Evaluation of Malolactic Bacteria Associated with Wines from Albariño Variety as Potential Starters: Screening for Quality and Safety
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Lactic Acid Bacteria (LAB) Isolation
2.2. Reference Strains
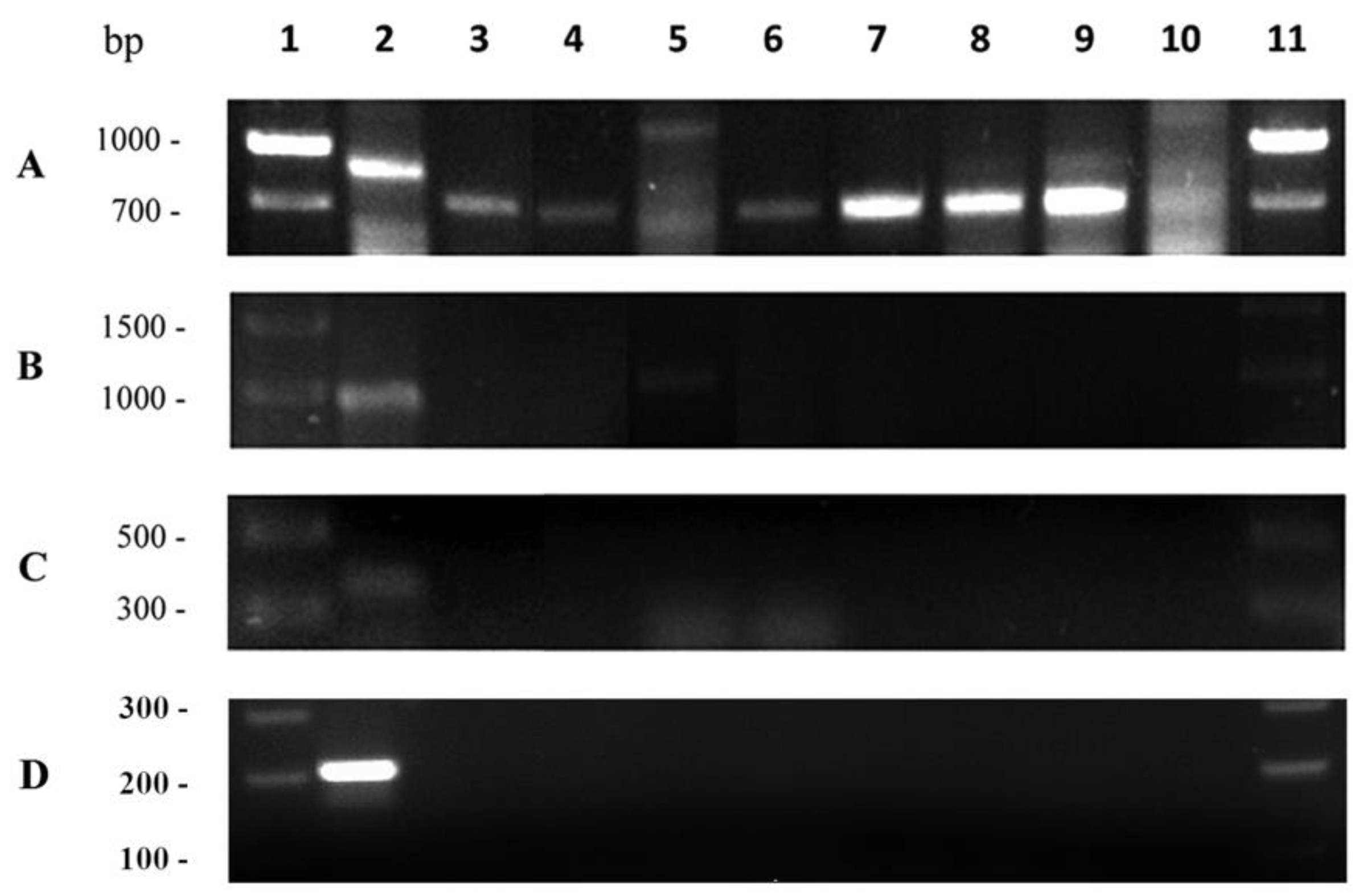
2.3. Identification of LAB Isolates
2.4. Characterization of LAB Strains
2.4.1. Microvinifications and Wine Analysis
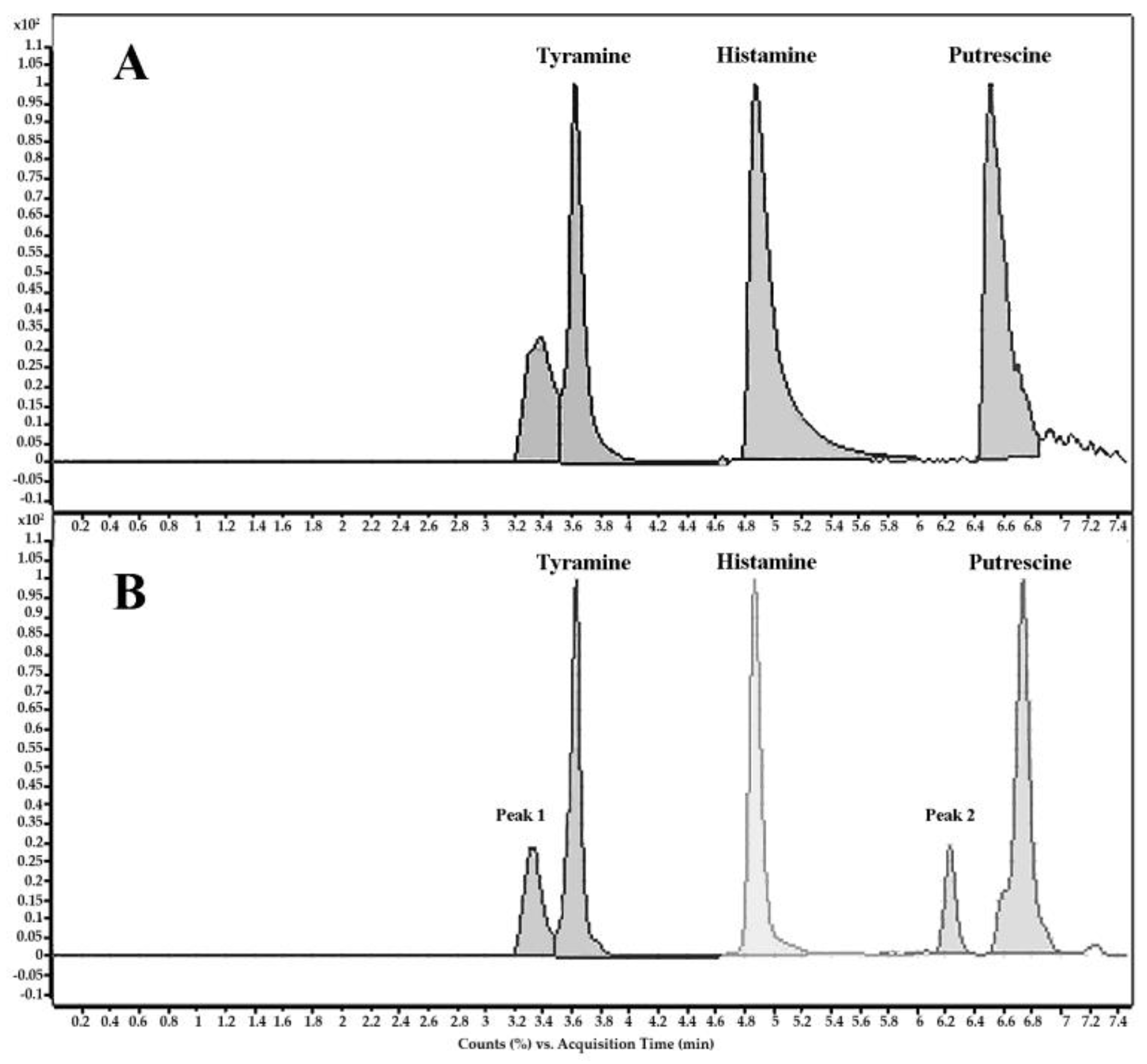
2.4.2. Biogenic Amines (BAs)-Forming Capacity
Molecular Determination of BAs Formation Capacity
Quantitative Determination of BAs
2.4.3. β-Glucosidase Activity
2.4.4. Phenolic Acid Decarboxylase Production Capacity
3. Results and Discussion
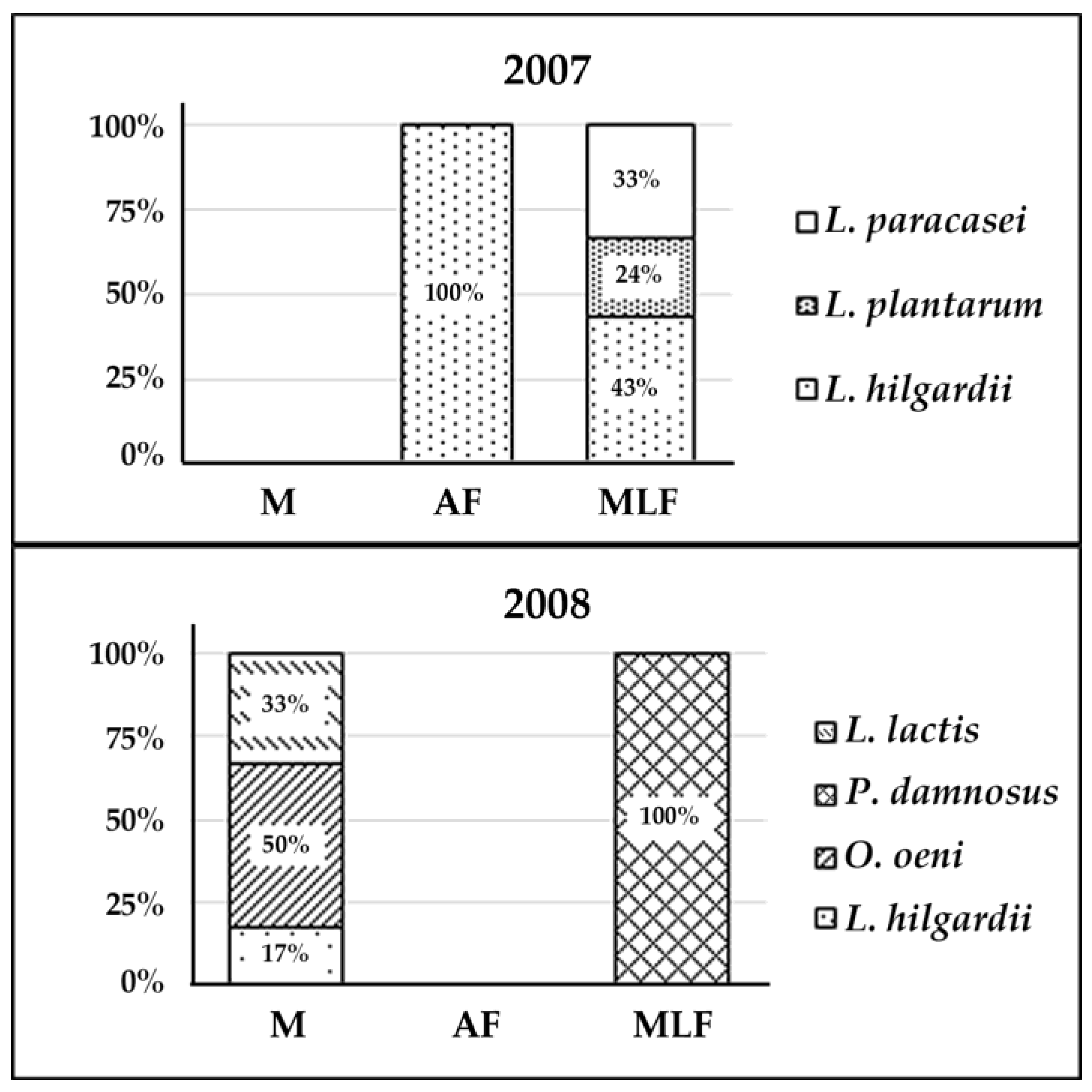
3.1. Isolation and Identification of the Isolates: Ecological Distribution
3.2. Oenological Characterization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lonvaud-Funel, A. Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie Leeuwenhoek 1999, 76, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Rodas, A.M.; Ferrer, S.; Pardo, I. 16S-ARDRA, a tool for identification of lactic acid bacteria isolated from grape must and wine. Syst. Appl. Microbiol. 2003, 26, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, P.; Izquierdo, P.M.; Seseña, S.; Palop, M.L. Analysis of lactic acid bacteria populations during spontaneous malolactic fermentation of Tempranillo wines at five wineries during two consecutive vintages. Food Control 2010, 21, 70–75. [Google Scholar] [CrossRef]
- González-Arenzana, L.; Santamaría, P.; López, R.; López-Alfaro, I. Indigenous lactic acid bacteria communities in alcoholic and malolactic fermentations of Tempranillo wines elaborated in ten wineries of La Rioja (Spain). Food Res. Int. 2013, 50, 438–445. [Google Scholar] [CrossRef]
