynski, D.; Mlcek, J.; Jurikova, T.; Sochor, J. Bioactive compounds and antioxidant activity in different types of berries. Int. J. Mol. Sci. 2015, 16, 24673–24706. [Google Scholar] [CrossRef]
- Herwaldt, B.L.; Ackers, M.L. An outbreak in 1996 of cyclosporiasis associated with imported raspberries. The Cyclospora Working Group. N. Engl. J. Med. 1997, 336, 1548–1556. [Google Scholar] [CrossRef] [PubMed]
- Nichols, G.L.; Freedman, J.; Pollock, K.G.; Rumble, C.; Chalmers, R.M.; Chiodini, P.; Hawkins, G.; Alexander, C.L.; Godbole, G.; Williams, C.; et al. Cyclospora infection linked to travel to Mexico, June to September 2015. Euro. Surveill. 2015, 20, 30048. [Google Scholar] [CrossRef] [PubMed]
- Chacín-Bonilla, L. Transmission of Cyclospora cayetanensis infection: A review focusing on soil-borne cyclosporiasis. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 215–216. [Google Scholar] [CrossRef]
- Bern, C.; Hernandez, B.; Lopez, M.B.; Arrowood, M.J.; Alvarez de Mejia, M.; Maria de Merida, A.; Hightower, A.W.; Venczel, L.; Herwaldt, B.L.; Klein, R.E. Epidemiologic studies of Cyclospora cayetanensis in Guatemala. Emerg. Infect. Dis. 1999, 5, 766–774. [Google Scholar] [CrossRef]
- Koumans, E.H.; Katz, D.J.; Malecki, J.M.; Kumar, S.; Wahlquist, S.P.; Arrowood, M.J.; Hightower, A.W.; Herwaldt, B.L. An outbreak of cyclosporiasis in Florida in 1995: A harbinger of multistate outbreaks in 1996 and 1997. Am. J. Trop. Med. Hyg. 1998, 59, 235–242. [Google Scholar] [CrossRef]
- Ortega, Y.R.; Sanchez, R. Update on Cyclospora cayetanensis, a food-borne and waterborne parasite. Clin. Microbiol. Rev. 2010, 23, 218–234. [Google Scholar] [CrossRef]
- Mundaca, C.C.; Torres-Slimming, P.A.; Araujo-Castillo, R.V.; Morán, M.; Bacon, D.J.; Ortega, Y.; Gilman, R.H.; Blazes, D.L. Use of PCR to improve diagnostic yield in an outbreak of cyclosporiasis in Lima, Peru. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 712–717. [Google Scholar] [CrossRef]
- Relman, D.A.; Schmidt, T.M.; Gajadhar, A.; Sogin, M.; Cross, J.; Yoder, K.; Sethabutr, O.; Echeverria, P. Molecular phylogenetic analysis of Cyclospora, the human intestinal pathogen, suggests that it is closely related to Eimeria species. J. Infect. Dis. 1996, 173, 440–445. [Google Scholar]
- Giangaspero, A.; Marangi, M.; Arace, E. Cyclospora cayetanensis travels in tap water on Italian trains. J. Water Health 2015, 13, 210–216. [Google Scholar] [CrossRef]
- Shapiro, K.; Kim, M.; Rajal, V.B.; Arrowood, M.J.; Packham, A.; Aguilar, B.; Wuertz, S. Simultaneous detection of four protozoan parasites on leafy greens using a novel multiplex PCR assay. Food Microbiol. 2019, 84, 103252. [Google Scholar] [CrossRef]
- Caradonna, T.; Marangi, M.; Del Chierico, F.; Ferrari, N.; Reddel, S.; Bracaglia, G.; Normanno, G.; Putignani, L.; Giangaspero, A. Detection and prevalence of protozoan parasites in ready-to-eat packaged salads on sale in Italy. Food Microbiol. 2017, 67, 67–75. [Google Scholar] [CrossRef]
