3.1. Analysis of Physicochemical Results of WSS
NH
3-N content, a key quality parameter, reflects protease-mediated protein hydrolysis efficiency. As shown in
Figure 2a, the
A. oryzae-inoculated group demonstrated significantly higher NH
3-N accumulation rates, indicating enhanced proteolysis. However, WSS NH
3-N levels remained substantially lower than the traditional soy sauce threshold (≥0.4 g/100 mL). Zhao et al. [
12] reported that walnut meal contains lower and structurally more complex protein content (less than half of soybean meal), predominantly composed of harder-to-hydrolyze glutelin and prolamins. The neutral/alkaline proteases from
A. oryzae show limited hydrolysis efficiency toward these proteins, whereas soybean proteins are mainly more easily hydrolyzable globulins. The most rapid NH
3-N increase occurred during the initial 7 days of fermentation, after which the accumulation rate progressively decreased.
Figure 2b reveals that the
A. oryzae group maintained significantly higher protease activity than
A. niger during the initial 30 fermentation days, followed by a sharp decline (Day 30–60). This pattern reflects progressive depletion of readily hydrolyzable soybean proteins. Meanwhile, walnut flour is rich in polyphenolic compounds such as tannins and tannic acid. It is hypothesized that these compounds can bind to proteins to form insoluble complexes that prevent proteases from entering the cleavage site and inhibit the growth of
A. oryzae. [
21]. Furthermore, Increasing salinity due to water evaporation progressively inhibited microbial and enzymatic activities [
22]. Consequently, the observed high protease activity during the initial 30 days drove protein hydrolysis, accounting for the significantly faster NH
3-N accumulation compared to subsequent phases. Notably,
A. niger exhibited minimal initial protease activity. This weakness is likely attributable to its proficiency in secreting glycoside hydrolases and organic acids; proteases constitute only a minor fraction of its enzymatic repertoire and are primarily acidic types adapted to a pH range of 2 to 5.0 [
23], Throughout the soy sauce fermentation process, the pH remained above the optimal range for these acidic proteases, resulting in persistently weak enzyme activity in the AN group. The inherent physiological characteristics of AN determine that it is not typically considered a highly efficient proteolytic microorganism; thus, it is generally more suitable as an auxiliary culture in soy sauce fermentation.
During soy sauce fermentation, the pH ultimately stabilizes within the range of approximately 4.5 to 5.5. A pH below 4.0 may indicate abnormal fermentation, whereas a pH exceeding 5.5 could suggest insufficient fermentation. The acidification, primarily driven by lactic acid bacteria, is the main cause of pH reduction. Crucially, Soy sauce’s umami and flavor compounds exhibit optimal stability within this mildly acidic environment [
23,
24,
25]. As shown in
Figure 2c, post fermentation revealed distinct pH dynamic, the AO group maintained significantly higher pH than the AN group, despite a shared decreasing trend. This divergence stemmed from
A. niger’s prolific secretion of organic acids, whereas
A. oryzae primarily generated proteases/amylases with limited acidogenesis. Correspondingly,
Figure 2d confirmed the AN group’s substantially higher total acidity (progressively increasing) versus AO and ON groups. It was reported that A. niger could utilize complex carbon sources in both soybean and walnut meals more thoroughly, leading to higher acid production and decreased pH value. However, the acid environment might be not beneficial for increasing NH
3-N levels by degrading proteins in walnut meal. This might be attributed to the inhibition of protease activities and the growth of protease-producing strains during post-fermentation of WSS [
26].
3.2. Impact of Fermentation with Different Aspergillus Species on Microbial Diversity
As shown in
Table 1, the ACE and Chao values of the AN group were higher than those of the AO and ON groups. This indicates that in the walnut fermentation system involving
A. niger, the microbial community had a greater total number of species and higher richness of rare species. The Shannon index of the ON group was lower than that of the AN and AO groups, while its Simpson index was relatively higher. This suggests that in the microbial community of the ON group, species richness was lower, the concentration of dominant species was higher, the community was dominated by a few species, and the evenness of species distribution was relatively poor. Among α-diversity indices, a high Shannon index coupled with a low Simpson index indicates that the community contains a large number of species with uniform distribution. Conversely, a low Shannon index together with a high Simpson index implies that the community has fewer species and that dominant species are concentrated.