- Franquès, J.; Araque, I.; Palahí, E.; Portillo, M.D.C.; Reguant, C.; Bordons, A. Presence of Oenococcus oeni and other lactic acid bacteria in grapes and wines from Priorat (Catalonia, Spain). LWT Food Sci. Technol. 2017, 81, 326–334. [Google Scholar] [CrossRef]
- Beneduce, L.; Spano, G.; Vernile, A.; Tarantino, D.; Massa, S. Molecular characterization of lactic acid populations associated with wine spoilage. J. Basic Microbiol. 2004, 44, 10–16. [Google Scholar] [CrossRef]
- Izquierdo, P.M.; García, E.; Martínez, J.; Chacón, J.L. Selection of lactic bacteria to induce malolactic fermentation in red wine of cv. Cencibel. Vitis 2004, 43, 149–153. [Google Scholar]
- Maicas, S. The use of alternative technologies to develop malolactic fermentation in wine. Appl. Microbiol. Biotechnol. 2001, 56, 35–39. [Google Scholar] [CrossRef]
- Bartowsky, E.J.; Costello, P.J.; Chambers, P.J. Emerging trends in the application of malolactic fermentation. Aust. J. Grape Wine Res. 2015, 21, 663–669. [Google Scholar] [CrossRef]
- Solieri, L.; Genova, F.; De Paola, M.; Giudici, P. Characterization and technological properties of Oenococcus oeni strains from wine spontaneous malolactic fermentations: A framework for selection of new starter cultures. J. Appl. Microbiol. 2010, 108, 285–298. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Sieiro, C.; Cansado, J.; Agrelo, D.; Velázquez, J.B.; Villa, T.G. Isolation and enological characterization of malolactic bacteria from the vineyards of northwestern Spain. Appl. Environ. Microbiol. 1990, 56, 2936–2938. [Google Scholar] [CrossRef] [PubMed]
- Testa, B.; Lombardi, S.J.; Tremonte, P.; Succi, M.; Tipaldi, L.; Pannella, G.; Coppola, R. Biodiversity of Lactobacillus plantarum from traditional Italian wines. World J. Microbiol. Biotechnol. 2014, 30, 2299–2305. [Google Scholar] [CrossRef] [PubMed]