- Shields, J.M.; Olson, B.H. PCR-restriction fragment length polymorphism method for detection of Cyclospora cayetanensis in environmental waters without microscopic confirmation. Appl. Environ. Microbiol. 2003, 69, 4662–4669. [Google Scholar] [CrossRef][Green Version]
- Orlandi, P.A.; Carter, L.; Brinker, A.M.; da Silva, A.J.; Chu, D.-M.; Lampel, K.A.; Monday, S.R. Targeting single-nucleotide polymorphisms in the 18S rRNA gene to differentiate Cyclospora species from Eimeria species by multiplex PCR. Appl. Environ. Microbiol. 2003, 69, 4806–4813. [Google Scholar] [CrossRef]
- Chu, D.-M.T.; Sherchand, J.B.; Cross, J.H.; Orlandi, P.A. Detection of Cyclospora cayetanensis in animal fecal isolates from Nepal using an FTA filter-base polymerase chain reaction method. Am. J. Trop. Med. Hyg. 2004, 71, 373–379. [Google Scholar] [CrossRef]
- Lalonde, L.F.; Gajadhar, A.A. Optimization and validation of methods for isolation and real-time PCR identification of protozoan oocysts on leafy green vegetables and berry fruits. Food Waterborne Parasitol. 2016, 2, 1–7. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Cavalli-Sforza, L.L.; Edwards, A.W.F. Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Genet. 1967, 19, 233–257. [Google Scholar] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. Mamm. Protein Metab. 1969, 3, 132. [Google Scholar]
- Eberhard, M.L.; da Silva, A.J.; Lilley, B.G.; Pieniazek, N.J. Morphologic and molecular characterization of new Cyclospora species from Ethiopian monkeys: C. cercopitheci sp.n., C. colobi sp.n., and C. papionis sp.n. Emerg. Infect. Dis. 1999, 5, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lv, B.; Wang, Q.; Wang, R.; Jian, F.; Zhang, L.; Ning, C.; Fu, K.; Wang, Y.; Qi, M.; et al. Prevalence and molecular characterization of Cyclospora cayetanensis, Henan, China. Emerg. Infect. Dis. 2011, 17, 1887–1890. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-R.; Sohn, W.-M. A case of human cyclosporiasis causing traveler’s diarrhea after visiting Indonesia. J. Korean Med. Sci. 2003, 18, 738–741. [Google Scholar] [CrossRef]
- Sulaiman, I.M.; Ortega, Y.; Simpson, S.; Kerdahi, K. Genetic characterization of human-pathogenic Cyclospora cayetanensis parasites from three endemic regions at the 18S ribosomal RNA locus. Infect. Genet. Evol. 2014, 22, 229–234. [Google Scholar] [CrossRef]
- Bednarska, M.; Bajer, A.; Welc-Falęciak, R.; Pawełas, A. Cyclospora cayetanensis infection in transplant traveller: A case report of outbreak. Parasit. Vectors. 2015, 8, 411. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chang, Y.; Shi, K.E.; Wang, R.; Fu, K.; Li, S.; Xu, J.; Jia, L.; Guo, Z.; Zhang, L. Multilocus sequence typing and clonal population genetic structure of Cyclospora cayetanensis in humans. Parasitology 2017, 144, 1890–1897. [Google Scholar] [CrossRef]