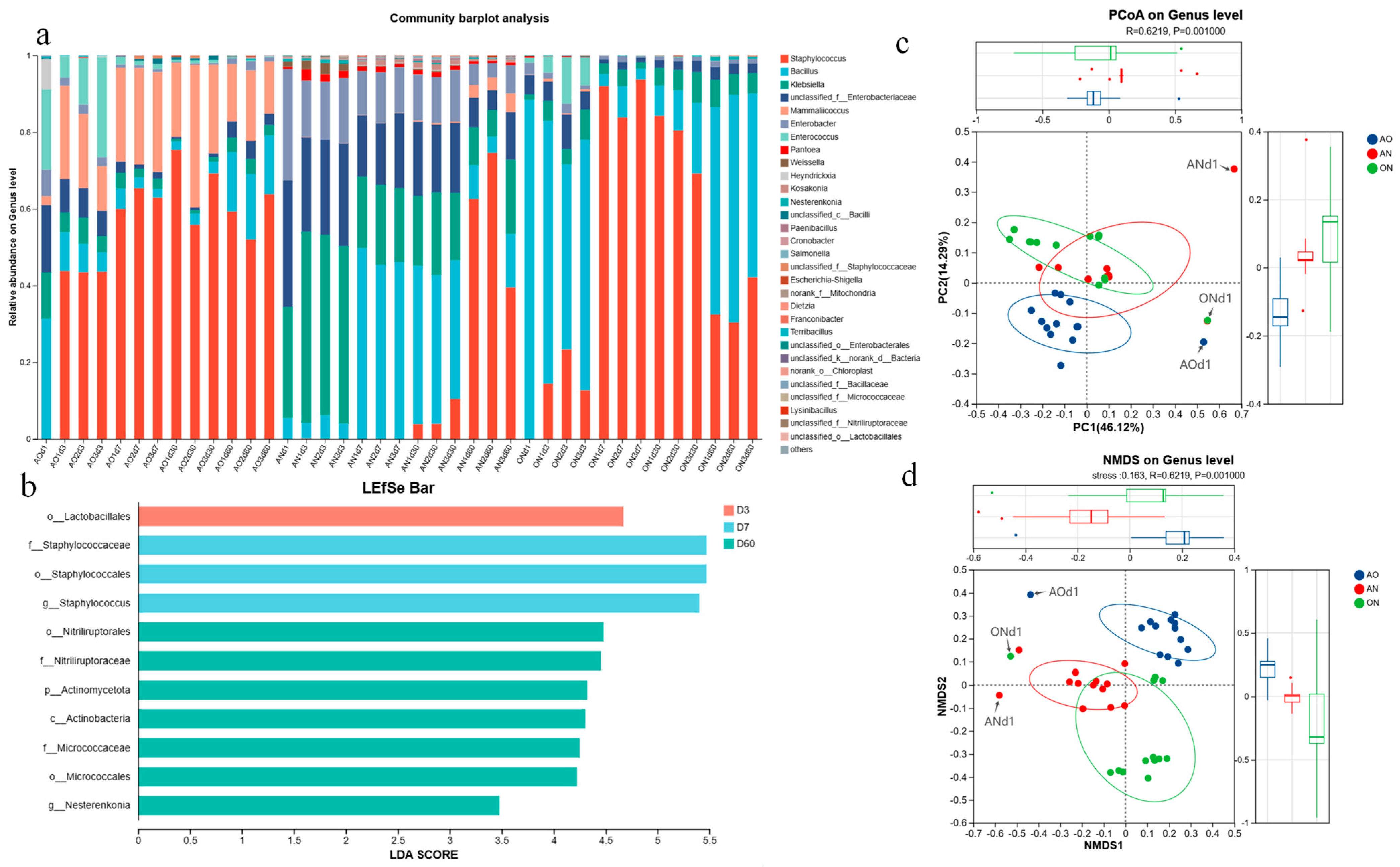
Figure 3 illustrated the differences in the average relative abundance of the same species among different groups, with annotations indicating whether the differences were significant. It intuitively presented the significance of differences in the same species across multiple groups. In general, as shown in
Figure 3d,
Staphylococcus and
Enterococcus accounted for a larger proportion in WSS samples supplemented with
A. oryzae, which were significantly higher than those in the AN group. The proportion of
Bacillus in the group supplemented with
A. niger was higher than that in the AO group; meanwhile, the contents of
Klebsiella,
Enterobacter,
Pantoea,
Weissella,
Kosakonia, and
Nesterenkonia in the AN group were all higher than those in the AO and ON groups. The abundance of
Mammaliicoccus in the AO group was higher than that in the group supplemented with
A. niger.
As shown in
Figure 3, the high abundance of
Staphylococcus in the AO group may be attributed to the fact that proteolytic products (e.g., short peptides, amino acids) secreted by
A. oryzae meet their nutritional requirements, and they are potentially involved in the synthesis of soy sauce flavor compounds such as thiols and volatile acids. Liu Hua et al. found that during the brewing of traditional soy sauce, the relative abundance of dominant genera including
Weissella and
Staphylococcus remains relatively constant in the late stage of fermentation [
27], which is consistent with the results of this study. Kong, F et al. employed three salt-tolerant
Staphylococcus strains in soy sauce brewing; through analyzing the physicochemical properties, organic acid composition, volatile flavor compounds (VFCs), and sensory characteristics during fermentation, they confirmed that
Staphylococcus contributes to the acidity, ester aroma, and mellow aroma of soy sauce [
28].