- Valdés La Hens, D.; Bravo-Ferrada, B.M.; Delfederico, L.; Caballero, A.C.; Semorile, L.C. Prevalence of Lactobacillus plantarum and Oenococcus oeni during spontaneous malolactic fermentation in Patagonian red wines revealed by polymerase chain reaction-denaturing gradient gel electrophoresis with two targeted genes. Aust. J. Grape Wine Res. 2015, 21, 49–56. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Moreira, J.L.S.; Mota, R.M.; Horta, M.F.; Teixeira, S.M.R.; Neumann, E.; Nicoli, J.R.; Nunes, A.C. Identification to the species level of Lactobacillus isolated in probiotic prospecting studies of human, animal or food origin by 16S-23S rRNA restriction profiling. BMC Microbiol. 2005, 5, 15. [Google Scholar] [CrossRef]
- Torriani, S.; Felis, G.E.; Dellaglio, F. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 2001, 67, 3450–3454. [Google Scholar] [CrossRef]
- De Las Rivas, B.; Marcobal, Á.; Muñoz, R. Improved multiplex-PCR method for the simultaneous detection of food bacteria producing biogenic amines. FEMS Microbiol. Lett. 2005, 244, 367–372. [Google Scholar] [CrossRef]
- Self, R.L.; Wu, W.H.; Marks, H.S. Simultaneous quantification of eight biogenic amine compounds in tuna by matrix solid-phase dispersion followed by HPLC-orbitrap mass spectrometry. J. Agric. Food Chem. 2011, 59, 5906–5913. [Google Scholar] [CrossRef]
- Grimaldi, A.; Bartowsky, E.; Jiranek, V. A survey of glycosidase activities of commercial wine strains of Oenococcus oeni. Int. J. Food Microbiol. 2005, 105, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Mtshali, P.S.; Divol, B.; van Rensburg, P.; du Toit, M. Genetic screening of wine-related enzymes in Lactobacillus species isolated from South African wines. J. Appl. Microbiol. 2010, 108, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Henríquez-Aedo, K.; Durán, D.; Garcia, A.; Hengst, M.B.; Aranda, M. Identification of biogenic amines-producing lactic acid bacteria isolated from spontaneous malolactic fermentation of Chilean red wines. LWT Food Sci. Technol. 2016, 68, 183–189. [Google Scholar] [CrossRef]
