Isolation of Penicillium citrinum from Roots of Clerodendron cyrtophyllum and Application in Biosynthesis of Aglycone Isoflavones from Soybean Waste Fermentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Isolation of Microorganisms
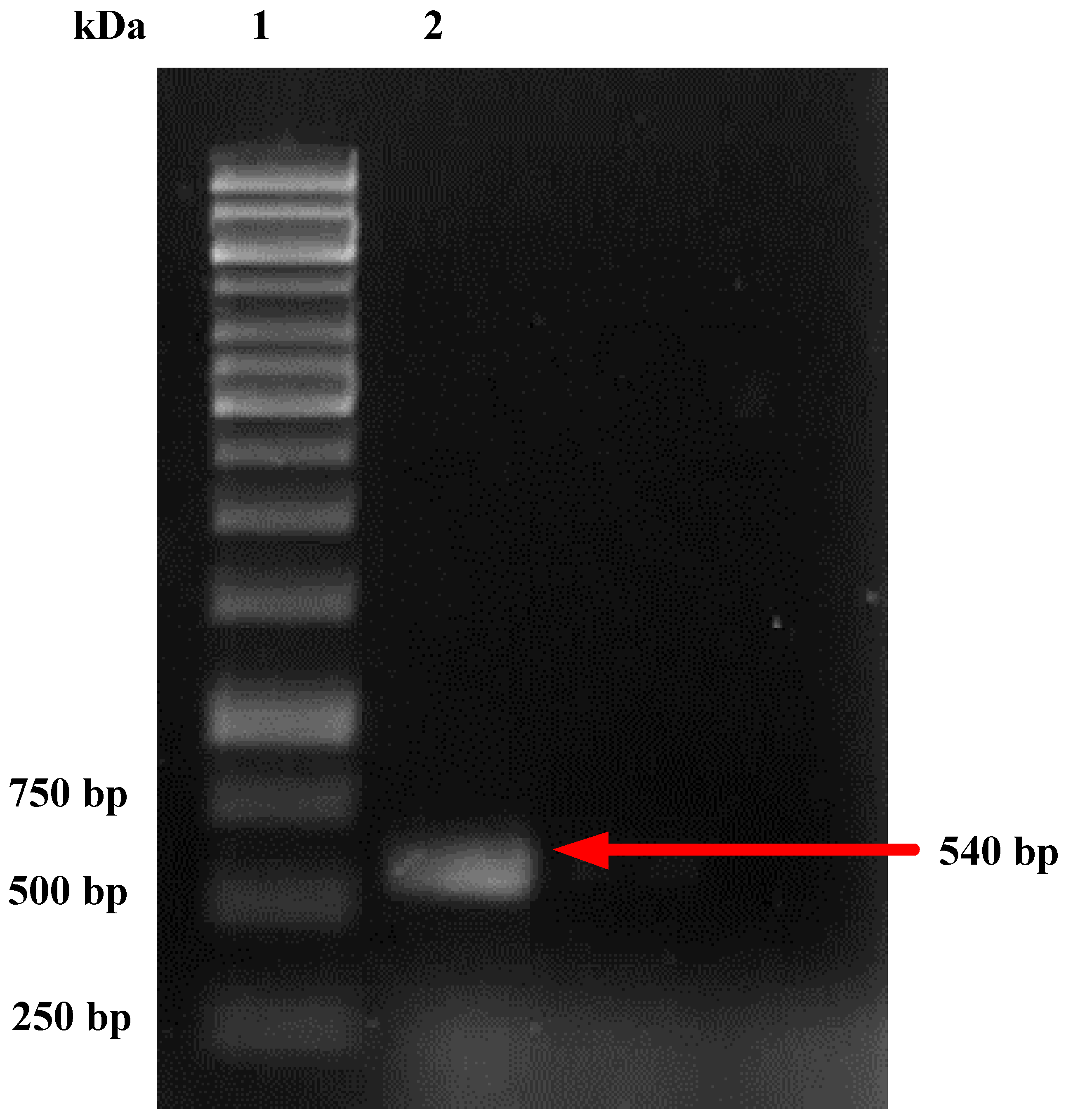
2.3. Fungal Identification
2.4. Enzymatic Activity
2.5. Optimization of the β-Glucosidase Production
2.6. Fermentation of Soybean Residual Using P. citrinium
2.7. Determination of Glucosides and Aglycones Using TLC, LC-MS/MS, NMR
2.8. Statistical Anaylsis
3. Results
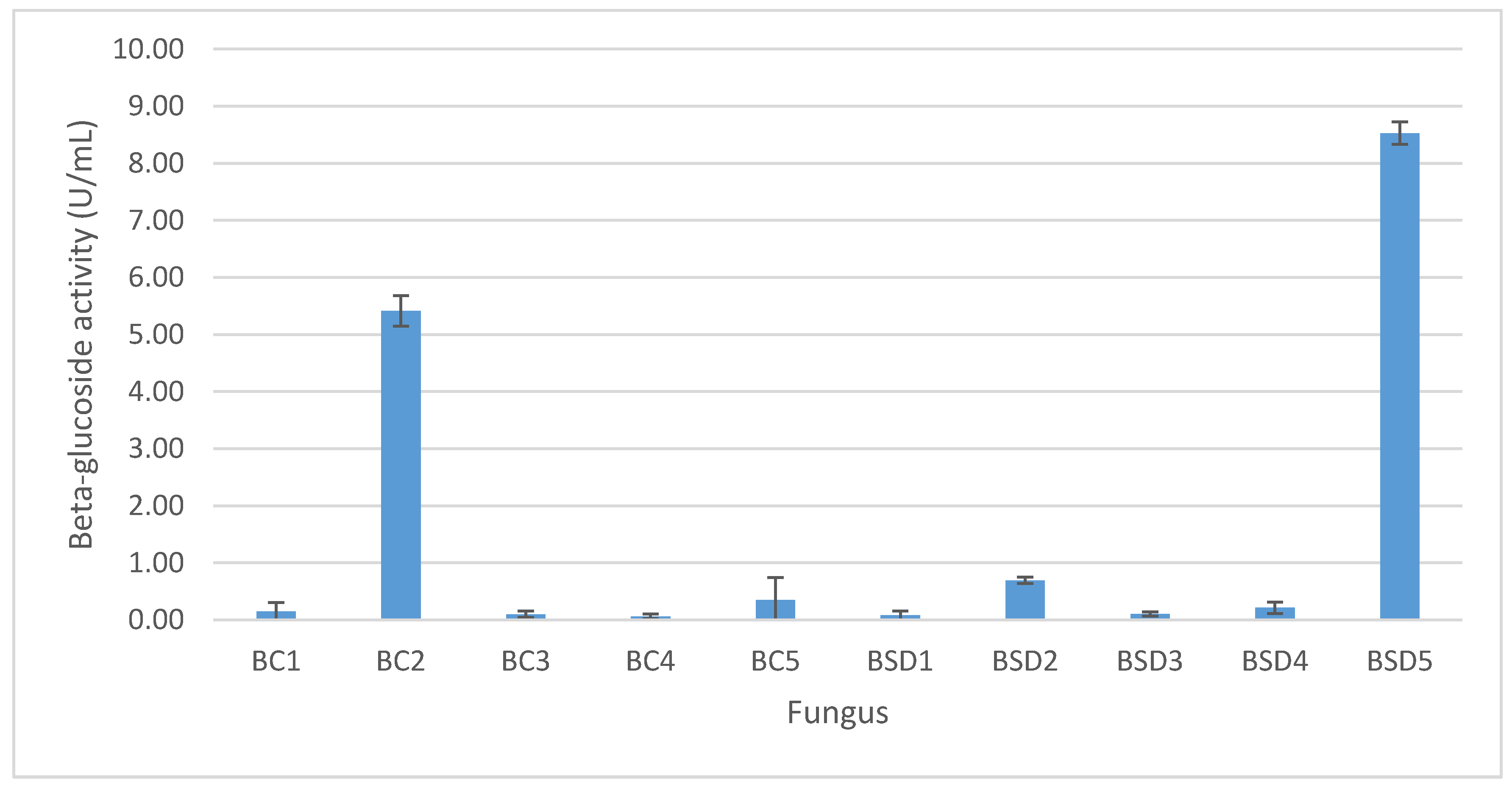
3.1. Isolation and Screening of β-Glucosidase-Producing Microorganisms
3.2. Isolation and Screening of β-Glucosidase-Producing Microorganisms
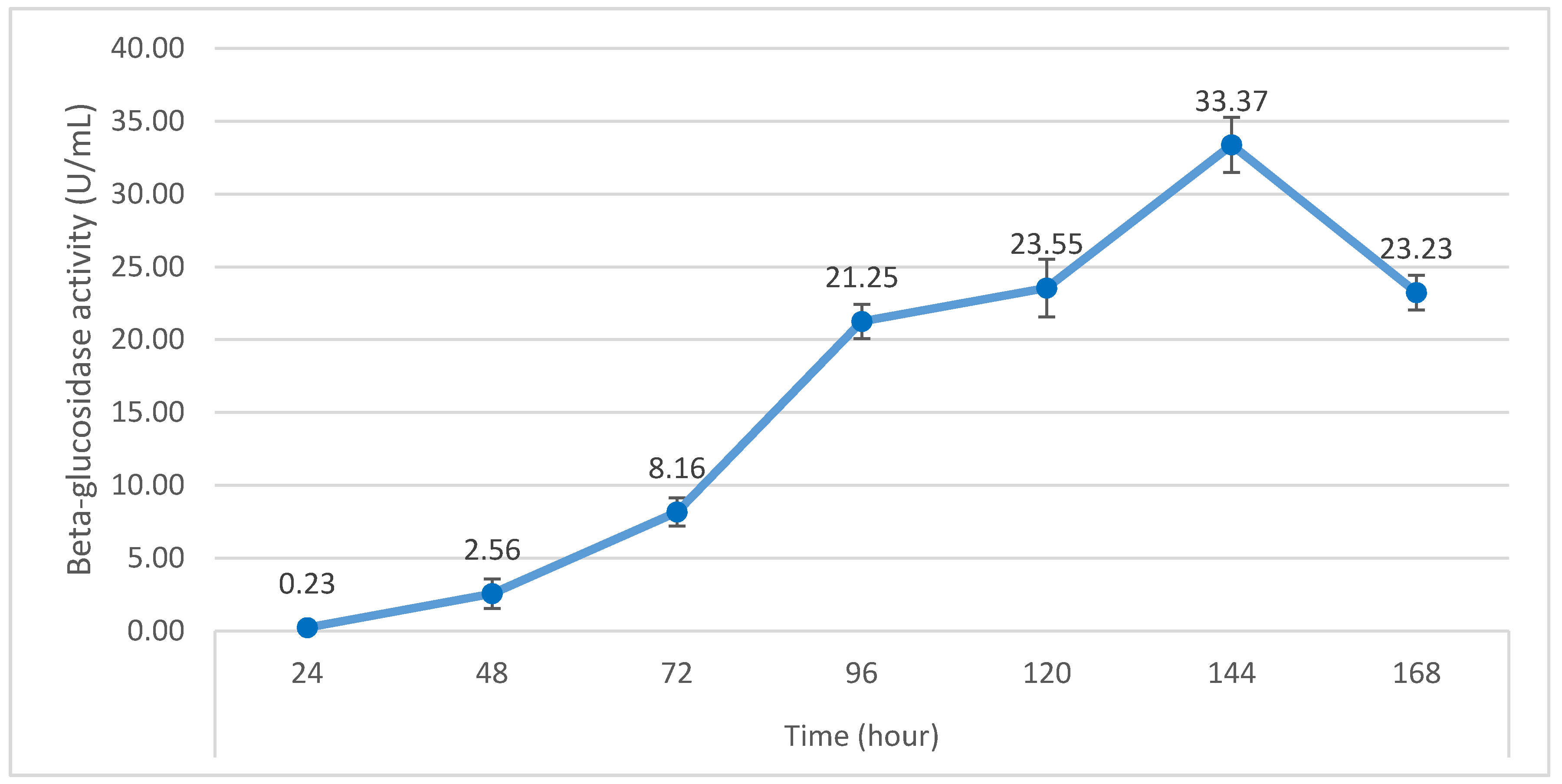
3.3. Optimization of β-Glucosidase Production by P. citrinum
3.4. Fermentation of Soybean Residual Extract by P. citrinum for Aglycones (Genistein, Daidzein) Production
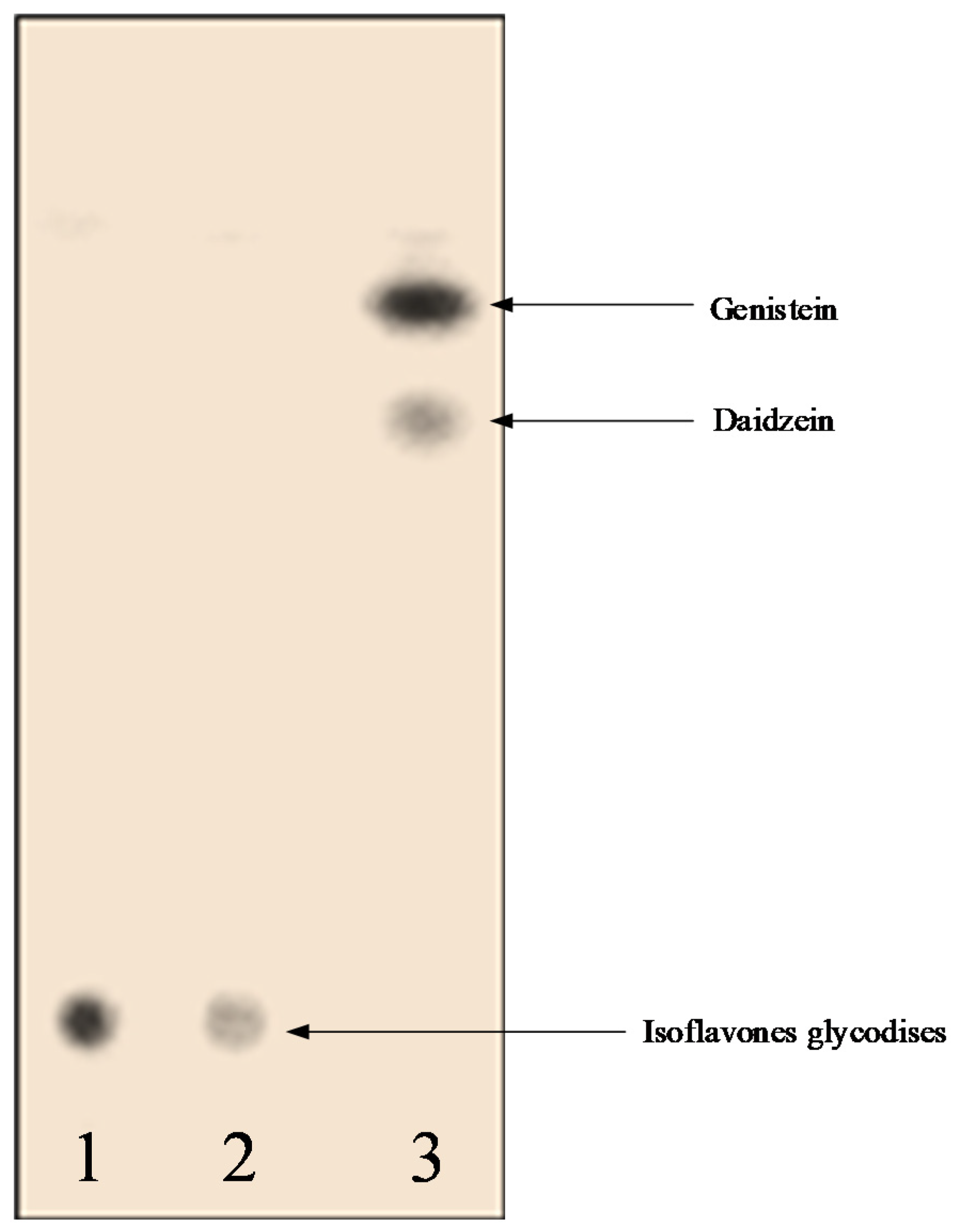
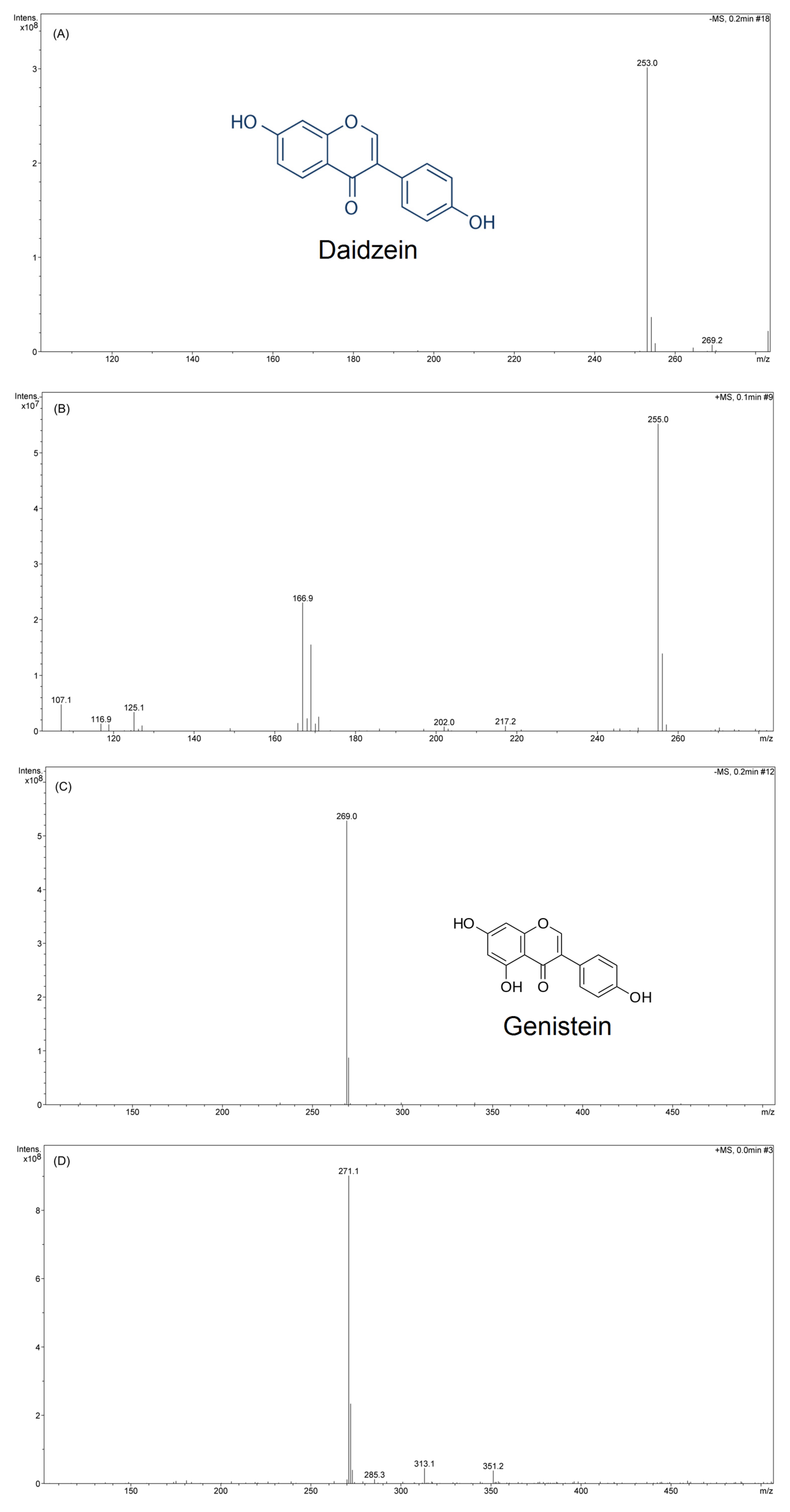
3.5. Identification and Characterization of Aglycones Using LC-MS/MS, TLC, and NMR
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Anderson, J.J.B.; Garner, S.C. Phytoestrogens and human function. Nutr. Today 1997, 32, 232. [Google Scholar] [CrossRef]
- Markovic, R.; Baltic, M.Z.; Pavlovic, M.; Glisic, M.; Radulovic, S.; Djordjevic, V.; Sefer, D. Isoflavones-from biotechnology to functional foods. Procedia Food Sci. 2015, 5, 176–179. [Google Scholar] [CrossRef]
- Yatsu, F.K.J.; Koester, L.S.; Bassani, V.L. Isoflavone-aglycone fraction from Glycine max: A promising raw material for isoflavone-based pharmaceutical or nutraceutical products. Rev. Bras. Farmacogn. 2016, 26, 259–267. [Google Scholar] [CrossRef]