Bacillus abundance was significantly higher in the
A. niger-added WWS fermentation system than in the group without
A. niger. Beyond secreting proteases and amylases,
A. niger also produces enzymes such as cellulases and pectinases. These additional enzymes hydrolyze polysaccharides present in the raw materials, including cellulose and pectin, into smaller sugar molecules like galacturonic acid and glucuronic acid. This process provides
Bacillus with a more diverse array of carbon sources.
Bacillus efficiently utilizes these varied sugars for growth and metabolism. Zheng et al. observed in their research that fermentation with
A. niger significantly increased the abundance of
Bacillus [
29]. Concurrently,
Bacillus facilitates the rapid decomposition of starch within the fermentation system, generating organic acids and aromatic compounds such as diacetyl, while also enhancing the antioxidant capacity of the final fermented product [
30].
Figure 3e presented the LDA (Linear Discriminant Analysis) scores of different discriminant species, visually demonstrating the relative influence of biomarker taxa identified between groups on the observed differences. The LDA discriminant bar chart statistically identified microbial taxa with significant discriminatory power among multiple groups. Higher LDA scores indicated a greater contribution of the species’ abundance to the observed group differences.
The AO group showed significant enrichment of biomarker taxa including
Mammalicoccus (Staphylococcaceae), Staphylococcales,
Enterococcus, and Enterococcaceae.
Mammalicoccus dominated the
A. oryzae system, likely due to abundant peptides/carbohydrates from efficient substrate hydrolysis by fungal proteases/amylases. As reported, food-associated
Mammalicoccus contributes to sensory profiles through carbohydrate/amino acid catabolism and ester synthesis. Flavor-associated small molecular compounds are also generated through aspects of their proteolytic and lipolytic activities [
31]. The taxa Staphylococcales and Staphylococcaceae were highly enriched in the AO group, reflecting a microenvironment shaped by
Aspergillus oryzae that favors the growth of microorganisms within this order and family. Their metabolism produces short-chain fatty acids (SCFAs), such as acetic acid and propionic acid, which contribute to the foundational flavor profile of the sauce (perceived as sour aroma and mellowness) [
32]. The AO group exhibited significant enrichment of stress-tolerant
Enterococcus and Enterococcaceae, compatible with the
A. oryzae fermentation environment. In contrast,
Klebsiella was uniquely enriched in the AN group, utilizing polysaccharides/oligosaccharides released by
A. niger cellulases/pectinases from plant cell walls. Its metabolism produced flavor-active volatiles (e.g., 2,3-butanediol, acetoin), contributing “creamy” and “fruity” notes to the sauce [
33].
In general, the fermentation efficiency of WSS under natural fermentation conditions was much lower than that of traditional soybean fermentation. The natural fermentation system had rich microbial diversity, and the growth of
A. oryzae and
A. niger might be antagonized by other microorganisms. Meanwhile, the impact of polyphenols in walnut meal on fermentation efficiency could not be ignored. According to Wang Yuzhen et al., the total phenol content in walnut meal reached 2943.12 mg GAE/g, and their study indicated that phenolic acid substances in walnuts (such as ellagic acid, (+)-catechin, chlorogenic acid, and epigallocatechin gallate) all inhibited protease activity [
34]. Among them, ellagic acid (EA) had the highest content among polyphenol monomers. Research by Guowan Su et al. showed that polyphenols induced changes in the secondary structure and amino acid composition of walnut protein. These changes led to hindered hydrolysis and enhanced acetylcholinesterase (AChE) inhibition [
35], resulting in reduced utilization of walnut protein by microorganisms and thus affecting the efficiency of soy sauce fermentation. High concentrations of polyphenols in walnut meal may directly inhibit the expression of protease genes in
Aspergillus or bind to the active sites of proteases, altering enzyme conformation and leading to decreased protease activity. The mechanism of interaction between polyphenols and proteases remains to be further studied.
3.3. Analysis of the Impact of Different Fermentation Times on Microbial Communities
Microbial composition and abundance varied significantly across fermentation stages and treatments.