- Rodríguez, H.; de las Rivas, B.; Muñoz, R. Efficacy of recA gene sequence analysis in the identification and discrimination of Lactobacillus hilgardii strains isolated from stuck wine fermentations. Int. J. Food Microbiol. 2007, 115, 70–78. [Google Scholar] [CrossRef][Green Version]
- Vigentini, I.; Praz, A.; Domeneghetti, D.; Zenato, S.; Picozzi, C.; Barmaz, A.; Foschino, R. Characterization of malolactic bacteria isolated from Aosta Valley wines and evidence of psychrotrophy in some strains. J. Appl. Microbiol. 2016, 120, 934–945. [Google Scholar] [CrossRef]
- Bravo-Ferrada, B.M.; Hollmann, A.; Delfederico, L.; Valdés La Hens, D.; Caballero, A.; Semorile, L. Patagonian red wines: Selection of Lactobacillus plantarum isolates as potential starter cultures for malolactic fermentation. World J. Microbiol. Biotechnol. 2013, 29, 1537–1549. [Google Scholar] [CrossRef]
- Nisiotou, A.; Sgouros, G.; Mallouchos, A.; Nisiotis, C.-S.; Michaelidis, C.; Tassou, C.; Banilas, G. The use of indigenous Saccharomyces cerevisiae and Starmerella bacillaris strains as a tool to create chemical complexity in local wines. Food Res. Int. 2018, 111, 498–508. [Google Scholar] [CrossRef]
- Du Toit, M.; Engelbrecht, L.; Lerm, E.; Krieger-Weber, S. Lactobacillus: The next generation of malolactic fermentation starter cultures—An overview. Food Bioprocess Technol. 2011, 4, 876–906. [Google Scholar] [CrossRef]
- Barbieri, F.; Montanari, C.; Gardini, F.; Tabanelli, G. Biogenic Amine Production by Lactic Acid Bacteria: A Review. Foods 2019, 8, 17. [Google Scholar] [CrossRef]
- Niu, T.; Li, X.; Guo, Y.; Ma, Y. Identification of a Lactic Acid Bacteria to Degrade Biogenic Amines in Chinese Rice Wine and Its Enzymatic Mechanism. Foods 2019, 8, 312. [Google Scholar] [CrossRef]