- Hong, J.L.; Qin, X.Y.; Shu, P.; Wang, Q.; Zhou, Z.F.; Wang, G.K.; Lin, B.B.; Wang, Q.; Qin, M.J. Comparative study of isoflavones in wild and cultivated soybeans as well as bean products by high-performance liquid chromatography coupled with mass spectrometry and chemometric techniques. Eur. Food Res. Technol. 2011, 233, 869. [Google Scholar] [CrossRef]
- Hu, S.C.; Hong, K.; Song, Y.C.; Liu, J.Y.; Tan, R.X. Biotransformation of soybean isoflavones by a marine Streptomyces sp. 060524 and cytotoxicity of the products. World J. Microbiol. Biotechnol. 2008, 25, 115. [Google Scholar] [CrossRef]
- Baú, T.R.; Ida, E.I. Soymilk processing with higher isoflavone aglycone content. Food Chem. 2015, 183, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Delmonte, P.; Perry, J.; Rader, J.I. Determination of isoflavones in dietary supplements containing soy, red clover and kudzu: Extraction followed by basic or acid hydrolysis. J. Chromatogr. A 2006, 1107, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Rostagno, M.A.; Villares, A.; Guillamón, E.; García-Lafuente, A.; Martínez, J.A. Sample preparation for the analysis of isoflavones from soybeans and soy foods. J. Chromatogr. A 2009, 1216, 2–29. [Google Scholar] [CrossRef] [PubMed]
- Handa, C.L.; Couto, U.R.; Vicensoti, A.H.; Georgetti, S.R.; Ida, E.I. Optimisation of soy flour fermentation parameters to produce β-glucosidase for bioconversion into aglycones. Food Chem. 2014, 152, 56–65. [Google Scholar] [CrossRef]
- Chun, J.; Kim, G.M.; Lee, K.W.; Choi, I.D.; Kwon, G.H.; Park, J.Y.; Jeong, S.J.; Kim, J.S.; Kim, J.H. Conversion of isoflavone glucosides to aglycones in soymilk by fermentation with lactic acid bacteria. J. Food Sci. 2007, 72, M39–M44. [Google Scholar] [CrossRef]
- Aguieiras, E.C.G.; Cavalcanti-Oliveira, E.D.; de Castro, A.M.; Langone, M.A.P.; Freire, D.M.G. Simultaneous enzymatic transesterification and esterification of an acid oil using fermented solid as biocatalyst. J. Am. Oil Chem. Soc. 2017, 94, 551–558. [Google Scholar] [CrossRef]