Figure 4a displays the genus-level bacterial community structure, featuring the top 30 species by relative abundance per sample. The
x-axis denotes fermentation conditions/durations, while the
y-axis indicates genus-level relative abundance.
Figure 4 reveals temporally dynamic microbial community structures across all fermentation conditions, with significant intergroup variations in bacterial composition and abundance. While
Staphylococcus was undetectable at Day 1, its abundance surged during fermentation, peaking at Day 30 in AO/ON groups but delaying until Day 60 in AN group. This pattern can be attributed to substrate availability dynamics. In the early fermentation phase,
A. oryzae initiates the breakdown of macromolecules in walnut and soybean meal. Nevertheless, limited degradation occurs initially, resulting in insufficient bioavailable nutrients to support substantial
Staphylococcus growth, hence its low abundance on Day 1. As fermentation progressed, sustained enzymatic activity from
A. oryzae progressively liberated higher concentrations of small-molecule nutrients. By Day 30, nutrient availability reached levels conducive for rapid
Staphylococcus proliferation, leading to its peak abundance in AO and ON groups. Conversely, in the AN group,
Staphylococcus initially faced limited accessible nutrients. Although polysaccharide degradation products accumulated over time,
Staphylococcus demonstrated relatively low efficiency in utilizing these substrates, requiring extended adaptation periods. Ultimately, by Day 60, more comprehensive substrate decomposition by
A. niger—yielding higher concentrations of polysaccharide derivatives coupled with sufficient proteolysis-derived nutrients—created optimal conditions for substantial
Staphylococcus proliferation, resulting in peak abundance.
Throughout the fermentation process, Bacillus—the predominant genus in the AO and ON group soy sauces—exhibited an initial decline, reaching its lowest abundance at Day 7, followed by subsequent increase. Conversely, in the AN group, Bacillus abundance peaked at Day 7 and gradually decreased thereafter. During early fermentation, A. oryzae initiated decomposition of macromolecules in walnut and soybean meals, releasing limited nutrients. At this stage, genera like Staphylococcus demonstrated stronger competitive capabilities for these resources, while Bacillus was competitively inferior, resulting in its declining abundance. Additionally, evolving environmental parameters further shaped microbial dynamics. Bacillus adapted effectively to the progressively acidic conditions generated by A. oryzae metabolism and utilized the enriched nutrient pool in later phases for substantial proliferation, driving its resurgence. In contrast, A. niger in the AN group rapidly degraded polysaccharides via pectinases and cellulases, generating abundant sugar derivatives (e.g., glucose, galacturonic acid) early in fermentation. Bacillus efficiently assimilated these hydrolysis products, enabling rapid growth and maximal abundance by Day 7. However, the AN group hosted richer distributions of Klebsiella, Enterobacteriaceae, and Enterobacter. As fermentation progressed, diminishing nutrients intensified competition. Bacillus gradually declined due to competitive disadvantage against these copiotrophic taxa.
Figure 4b highlighted microbial community differences by quantifying the contribution of microbial taxa at various taxonomic levels to inter-group variations through Linear Discriminant Analysis (LDA) scores. During initial fermentation (Day 3), abundant carbohydrates and a near-neutral microenvironment favored Lactobacillales, which rapidly fermented sugars to produce lactic acid. This activity acidified the system, inhibiting pathogenic bacteria growth. By Day 7, the highest LDA scores were observed for family Staphylococcaceae, order Staphylococcales, and genus
Staphylococcus. At this stage, substantial small-molecule nutrients (e.g., amino acids, monosaccharides) had been liberated through substrate decomposition.
Staphylococcus populations expanded significantly due to superior nutrient competitiveness and environmental adaptability, establishing these taxa as key discriminators of microbial differences at Day 7. Concurrently, they generated characteristic flavor compounds of soy sauce. Xuefei Shao et al. reported analogous dominance of staphylococci during later fermentation stages in fermented sausages, where they similarly promoted the formation of taste and aroma compounds [
36]. At Day 60, taxa including order Nitriliruptorales, family Nitriliruptoraceae, phylum Actinomycetota, and class Actinobacteria exhibited LDA scores approaching 4.5, identifying them as signature microbial groups for this terminal fermentation phase.
As evidenced by
Figure 4c, ANOSIM (Analysis of Similarities) revealed significant differences in microbial communities among
Aspergillus treatment groups (R = 0.6219,
p = 0.001;
p < 0.05). This statistically robust distinction demonstrates that
Aspergillus species substantially altered the microbial composition of the soy sauce fermentation system. The R-value of 0.6219 indicates substantial dissimilarity between groups, signifying that different
Aspergillus strains exerted distinct effects on microbial communities, ultimately driving divergent community structures. Among the three groups, The AO group exhibited high microbial consistency and structural stability, while the AN group showed greater community discreteness with weaker stability. The ON group demonstrated intermediate stability with moderately conserved community structure. Throughout fermentation, distinct microbial profiles emerged initially across different
Aspergillus treatments. As fermentation progressed, communities underwent significant restructuring, ultimately converging toward relatively stable configurations in all three groups.
