Lactiplantibacillus plantarum Z45 from Sour Soup Improves Flavor and Safety of Fermented Corn: Insights from Genomic and Metabolomic Approaches
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Strains
2.2. Antibacterial Activity
2.3. Identification of Strains
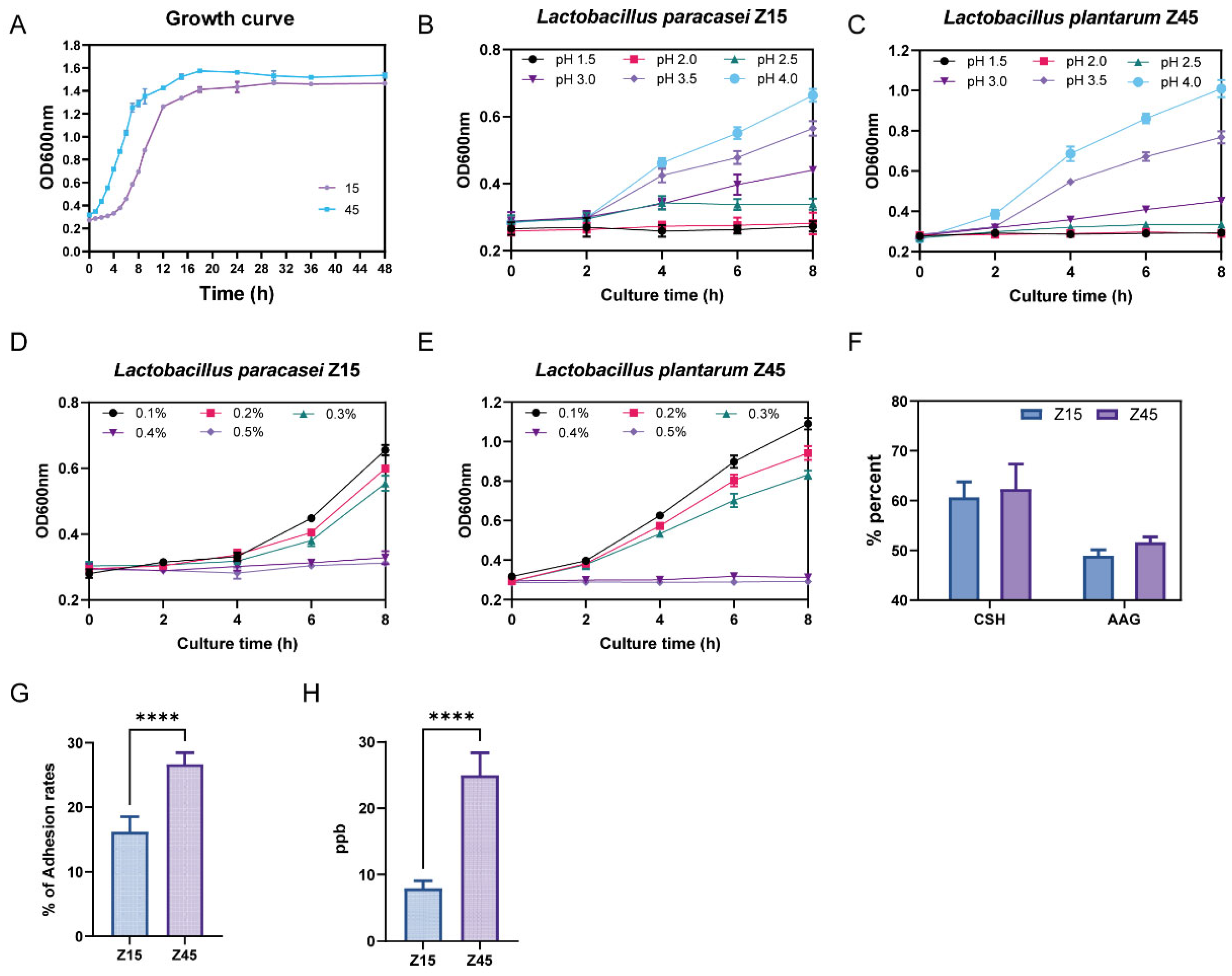
2.4. Growth Curves of Isolated Strains
2.5. Evaluation of Probiotic Potential
2.5.1. Antioxidant Activity
2.5.2. Bile Salt Resistance and Acid Resistance
2.5.3. Gastrointestinal Fluid Tolerance, Auto-Aggregation Activity and Cell Surface Hydrophobicity
2.5.4. Detection of Adhesion to Caco2 Cells and Folate Content
2.6. WGS and Annotation of Z45
2.7. In Vitro Evaluation
2.7.1. Haemolytic Activity
2.7.2. Antibiotic Susceptibility Assay
2.8. Experimental Design for Sour Soup Fermentation with Bacterial Strains
2.9. Sensory Evaluation, Safety Assessment and Physicochemical Parameters
2.9.1. Sensory Evaluation
2.9.2. Biogenic Amine Detection
2.9.3. Physicochemical Parameters
2.10. 16S rRNA Sequencing
2.11. Non-Targeted Metabolomics Analysis of Sour Soup
2.12. Statistical Analysis
3. Results and Discussion
3.1. Antibacterial Activity of Isolated Strains
3.2. Probiotic Potential Testing Results of the Strain
3.3. WGS and Annotation
3.4. In Vitro Safety Evaluation
3.5. Secondary Metabolites and Comparative Genomes of Z45
3.6. Folate Metabolic Pathway of Z45
3.7. Experimental Results of Sensory Evaluation, Safety Assessment, and Physicochemical Parameters
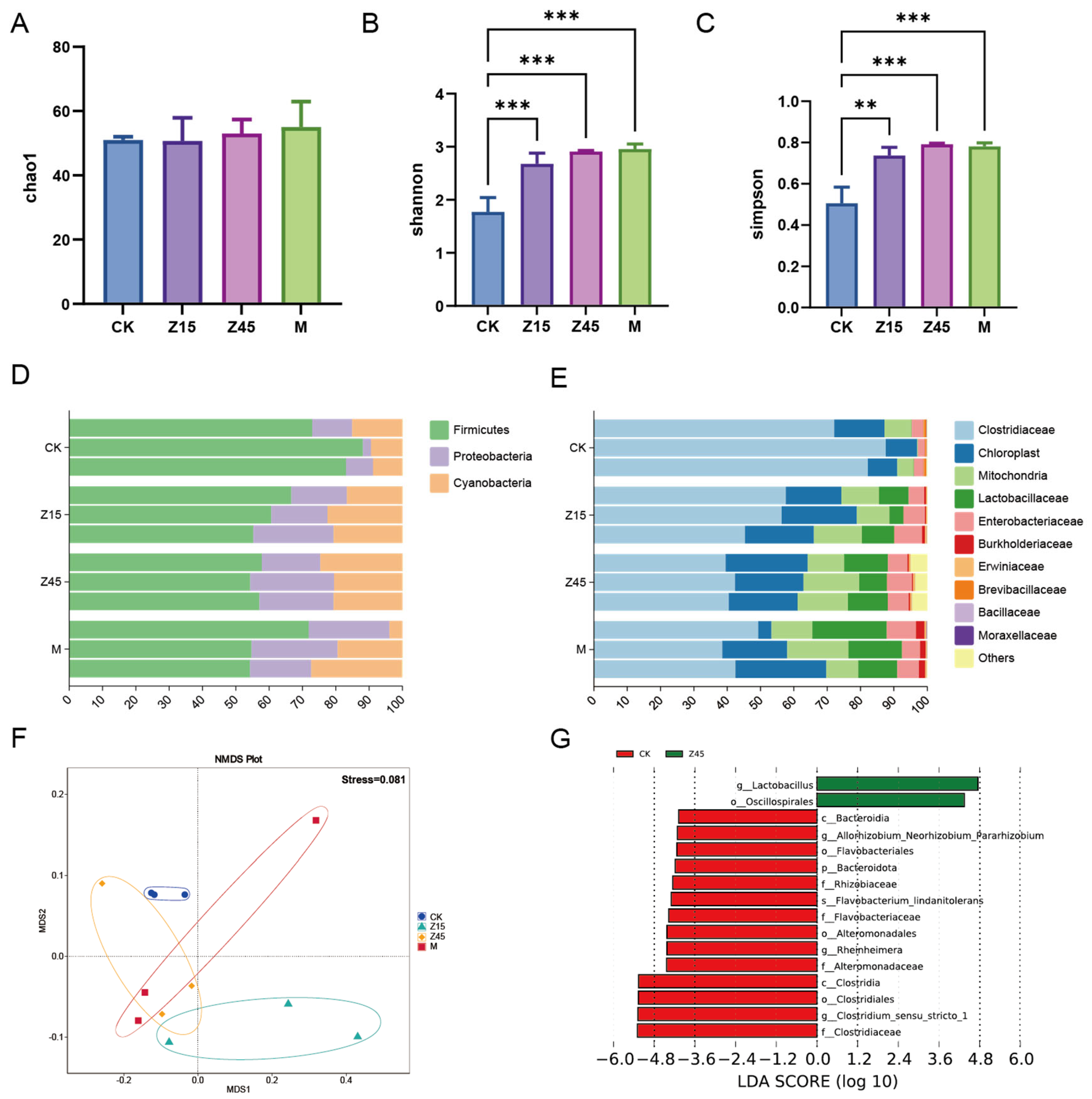
3.8. Bacterial Community Composition
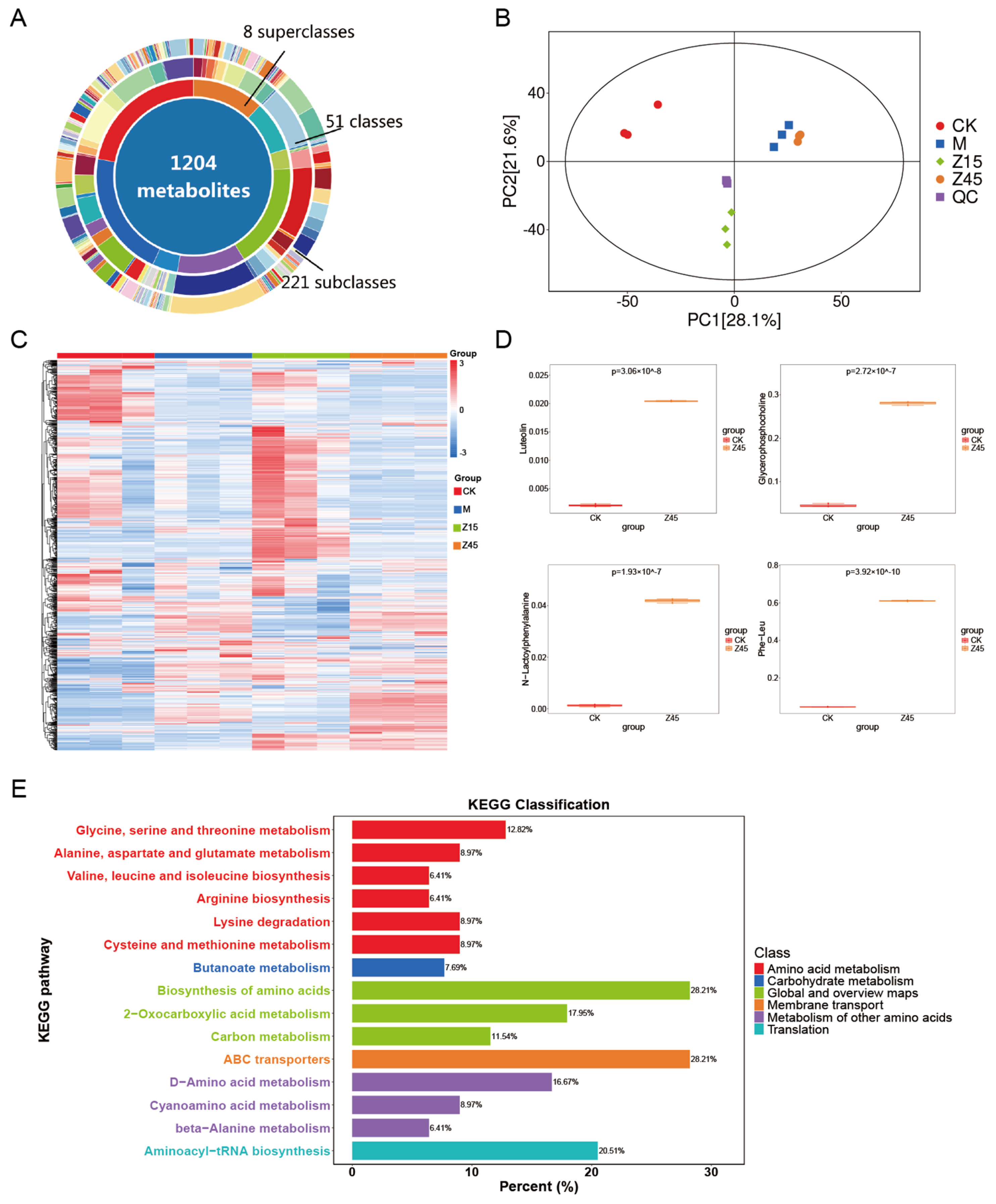