- Guo, Y.Y.; Yang, Y.P.; Peng, Q.; Han, Y. Biogenic amines in wine: A review. Int. J. Food Sci. Technol. 2015, 50, 1523–1532. [Google Scholar] [CrossRef]
- Nisiotou, A.A.; Dourou, D.; Filippousi, M.; Diamantea, E.; Fragkoulis, P.; Tassou, C.; Banilas, G. Genetic and technological characterisation of vineyard- and winery-associated lactic acid bacteria. BioMed Res. Int. 2015, 2015, 508254. [Google Scholar] [CrossRef] [PubMed]
- Lonvaud-Funel, A. Biogenic amines in wines: Role of lactic acid bacteria. FEMS Microbiol. Lett. 2001, 199, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Bauza, T.; Blaise, A.; Daumas, F.; Cabanis, J.C. Determination of biogenic amines and their precursor amino acids in wines of the Vallée du Rhône by high-performance liquid chromatography with precolumn derivatization and fluorimetric detection. J. Chromatogr. A 1995, 707, 373–379. [Google Scholar] [CrossRef]
- Moreno-Arribas, M.V.; Polo, M.C.; Jorganes, F.; Munoz, R. Screening of biogenic amine production by lactic acid bacteria isolated from grape must and wine. Int. J. Food Microbiol. 2003, 84, 117–123. [Google Scholar] [CrossRef]
- Elsanhoty, R.M.; Ramadan, M.F. Genetic screening of biogenic amines production capacity from some lactic acid bacteria strains. Food Control 2016, 68, 220–228. [Google Scholar] [CrossRef]
- Pérez-Martín, F.; Seseña, S.; Izquierdo, P.M.; Martín, R.; Palop, M.L. Screening for glycosidase activities of lactic acid bacteria as a biotechnological tool in oenology. World J. Microbiol. Biotechnol. 2012, 28, 1423–1432. [Google Scholar] [CrossRef]
- Sieiro, C.; Villa, T.G.; da Silva, A.F.; García-Fraga, B.; Vilanova, M. Albariño wine aroma enhancement through the use of a recombinant polygalacturonase from Kluyveromyces marxianus. Food Chem. 2014, 145, 179–185. [Google Scholar] [CrossRef]
- Couto, J.A.; Campos, F.M.; Figueiredo, A.R.; Hogg, T.A. Ability of lactic acid bacteria to produce volatile phenols. Am. J. Enol. Vitic. 2006, 57, 166–171. [Google Scholar]