- Habets, M.N.; Cremers, A.J.H.; Bos, M.P.; Savelkoul, P.; Eleveld, M.J.; Meis, J.F.; Hermans, P.W.M.; Melchers, W.J.; de Jonge, M.I.; Diavatopoulos, D.A. A novel quantitative PCR assay for the detection of Streptococcus pneumoniae using the competence regulator gene target comX. J. Med. Microbiol. 2016, 65, 129–136. [Google Scholar] [CrossRef]
- Murphy, H.R.; Cinar, H.N.; Gopinath, G.; Noe, K.E.; Chatman, L.D.; Miranda, N.E.; Wetherington, J.H.; Neal-McKinney, J.; Pires, G.S.; Sachs, E.; et al. Interlaboratory validation of an improved method for detection of Cyclospora cayetanensis in produce using a real-time PCR assay. Food Microbiol. 2018, 69, 170–178. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors-occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Fryauff, D.J.; Krippner, R.; Prodjodipuro, P.; Ewald, C.; Kawengian, S.; Pegelow, K.; Yun, T.; von Heydwolff-Wehnert, C.; Oyofo, B.; Gross, R. Cyclospora cayetanensis among expatriate and indigenous populations of West Java, Indonesia. Emerg. Infect. Dis. 1999, 5, 585–588. [Google Scholar] [CrossRef]
- Karanja, R.M.; Gatei, W.; Wamae, N. Cyclosporiasis: An emerging public health concern around the world and in Africa. Afr. Health. Sci. 2007, 7, 62–67. [Google Scholar]
- Dixon, B.R.; Bussey, J.M.; Parrington, L.J.; Parenteau, M. Detection of Cyclospora cayetanensis oocysts in human fecal specimens by flow cytometry. J. Clin. Microbiol. 2005, 43, 2375–2379. [Google Scholar] [CrossRef]
- Jinneman, K.C.; Wetherington, J.H.; Hill, W.E.; Adams, A.M.; Johnson, J.M.; Tenge, B.J.; Dang, N.L.; Manger, R.L.; Wekell, M.M. Template preparation for PCR and RFLP of amplification products for the detection and identification of Cyclospora sp. and Eimeria spp. oocysts directly from raspberries. J. Food Prot. 1998, 61, 1497–1503. [Google Scholar] [CrossRef]
- Steele, M.; Unger, S.; Odumeru, J. Sensitivity of PCR detection of Cyclospora cayetanensis in raspberries, basil, and mesclun lettuce. J. Microbiol. Methods 2003, 54, 277–280. [Google Scholar] [CrossRef]
- Sim, S.; Won, J.; Kim, J.-W.; Kim, K.; Park, W.-Y.; Yu, J.-R. Simultaneous molecular detection of Cryptosporidium and Cyclospora from raw vegetables in Korea. Korean J. Parasitol. 2017, 55, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Allard, M.W.; Strain, E.; Melka, D.; Bunning, K.; Musser, S.M.; Brown, E.W.; Timme, R. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J. Clin. Microbiol. 2016, 54, 1975–1983. [Google Scholar] [CrossRef]
- Chavatte, J.-M.; Roland, J. Incidental detection of Cyclospora cayetanensis during general health screening: A case study from Singapore. J. Trop. Dis. Public Health 2016, 4, 1–5. [Google Scholar] [CrossRef]