3.9. Non-Targeted Metabolome Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khan, M.T.; Dwibedi, C.; Sundh, D.; Pradhan, M.; Kraft, J.D.; Caesar, R.; Tremaroli, V.; Lorentzon, M.; Bäckhed, F. Synergy and oxygen adaptation for development of next-generation probiotics. Nature 2023, 620, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Pang, L.; Huang, Y.; Li, R.; Guo, L.; Man, C.; Yang, X.; Jiang, Y. Effects of postbiotics produced by Lactobacillus plantarum JM015 isolated from traditional fermented dairy products on Salmonella-induced intestinal inflammation: A preventive strategy. Food Chem. 2025, 469, 142549. [Google Scholar] [CrossRef] [PubMed]
- Mary Ellen, S.; Daniel, J.M.; Gregor, R.; Glenn, R.G.; Robert, A.R. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef]
- Ouarabi, L.; Chait, Y.A.; Seddik, H.A.; Drider, D.; Bendali, F. Newly Isolated Lactobacilli strains from Algerian Human Vaginal Microbiota: Lactobacillus fermentum Strains Relevant Probiotic’s Candidates. Probiotics Antimicrob. Proteins 2019, 11, 43–54. [Google Scholar] [CrossRef]
- Rajoka, M.S.R.; Zhao, H.; Lu, Y.; Lian, Z.; Li, N.; Hussain, N.; Shao, D.; Jin, M.; Li, Q.; Shi, J. Anticancer potential against cervix cancer (HeLa) cell line of probiotic Lactobacillus casei and Lactobacillus paracasei strains isolated from human breast milk. Food Funct. 2018, 9, 2705–2715. [Google Scholar] [CrossRef]
- Ilango, S.; Antony, U. Probiotic microorganisms from non-dairy traditional fermented foods. Trends Food Sci. Technol. 2021, 118, 617–638. [Google Scholar] [CrossRef]
- Song, W.; Yi, H.; Lu, F.; Deng, Y.; Zhu, M.; Wang, J.; Zhao, X.; Xiao, Z.; Zhang, Y. Correlation between microbial communities and flavor compounds in Suantangzi dough from Liaoning Province, China. Food Chem. 2025, 464, 141892. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, L.; Wen, R.; Chen, Q.; Kong, B. Role of lactic acid bacteria in flavor development in traditional Chinese fermented foods: A review. Crit. Rev. Food Sci. Nutr. 2022, 62, 2741–2755. [Google Scholar] [CrossRef]