Figure 4d shows NMDS results aligned with PCoA outcomes, confirming significant inter-group differences and consistent
Aspergillus-driven community variation. Furthermore, microbial composition differed significantly between initial (Day 1) and terminal fermentation stages. Both fermentation duration and
Aspergillus selection significantly influenced walnut sauce microbial dynamics.
3.4. Correlation Analysis Results of Physicochemical Properties and Microbial Diversity
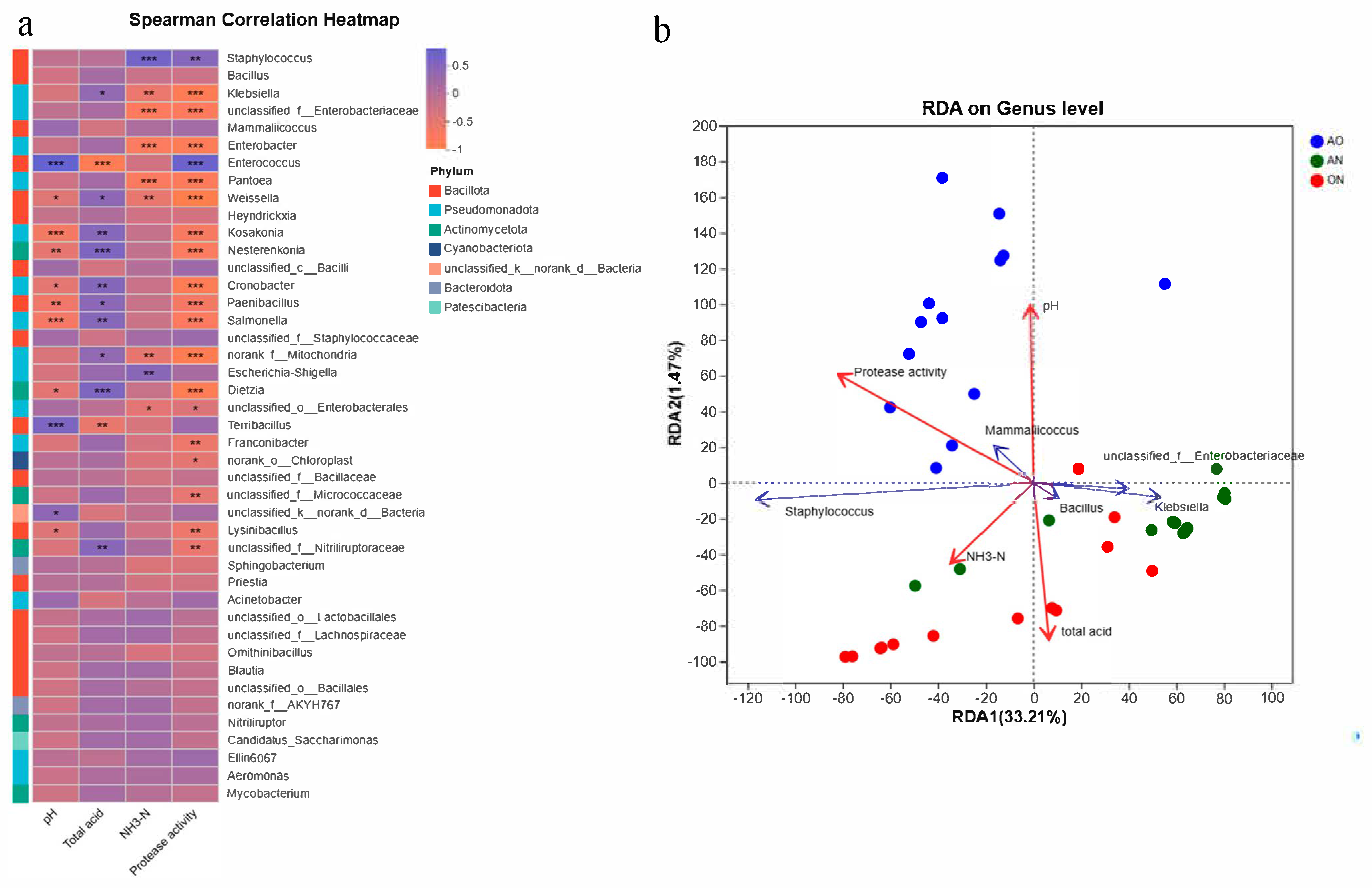
Figure 5 illustrated the correlation between microbial taxa and environmental factors, visually presenting both the magnitude and statistical significance of associations between multiple environmental parameters and various species. As shown in
Figure 5a,
Enterococcus,
Terribacillus, and unidentified
Bacteria exhibited significantly positive correlations with environmental pH, whereas
Kosakonia,
Nesterenkonia,
Cronobacter,
Paenibacillus,
Salmonella, and
Dietzia showed significantly negative correlations with pH. Conversely, their correlation patterns with total acidity demonstrated an inverse relationship. In fermentation systems, higher total acidity corresponded to lower pH values.