- De Las Rivas, B.; Rodríguez, H.; Curiel, J.A.; Landete, J.M.; Muñoz, R. Molecular screening of wine lactic acid bacteria degrading hydroxycinnamic acids. J. Agric. Food Chem. 2008, 57, 490–494. [Google Scholar] [CrossRef]
| Name | Isolation Year | Sampling Stage * | API® 50 CHL | rDNA 16S Sequencing | 16S-ARDRA (Enzyme/s) | ISR Restriction Analysis (Enzyme) | recA Gene Sequencing |
|---|---|---|---|---|---|---|---|
| L. plantarum CECT 220 | |||||||
| L. paraplantarum CECT 5787 | |||||||
| UVI-6 | 2007 | MLF | L. plantarum/L. paraplantarum/L. pentosus | L. plantarum/L. paraplantarum/L. pentosus | L. plantarum/L. paraplantarum/L. pentosus (BfaI) | - | L. plantarum |
| UVI-8 | L. plantarum/L. paraplantarum/L. pentosus | ||||||
| UVI-9 | L. plantarum/L. paraplantarum/L. pentosus | ||||||
| UVI-10 | L. plantarum/L. paraplantarum/L. pentosus | ||||||
| UVI-11 | - | ||||||
| l. casei CECT 475 | |||||||
| L. paracasei CECT 4175 | |||||||
| L. rhamnosus CECT 288 | |||||||
| UVI-1 | 2007 | MLF | L. casei/L. paracasei/L. rhamnosus | L. casei/L. paracasei/L. rhamnosus | L. casei/L. paracasei/L. rhamnosus (BfaI, BtgZI) | L. paracasei (VspI) | - |
| UVI-2 | L. casei/L. paracasei/L. rhamnosus | ||||||
| UVI-3 | L. casei/L. paracasei/L. rhamnosus | ||||||
| UVI-5 | L. casei/L. paracasei/L. rhamnosus | ||||||
| UVI-7 | L. casei/L. paracasei/L. rhamnosus | ||||||
| UVI-12 | - | ||||||
| UVI-19 | L. casei/Lb. paracasei/Lb. rhamnosus | ||||||
| L. hilgardii CECT 4786 | |||||||
| UVI-4 | 2007 | MLF | L. hilgardii | L. hilgardii | L. hilgardii (MseI, AluI, HaeIII) | - | - |
| UVI-13 | |||||||
| UVI-14 | |||||||
| UVI-15 | |||||||
| UVI-16 | |||||||
| UVI-17 | |||||||
| UVI-18 | |||||||
| UVI-20 | |||||||
| UVI-21 | |||||||
| UVI-22 | AF | ||||||
| UVI-23 | AF | ||||||
| UVI-26 | 2008 | M | |||||
| O. oeni CECT 217 | |||||||