- Yuan, Y.; Gao, R.; Liang, Q.; Song, L.; Huang, J.; Lang, N.; Zhou, J. A Foodborne Bongkrekic Acid Poisoning Incident—Heilongjiang Province, 2020. China CDC Wkly. 2020, 2, 975–978. [Google Scholar] [CrossRef]
- Stefanovic, E.; Fitzgerald, G.; McAuliffe, O. Advances in the genomics and metabolomics of dairy lactobacilli: A review. Food Microbiol. 2017, 61, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Kiousi, D.E.; Rathosi, M.; Tsifintaris, M.; Chondrou, P.; Galanis, A. Pro-biomics: Omics Technologies to Unravel the Role of Probiotics in Health and Disease. Adv. Nutr. 2021, 12, 1802–1820. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Metabolomics: Applications to food science and nutrition research. Trends Food Sci. Technol. 2008, 19, 482–493. [Google Scholar] [CrossRef]
- Zheng, X.; Liang, Q.; Zhao, B.; Song, X.; Zhang, Y. Whole genome sequencing and analysis of probiotic characteristics for Lactiplantibacillus plantarum EL2 isolated from yak yogurt. LWT 2024, 198, 116039. [Google Scholar] [CrossRef]
- Oh, J.-Y.; Chae, J.-C.; Han, J.-I.; Song, W.-K.; Lee, C.-M.; Park, H.-M. Distribution and epidemiological relatedness of methicillin-resistant Staphylococcus aureus isolated from companion dogs, owners, and environments. J. Vet. Med. Sci. 2020, 82, 1379–1386. [Google Scholar] [CrossRef]
- Zhao, M.; Zhang, Y.; Li, Y.; Liu, K.; Zhang, C.; Li, G. Complete Genome Sequence and Probiotic Properties of Pediococcus acidilactici CLP03 Isolated from Healthy Felis catus. Probiotics Antimicrob. Proteins 2023, 17, 903–917. [Google Scholar] [CrossRef]
- Shu, H.; He, X.; Hong, Z.; Dong, K.; Zou, Y.; Cao, M.; Wang, R.; Xu, Y.; Liao, L.; Zuo, H.; et al. Screening and genome analysis of potential probiotic lactic acid bacteria with broad-spectrum antibacterial activity from Sichuan sun-dried vinegar grains (Cupei). LWT 2024, 202, 116288. [Google Scholar] [CrossRef]