Enterococcus and
Terribacillus exhibited positive correlations with pH, potentially because their enzymatic systems demonstrated higher stability and activity in neutral to slightly alkaline environments [
37]. These pH conditions aligned with their physiological requirements, facilitating cellular growth and proliferation, thereby driving positive correlations. Conversely,
Kosakonia and
Nesterenkonia showed negative correlations with pH. Such taxa likely possessed acid-adaptation mechanisms, such as specialized proton pumps in cellular membranes that regulated intracellular pH homeostasis. Alternatively, their metabolic pathways (e.g., enzymes involved in anaerobic acid-producing fermentation) might have functioned more actively under acidic conditions. Acidic environments could have provided competitive advantages, consequently increasing their abundance in low-pH settings and thus exhibiting negative correlations.
NH3-N represented a crucial quality indicator in soy sauce fermentation. Staphylococcus and Escherichia-Shigella demonstrated increasing abundance with rising NH3-N levels, whereas Klebsiella, Enterobacteriaceae, Enterobacter, Pantoea, Weissella, and Mitochondria exhibited declining populations despite progressive accumulation of NH3-N during fermentation. Staphylococcus and Escherichia-Shigella possessed metabolic adaptations enabling resilience to fermentation environment shifts. As physicochemical parameters (e.g., pH, osmotic pressure) evolved throughout fermentation, these taxa-maintained functionality within tolerance thresholds. The increasing NH3-N availability synergized with their metabolic competencies, thereby promoting population expansion. Conversely, negatively correlated taxa likely possessed divergent nitrogen source preferences. They preferentially utilized alternative nitrogen forms rather than competing directly for ammonia nitrogen. When NH3-N concentrations increased, these organisms faced competitive exclusion by Staphylococcus and Escherichia-Shigella, resulting in insufficient nitrogen acquisition to sustain growth and consequent population decline.
Regarding protease activity, Staphylococcus and Enterococcus exhibited significantly positive correlations with protease activity levels. In contrast, Klebsiella, Enterobacteriaceae, Enterobacter, Pantoea, and Weissella demonstrated significantly negative correlations with protease activity. Staphylococcus and Enterococcus functioned as protease-producing bacteria that secreted extracellular proteases. These proteases might directly decompose fermentation substrates, thereby enhancing overall protease activity in the system. Certain negatively correlated taxa (e.g., Pantoea) secreted protease inhibitors or reduced enzymatic activity through intracellular metabolic consumption of environmental free proteases.
RDA analysis (
Figure 5b) clearly revealed differences in the associations between microorganisms and physicochemical indices among different fermentation groups (AO/AN/ON). In the AO group, the arrow of
Staphylococcus highly overlapped with sample points, and there was a small angle (positive correlation) between its arrow and those of protease activity and NH
3-N. This indicated that
Staphylococcus was the core taxon in the AO group, which promoted protein decomposition and increased NH
3-N content via high protease activity, thereby driving fermentation quality. In the AN group, the arrows of
Klebsiella and
Bacillus overlapped with sample points; there was a small angle (positive correlation) between their arrows and the pH arrow, and a large angle (negative correlation) with the total acid arrow. That is, these two genera were the core taxa in the AN group, which shaped the fermentation microenvironment by alkalizing the environment and consuming acids. In the ON group, the arrows of Enterobacteriaceae and
Mammaliicoccus overlapped with sample points; there was a small angle (positive correlation) between their arrows and the total acid arrow, and a large angle (negative correlation) with the pH arrow. This meant that taxa such as Enterobacteriaceae were the core taxa in the ON group, which influenced the fermentation process by producing acids, acidifying the environment, and reducing pH.