- Papadaki, A.; Kopsahelis, N.; Mallouchos, A.; Mandala, I.; Koutinas, A.A. Bioprocess development for the production of novel oleogels from soybean and microbial oils. Food Res. Int. 2019, 126, 108684. [Google Scholar] [CrossRef]
- Papadaki, A.; Androutsopoulos, N.; Patsalou, M.; Koutinas, M.; Kopsahelis, N.; Castro, A.; Papanikolaou, S.; Koutinas, A. Biotechnological production of fumaric acid: The effect of morphology of rhizopus arrhizus NRRL 2582. Fermentation 2017, 3, 33. [Google Scholar] [CrossRef]
- Papadaki, A.; Papapostolou, H.; Alexandri, M.; Kopsahelis, N.; Papanikolaou, S.; de Castro, A.M.; Freire, D.M.G.; Koutinas, A.A. Fumaric acid production using renewable resources from biodiesel and cane sugar production processes. Environ. Sci. Pollut. Res. 2018, 25, 35960–35970. [Google Scholar] [CrossRef]
- Haas, M.J. Improving the economics of biodiesel production through the use of low value lipids as feedstocks: Vegetable oil soapstock. Fuel Process. Technol. 2005, 86, 1087–1096. [Google Scholar] [CrossRef]
- Aguieiras, E.C.G.; Papadaki, A.; Mallouchos, A.; Mandala, I.; Sousa, H.; Freire, D.M.G.; Koutinas, A.A. Enzymatic synthesis of bio-based wax esters from palm and soybean fatty acids using crude lipases produced on agricultural residues. Ind. Crops Prod. 2019, 139, 111499. [Google Scholar] [CrossRef]
- Fujita, A.; Alencar, S.M.; Park, Y.K. Conversion of isoflavone glucosides to aglycones by partially purified β-glucosidases from microbial and vegetable sources. Appl. Biochem. Biotechnol. 2015, 176, 1659–1672. [Google Scholar] [CrossRef]
- Ahn-Jarvis, J.H.; Teegarden, M.D.; Schwartz, S.J.; Lee, K.; Vodovotz, Y. Modulating conversion of isoflavone glycosides to aglycones using crude beta-glycosidase extracts from almonds and processed soy. Food Chem. 2017, 237, 685–692. [Google Scholar] [CrossRef]