- Zhao, M.; Li, Y.; Zhang, Y.; Li, G. Genomic analysis and functional properties of Lactobacillus johnsonii GJ231 isolated from healthy beagles. Front. Microbiol. 2024, 15, 1437036. [Google Scholar] [CrossRef]
- Zhao, M.; Liu, K.; Zhang, Y.; Li, Y.; Zhou, N.; Li, G. Probiotic characteristics and whole-genome sequence analysis of Pediococcus acidilactici isolated from the feces of adult beagles. Front. Microbiol. 2023, 14, 1179953. [Google Scholar] [CrossRef]
- Adimpong, D.B.; Nielsen, D.S.; Sørensen, K.I.; Derkx, P.M.; Jespersen, L. Genotypic characterization and safety assessment of lactic acid bacteria from indigenous African fermented food products. BMC Microbiol. 2012, 12, 75. [Google Scholar] [CrossRef]
- Zhou, J.S.; Pillidge, C.J.; Gopal, P.K.; Gill, H.S. Antibiotic susceptibility profiles of new probiotic Lactobacillus and Bifidobacterium strains. Int. J. Food Microbiol. 2005, 98, 211–217. [Google Scholar] [CrossRef]
- Zhao, M.; Zhang, Y.; Li, Y.; Li, G. Developing Gut-Healthy Strains for Pets: Probiotic Potential and Genomic Insights of Canine-Derived Lactobacillus acidophilus GLA09. Microorganisms 2025, 13, 350. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Q.; He, L.; Li, C. Determination of the microbial communities of Guizhou Suantang, a traditional Chinese fermented sour soup, and correlation between the identified microorganisms and volatile compounds. Food Res. Int. 2020, 138, 109820. [Google Scholar] [CrossRef]