The arrow of protease activity was long and pointed to the AO group, indicating that it was mainly driven by Staphylococcus in the AO group, and single-strain fermentation with A. oryzae was more conducive to protease synthesis and protein decomposition. The small angle between the NH3-N content arrow and those of the AO group and Staphylococcus showed that its accumulation was directly related to the protein decomposition by Staphylococcus in the AO group, and fermentation with A. oryzae was the main factor increasing NH3-N content. The small angle between the total acid arrow and those of the ON group and Enterobacteriaceae indicated that its accumulation was mainly caused by Enterobacteriaceae in the ON group, and mixed fermentation increased total acid content via synergistic acid production.
3.5. Molecular Docking of Protease with Walnut Protein and Walnut Polyphenols
The template used for walnut protein modeling was Q2TPW5.1. A (11S globulin seed storage protein, derived from plants of the genus Juglans). The sequence identity between the target sequence and the template sequence was 100.00%, with a GMQE value of 0.85, indicating that the modeling result was highly reliable. The template used for modeling the neutral protease from A. oryzae was P46076.1.A (Neutral protease 2, derived from A. oryzae), with a GMQE value of 0.91. This indicated that the model had high reliability and could be used for subsequent structural and functional analyses. The acid protease from A. niger was modeled using A0A319DLK0.1.A (Aspergillopepsin, derived from Aspergillus ellipticus CBS 70779), with a GMQE value of 0.90, confirming successful modeling.
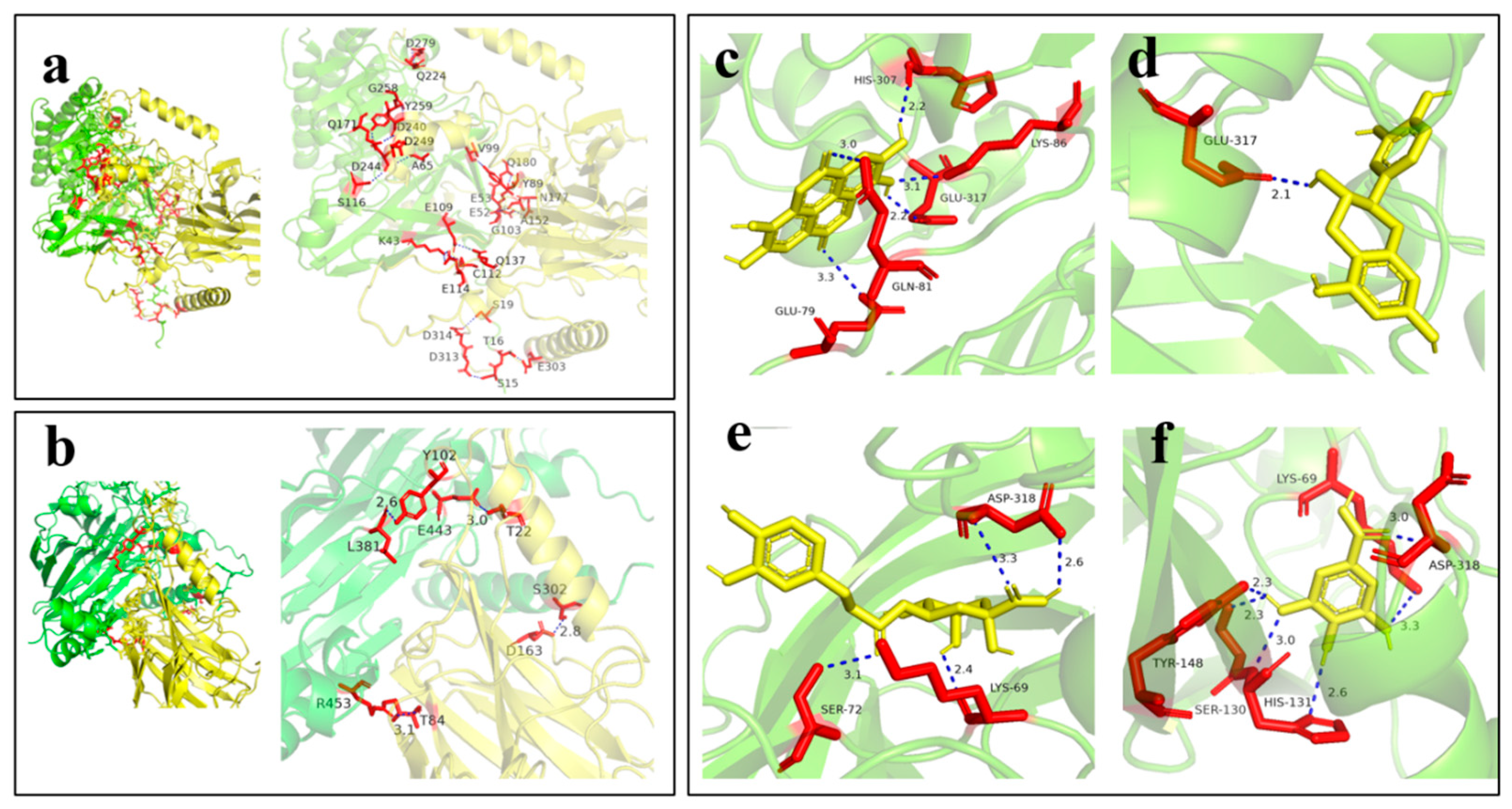
Molecular docking results showed that in the docking of neutral protease from
A. oryzae with walnut protein (
Figure 6a), the interaction sites involved numerous amino acids, with interactions distributed widely and forming a complex binding network. This indicated that its binding modes with walnut protein were diverse, and binding was achieved through the synergistic effect of multiple amino acids. The complex binding mode and extensive interaction sites might have caused the structure of walnut protein to be destroyed at multiple points, promoting the gradual hydrolysis of the protein into various small-molecule peptides and amino acids, which was conducive to the diverse production of flavor substances in soy sauce fermentation. For the acid protease from
A. niger (
Figure 6b), the docking involved relatively fewer amino acids, and the binding regions were relatively concentrated. This might have limited its hydrolytic effect on walnut protein, resulting in an overall fermentation rate significantly lower than that of the