- da Silva, L.H.; Celeghini, R.M.S.; Chang, Y.K. Effect of the fermentation of whole soybean flour on the conversion of isoflavones from glycosides to aglycones. Food Chem. 2011, 128, 640–644. [Google Scholar] [CrossRef]
- Xie, L.; Hettiarachchy, N.S.; Cai, R.; Tsuruhami, K.; Koikeda, S. Conversion of Isoflavone Glycosides to Aglycones in SoyLife and Soymeal Using β-glycosidase. J. Food Sci. 2003, 68, 427–430. [Google Scholar] [CrossRef]
- Chang, F.; Xue, S.; Xie, X.; Fang, W.; Fang, Z.; Xiao, Y. Carbohydrate-binding module assisted purification and immobilization of β-glucosidase onto cellulose and application in hydrolysis of soybean isoflavone glycosides. J. Biosci. Bioeng. 2018, 125, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Molina, G.; de Lima, E.A.; Borin, G.P.; de Barcelos, M.C.S.; Pastore, G.M. Chapter 7—Beta-Glucosidase From Penicillium. In New and Future Developments in Microbial Biotechnology and Bioengineering; Gupta, V.K., Rodriguez-Couto, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 137–151. [Google Scholar]
- Yan, F.; Xia, W.; Zhang, X.; Chen, S.; Nie, X.; Qian, L. Characterization of β-glucosidase from Aspergillus terreus and its application in the hydrolysis of soybean isoflavones. J. Zhejiang Univ. Sci. B 2016, 17, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.H.; Jo, M.N.; Kim, K.T.; Paik, H.D. Evaluation of glucosidases of Aspergillus niger strain comparing with other glucosidases in transformation of ginsenoside Rb1 to ginsenosides Rg3. J. Ginseng Res. 2014, 38, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Baraldo Junior, A.; Borges, D.G.; Tardioli, P.W.; Farinas, C.S. Characterization of β-Glucosidase Produced by Aspergillus niger under Solid-State Fermentation and Partially Purified Using MANAE-Agarose. Biotechnol. Res. Int. 2014, 2014, 317092. [Google Scholar] [CrossRef] [PubMed]