| UVI-24 | 2008 | M | - | O. oeni | O. oeni (AluI, BfaI) | - | - |
| UVI-25 | |||||||
| UVI-29 | |||||||
| P. damnosus CECT 793 | |||||||
| UVI-28 | 2008 | MLF | - | P. damnosus | P. damnosus (AluI, HaeIII) | - | - |
| UVI-31 | P. damnosus | ||||||
| UVI-32 | - | ||||||
| UVI-33 | P. damnosus | ||||||
| UVI-34 | - | ||||||
| UVI-35 | P. damnosus | ||||||
| L. lactis CECT 185 | |||||||
| UVI-27 | 2008 | M | Lc. lactis | Lc. lactis | - | - | - |
| UVI-30 | - |
| Strain | Malic Acid | Lactic Acid | Acetic Acid |
|---|---|---|---|
| Control a | 3.95 (±0.39) | 0.67 (±0.43) | 0.21(±0.01) |
| UVI-2 (L. paracasei) | 0.98 (±0.11) | 3.40 (±0.07) | 0.12 (±0.01) |
| UVI-3 (L. paracasei) | 3.65 (±0.07) | 0.08 (±0.04) | 0.22 (±0.00) |
| UVI-5 (L. paracasei) | 2.48 (±0.25) | 1.95 (±0.28) | 0.18 (±0.00) |
| UVI-9 (L. plantarum) | 3.48 (±0.04) | 0.10 (±0.00) | 0.23 (±0.01) |
| UVI-13 (L. hilgardii) | 3.58 (±0.04) | 0.08 (±0.04) | 0.22 (±0.01) |
| UVI-14 (L. hilgardii) | 1.23 (±0.04) | 3.68 (±0.04) | 0.21 (±0.03) |
| UVI-15 (L. hilgardii) | 3.55 (±0.07) | 0.03 (±0.04) | 0.23 (±0.01) |
| UVI-16 (L. hilgardii) | 3.55 (±0.07) | 0.03 (±0.04) | 0.22 (±0.01) |
| UVI-17 (L. hilgardii) | 1.03 (±0.04) | 4.00 (±0.07) | 0.22 (±0.01) |
| UVI-18 (L. hilgardii) | 1.05 (±0.00) | 3.35 (±0.14) | 0.11 (±0.03) |
| UVI-20 (L. hilgardii) | 1.18 (±0.04) | 3.93 (±0.04) | 0.23 (±0.01) |
| UVI-21 (L. hilgardii) | 1.10 (±0.14) | 3.78 (±0.25) | 0.16 (±0.02) |
| UVI-23 (L. hilgardii) | 1.08 (±0.11) | 3.90 (±0.00) | 0.19 (±0.01) |
| UVI-24 (O. oeni) | 3.55 (±0.07) | 0.03 (±0.04) | 0.23 (±0.00) |
| UVI-25 (O. oeni) | 3.48 (±0.04) | 0.18 (±0.04) | 0.23 (±0.01) |
| UVI-26 (L. hilgardii) | 4.03 (±0.25) | 0.83 (±0.18) | 0.33 (±0.01) |
| Strain | Species | β-Glucosidase Activity (U) * | Biogenic Amines Production: Genes (Concentration) † | Phenolic Acid Decarboxylase Gene †† | ||
|---|---|---|---|---|---|---|