A. oryzae group.
In this study, four relatively abundant polyphenols in walnuts—ellagic acid (EA), catechin (CAT), chlorogenic acid (CA), and gallic acid (GA)—were selected to dock with the neutral protease from
A. oryzae to investigate their effects on WSS fermentation. As shown in
Figure 6c–f, polyphenols bound tightly to the amino acids in the active center of the neutral protease from
A. oryzae through hydrogen bonds and hydrophobic interactions. The polyphenols occupied the substrate-binding sites in the active center of the protease, directly preventing the binding of walnut protein to the enzyme and inhibiting protein hydrolysis. Meanwhile, the multi-site interactions between polyphenols and the enzyme might have altered the conformation of the enzyme’s active center, reduced its catalytic efficiency, and decreased the production of flavor precursors such as amino acids and peptides.
Molecular docking results showed that there were differences in binding energies between four polyphenols (ellagic acid, catechin, chlorogenic acid, and gallic acid) and the neutral protease from
A. oryzae (AO). The binding energy data (
Table 2) indicated that the absolute value of the binding energy for AO-CA was the largest overall; for example, the data in Group 1 reached −8.27 kcal/mol, suggesting that its binding to the protease was more stable. The average binding energy for AO-EA was also relatively high, indicating strong binding stability. From the docking diagrams, taking AO-EA as an example (
Figure 6a), ellagic acid formed multiple sets of hydrogen bonds with amino acid residues such as HIS-307, LYS-86, and GLU-317 of the protease (e.g., a 2.2 Å hydrogen bond between HIS-307 and ellagic acid). These hydrogen bond interactions enabled the polyphenol to bind to the active center region of the protease, hindering the binding of the substrate to the enzyme. In AO-CA (
Figure 6c), chlorogenic acid formed hydrogen bonds with amino acids such as ASP-318 and LYS-69, which also generated steric hindrance to the active center of the protease. In summary, different polyphenols stably bound to the neutral protease from
A. oryzae through interactions such as hydrogen bonds formed with amino acid residues in the enzyme’s active center. The lower the binding energy, the more stable the binding, which more easily hindered the catalysis of the substrate by the protease, affected the hydrolysis of proteins during
A. oryzae fermentation, and thus might have altered the fermentation quality and efficiency of WSS.
In addition, Cheng et al. used molecular docking results in their research to reveal the mechanism by which polyphenols inhibit the activity of glycosidases and proteases through stable binding to the active centers of the enzymes. This inhibitory effect indirectly inhibited fungal growth and protease secretion by reducing nutrient supply to fungi [
38]. Makarewicz et al. pointed out that polyphenols could bind to the active centers of microorganism-secreted enzymes (e.g., proteases, glycosidases), altering enzyme conformation and inhibiting their activity. Meanwhile, they could also affect community structure and metabolic functions by selectively inhibiting or promoting the growth of specific microorganisms [
39]. This mechanism provided a theoretical basis for analyzing the inhibition of
A. oryzae/
A. niger protease activity by walnut polyphenols and the regulation of dynamic changes in microbial communities during WSS fermentation. Specifically, polyphenols in walnuts might have affected the catalytic efficiency of enzymes and the growth metabolism of microorganisms in the fermentation system through similar modes of action, thereby altering the fermentation process and quality characteristics of soy sauce.