- Langston, J.; Sheehy, N.; Xu, F. Substrate specificity of Aspergillus oryzae family 3 β-glucosidase. Biochim. Biophys. Act. BBA Proteins Proteom. 2006, 1764, 972–978. [Google Scholar] [CrossRef]
- Krogh, K.B.R.M.; Harris, P.V.; Olsen, C.L.; Johansen, K.S.; Hojer-Pedersen, J.; Borjesson, J.; Olsson, L. Characterization and kinetic analysis of a thermostable GH3 β-glucosidase from Penicillium brasilianum. Appl. Microbiol. Biotechnol. 2010, 86, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, T.; Igarashi, K.; Yoshida, M.; Samejima, M. Molecular cloning and characterization of two intracellular β-glucosidases belonging to glycoside hydrolase family 1 from the basidiomycete Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 2006, 73, 807. [Google Scholar] [CrossRef]
- Kudo, K.; Watanabe, A.; Ujiie, S.; Shintani, T.; Gomi, K. Purification and enzymatic characterization of secretory glycoside hydrolase family 3 (GH3) aryl β-glucosidases screened from Aspergillus oryzae genome. J. Biosci. Bioeng. 2015, 120, 614–623. [Google Scholar] [CrossRef]
- Li, D.; Li, X.; Dang, W.; Tran, P.L.; Park, S.H.; Oh, B.C.; Hong, W.S.; Lee, J.S.; Park, K.H. Characterization and application of an acidophilic and thermostable β-glucosidase from Thermofilum pendens. J. Biosci. Bioeng. 2013, 115, 490–496. [Google Scholar] [CrossRef]
- Dekker, R.F.H. Kinetic, inhibition, and stability properties of a commercial β-D-glucosidase (cellobiase) preparation from aspergillus niger and its suitability in the hydrolysis of lignocellulose. Biotechnol. Bioeng. 1986, 28, 1438–1442. [Google Scholar] [CrossRef]