- Barratt, J.L.N.; Park, S.; Nascimento, F.S.; Hofstetter, J.; Plucinski, M.; Casillas, S.; Bradbury, R.S.; Arrowood, M.J.; Qvarnstrom, Y.; Talundzic, E. Genotyping genetically heterogeneous Cyclospora cayetanensis infections to complement epidemiological case linkage. Parasitology 2019, 146, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, Y.; Wang, X.; Zhang, L.; Ortega, Y.; Feng, Y. Mitochondrial genome sequence variation as a useful marker for assessing genetic heterogeneity among Cyclospora cayetanensis isolates and source-tracking. Parasit. Vectors 2019, 12, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hofstetter, J.N.; Nascimento, F.S.; Park, S.; Casillas, S.; Herwaldt, B.L.; Arrowood, M.J.; Qvarnstrom, Y. Evaluation of Multilocus Sequence Typing of Cyclospora cayetanensis based on microsatellite markers. Parasite 2019, 26, 3. [Google Scholar] [CrossRef] [PubMed]
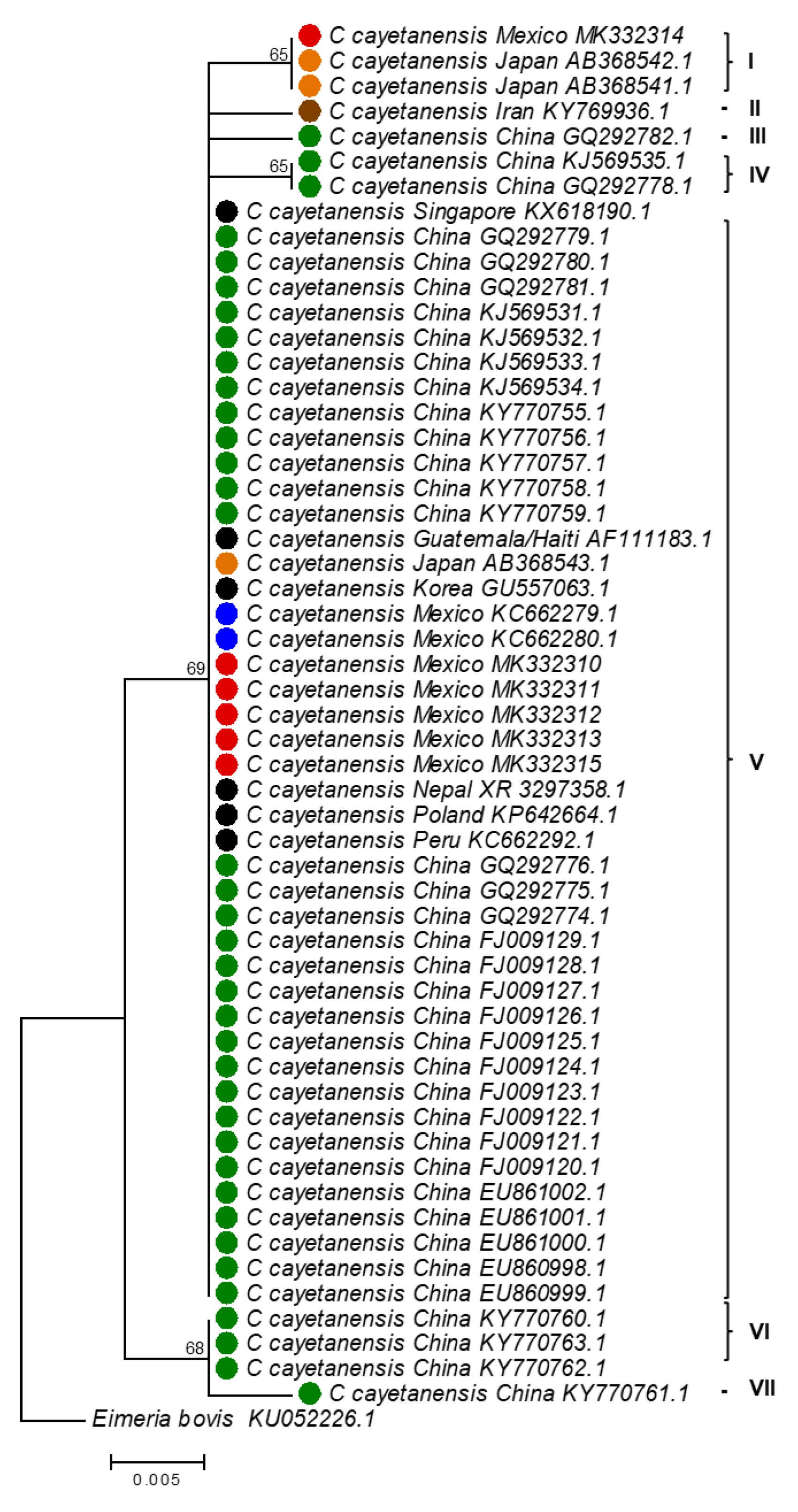

| Accession Numbers | Origin | Reference |
|---|---|---|
| AB368541–AB368543 | Japan | Unpublished |
| AF111183 | Guatemala-Haiti | [24] |
| EU860998–EU861002 | China | [25] |
| FJ009120–FJ009129 | China | [25] |
| GQ292774–GQ292776 | China | [25] |
| GQ292778–GQ292782 | China | [25] |
| GU557063 | Korea | [26] |
| KC662279–KC662280 | Mexico | [27] |
| KC662292 | Peru | [27] |
| KJ569531–KJ569535 | China | Unpublished |
| KP642664 | Poland | [28] |
| KY769936 | Iran | Unpublished |
| KX618190 | Singapore | [26] |
| KY770755–KY770763 | China | [29] |
| XR_003297358 | Nepal | Unpublished |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Resendiz-Nava, C.N.; Orozco-Mosqueda, G.E.; Mercado-Silva, E.M.; Flores-Robles, S.; Silva-Rojas, H.V.; Nava, G.M. A Molecular Tool for Rapid Detection and Traceability of Cyclospora cayetanensis in Fresh Berries and Berry Farm Soils. Foods 2020, 9, 261. https://doi.org/10.3390/foods9030261
Resendiz-Nava CN, Orozco-Mosqueda GE, Mercado-Silva EM, Flores-Robles S, Silva-Rojas HV, Nava GM. A Molecular Tool for Rapid Detection and Traceability of Cyclospora cayetanensis in Fresh Berries and Berry Farm Soils. Foods. 2020; 9(3):261. https://doi.org/10.3390/foods9030261
Chicago/Turabian StyleResendiz-Nava, Carolina N., Guadalupe E. Orozco-Mosqueda, Edmundo M. Mercado-Silva, Susana Flores-Robles, Hilda V. Silva-Rojas, and Gerardo M. Nava. 2020. "A Molecular Tool for Rapid Detection and Traceability of Cyclospora cayetanensis in Fresh Berries and Berry Farm Soils" Foods 9, no. 3: 261. https://doi.org/10.3390/foods9030261
APA StyleResendiz-Nava, C. N., Orozco-Mosqueda, G. E., Mercado-Silva, E. M., Flores-Robles, S., Silva-Rojas, H. V., & Nava, G. M. (2020). A Molecular Tool for Rapid Detection and Traceability of Cyclospora cayetanensis in Fresh Berries and Berry Farm Soils. Foods, 9(3), 261. https://doi.org/10.3390/foods9030261