| hdc | odc | tdc | pad | |||
| UVI-2 | L. paracasei | 36.9 (±1.7) | - | - | + (0.035) | - |
| UVI-5 | L. paracasei | 23.6 (±5.0) | - | + | + (0.065) | - |
| UVI-14 | L. hilgardii | 14.7 (±0.8) | - | - | + | - |
| UVI-17 | L. hilgardii | 20.6 (±0.8) | - | - | + | - |
| UVI-18 | L. hilgardii | 20.6 (±0.8) | - | - | + (0.152) | - |
| UVI-20 | L. hilgardii | 23.5 (±5.0) | - | - | + | - |
| UVI-21 | L. hilgardii | 28.9 (±2.5) | - | - | + | - |
| UVI-23 | L. hilgardii | 25.3 (±4.2) | - | - | + | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Seijas, J.; García-Fraga, B.; da Silva, A.F.; Zas-García, X.; Lois, L.C.; Gago-Martínez, A.; Leão-Martins, J.M.; Sieiro, C. Evaluation of Malolactic Bacteria Associated with Wines from Albariño Variety as Potential Starters: Screening for Quality and Safety. Foods 2020, 9, 99. https://doi.org/10.3390/foods9010099
López-Seijas J, García-Fraga B, da Silva AF, Zas-García X, Lois LC, Gago-Martínez A, Leão-Martins JM, Sieiro C. Evaluation of Malolactic Bacteria Associated with Wines from Albariño Variety as Potential Starters: Screening for Quality and Safety. Foods. 2020; 9(1):99. https://doi.org/10.3390/foods9010099
Chicago/Turabian StyleLópez-Seijas, Jacobo, Belén García-Fraga, Abigail F. da Silva, Xavier Zas-García, Lucía C. Lois, Ana Gago-Martínez, José Manuel Leão-Martins, and Carmen Sieiro. 2020. "Evaluation of Malolactic Bacteria Associated with Wines from Albariño Variety as Potential Starters: Screening for Quality and Safety" Foods 9, no. 1: 99. https://doi.org/10.3390/foods9010099
APA StyleLópez-Seijas, J., García-Fraga, B., da Silva, A. F., Zas-García, X., Lois, L. C., Gago-Martínez, A., Leão-Martins, J. M., & Sieiro, C. (2020). Evaluation of Malolactic Bacteria Associated with Wines from Albariño Variety as Potential Starters: Screening for Quality and Safety. Foods, 9(1), 99. https://doi.org/10.3390/foods9010099