- Tendulkar, S.R.; Saikumari, Y.K.; Patel, V.; Raghotama, S.; Munshi, T.K.; Balaram, P.; Chattoo, B.B. Isolation, purification and characterization of an antifungal molecule produced by Bacillus licheniformis BC98, and its effect on phytopathogen Magnaporthe grisea. J. Appl. Microbiol. 2007, 103, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- Kidd, S.; Halliday, C.L.; Alexiou, H.; Ellis, D.H. Descriptions of Medical Fungi; CutCut Digital: Richmond, VA, USA, 2016. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Cai, Y.J.; Buswell, J.A.; Chang, S.T. β-Glucosidase components of the cellulolytic system of the edible straw mushroom, Volvariella volvacea. Enzyme Microb. Technol. 1998, 22, 122–129. [Google Scholar] [CrossRef]
- So, J.H.; Kim, W.C.; Shin, J.H.; Yu, C.B.; Rhee, I.K. Purification and characterization of a β-glucosidase capable of hydrolyzing soybean isoflavone glycosides from Pichia guilliermondii K123-1. Food Sci. Biotechnol. 2010, 19, 1373–1379. [Google Scholar] [CrossRef]
- Lachke, A.H.; Bastawde, K.B.; Powar, V.K.; Srinivasan, M.C. Enhanced production of extracellular β-glucosidase by penicillium funiculosum in submerged culture. Biotechnol. Lett. 1983, 5, 649–652. [Google Scholar] [CrossRef]
- Beitel, S.M.; Knob, A. Penicillium miczynskii β -glucosidase: A Glucose-Tolerant Enzyme Produced Using Pineapple Peel as Substrate. Ind. Biotechnol. 2013, 9, 293–300. [Google Scholar] [CrossRef]
- Jeya, M.; Joo, A.R.; Lee, K.M.; Tiwari, M.K.; Lee, K.M.; Kim, S.H.; Lee, J.K. Characterization of β-glucosidase from a strain of Penicillium purpurogenum KJS506. Appl. Microbiol. Biotechnol. 2010, 86, 1473–1484. [Google Scholar] [CrossRef] [PubMed]
- Ng, I.S.; Li, C.W.; Chan, S.P.; Chir, J.L.; Chen, P.T.; Tong, C.G.; Yu, S.M.; Ho, T.H.D. High-level production of a thermoacidophilic β-glucosidase from Penicillium citrinum YS40-5 by solid-state fermentation with rice bran. Bioresour. Technol. 2010, 101, 1310–1317. [Google Scholar] [CrossRef]
- Xue, Y.; Song, X.; Yu, J. Overexpression of β-glucosidase from Thermotoga maritima for the production of highly purified aglycone isoflavones from soy flour. World J. Microbiol. Biotechnol. 2009, 25, 2165–2172. [Google Scholar] [CrossRef]
- Maitan-Alfenas, G.P.; de A Lage, L.G.; de Almeida, M.N.; Visser, E.M.; de Rezende, S.T.; Guimarães, V.M. Hydrolysis of soybean isoflavones by Debaryomyces hansenii UFV-1 immobilised cells and free β-glucosidase. Food Chem. 2014, 146, 429–436. [Google Scholar] [CrossRef]
- Yang, S.; Wang, L.; Yan, Q.; Jiang, Z.; Li, L. Hydrolysis of soybean isoflavone glycosides by a thermostable β-glucosidase from Paecilomyces thermophila. Food Chem. 2009, 115, 1247–1252. [Google Scholar] [CrossRef]
- Nemitz, M.C.; Teixeira, H.F.; von Poser, G.L. A new approach for the purification of soybean acid extract: Simultaneous production of an isoflavone aglycone-rich fraction and a furfural derivative-rich by-product. Ind. Crops Prod. 2015, 67, 414–421. [Google Scholar] [CrossRef]

| Fermentation Time (h) | Isoflavone Glucosides | Aglycones | Remark |
|---|---|---|---|
| 0 | ++++ | - | Original soybean extract without addition of enzyme |
| 24 | ++++ | - | |
| 48 | +++ | + | |
| 72 | - | ++++ | |
| 96 | - | ++++ | |
| 120 | - | ++++ |
| Soybean Residual Extract/Enzyme Ratio (v/v, %) | Isoflavone Glucosides | Aglycones | Remark |
|---|---|---|---|
| 0 | ++++ | - | Original soybean extract without addition of enzyme |
| 5 | - | + | |
| 10 | - | +++ | |
| 15 | + | +++ | |
| 20 | ++ | +++ |
| Fungal Strain | Fermentation Conditions | β-Glucosidase Activity (U/mL) | Ref. |
|---|---|---|---|
| P. funiculosum | Substrate: Rice bran, defatted oil cakes Temperature: 20 °C pH: 4.5 Fermentation time: 288 h | 30–36 | [34] |
| P. miczynskii | Substrate: Pineapple peel Temperature: 30 °C pH: 5.5 Fermentation time: 216 h | 2.82 | [35] |
| P. purpurogenum KJS506 | Substrate: Rice straw Temperature: 32 °C pH: 4 Fermentation time: 144 h | 26.4 | [36] |
| P. citrinium YS40-5 | Substrate: Rice bran Temperature: 70 °C pH: 6.0 Fermentation time: 96 h | 57.5 U/g | [37] |
| P. citrinium | Substrate: PDB Temperature: 30 °C pH: 4.5 Fermentation time: 144 h | 33.72 | Current study |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doan, D.T.; Luu, D.P.; Nguyen, T.D.; Hoang Thi, B.; Pham Thi, H.M.; Do, H.N.; Luu, V.H.; Pham, T.D.; Than, V.T.; Pham Thi, H.H.; et al. Isolation of Penicillium citrinum from Roots of Clerodendron cyrtophyllum and Application in Biosynthesis of Aglycone Isoflavones from Soybean Waste Fermentation. Foods 2019, 8, 554. https://doi.org/10.3390/foods8110554
Doan DT, Luu DP, Nguyen TD, Hoang Thi B, Pham Thi HM, Do HN, Luu VH, Pham TD, Than VT, Pham Thi HH, et al. Isolation of Penicillium citrinum from Roots of Clerodendron cyrtophyllum and Application in Biosynthesis of Aglycone Isoflavones from Soybean Waste Fermentation. Foods. 2019; 8(11):554. https://doi.org/10.3390/foods8110554
Chicago/Turabian StyleDoan, Duy Tien, Duc Phuong Luu, Thanh Duong Nguyen, Bich Hoang Thi, Hong Minh Pham Thi, Huu Nghi Do, Van Huyen Luu, The Dan Pham, Van Thai Than, Hai Ha Pham Thi, and et al. 2019. "Isolation of Penicillium citrinum from Roots of Clerodendron cyrtophyllum and Application in Biosynthesis of Aglycone Isoflavones from Soybean Waste Fermentation" Foods 8, no. 11: 554. https://doi.org/10.3390/foods8110554
APA StyleDoan, D. T., Luu, D. P., Nguyen, T. D., Hoang Thi, B., Pham Thi, H. M., Do, H. N., Luu, V. H., Pham, T. D., Than, V. T., Pham Thi, H. H., Pham, M. Q., & Tran, Q. T. (2019). Isolation of Penicillium citrinum from Roots of Clerodendron cyrtophyllum and Application in Biosynthesis of Aglycone Isoflavones from Soybean Waste Fermentation. Foods, 8(11), 554. https://doi.org/10.3390/foods8110554