- Zhao, M.; Zhang, Y.; Li, Y.; Liu, K.; Bao, K.; Li, G. Impact of Pediococcus acidilactici GLP06 supplementation on gut microbes and metabolites in adult beagles: A comparative analysis. Front. Microbiol. 2024, 15, 1369402. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hu, D.; Chen, Y.; Huang, H.; Zhang, H.; Zhao, J.; Gu, Z.; Chen, W. Strain-specific properties of Lactobacillus plantarum for prevention of Salmonella infection. Food Funct. 2018, 9, 3673–3682. [Google Scholar] [CrossRef] [PubMed]
- Ngamsomchat, A.; Kaewkod, T.; Konkit, M.; Tragoolpua, Y.; Bovonsombut, S.; Chitov, T. Characterisation of Lactobacillus plantarum of Dairy-Product Origin for Probiotic Chèvre Cheese Production. Foods 2022, 11, 934. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Wang, J. Oxidative stress tolerance and antioxidant capacity of lactic acid bacteria as probiotic: A systematic review. Gut Microbes 2020, 12, 1801944. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, D.; Zhao, S.; Huang, Y.; Yu, J.; Zhou, Q. Assessing the safety and probiotic characteristics of Bacillus coagulans 13002 based on complete genome and phenotype analysis. LWT 2022, 155, 112847. [Google Scholar] [CrossRef]
- Bezkorovainy, A. Probiotics: Determinants of survival and growth in the gut. Am. J. Clin. Nutr. 2001, 73, 399S–405S. [Google Scholar] [CrossRef]
- Lu, Y.; Liang, W.; Wang, R.; Liang, Q.; Zeng, X.-A.; Huang, Y. Assessment of the safety and probiotic properties of Lactiplantibacillus plantarum HYY-DB9 based on comprehensive genomic and phenotypic analysis. LWT 2024, 203, 116386. [Google Scholar] [CrossRef]
- Marco, M.L.; Pavan, S.; Kleerebezem, M. Towards understanding molecular modes of probiotic action. Curr. Opin. Biotechnol. 2006, 17, 204–210. [Google Scholar] [CrossRef]
- Zibaei-Rad, A.; Rahmati-Joneidabad, M.; Alizadeh Behbahani, B.; Taki, M. Assessing the protection mechanisms on Enterobacter aerogenes ATCC 13048 by potentially probiotic strain Lacticaseibacillus casei XN18: An experimental and modeling study. Microb. Pathog. 2023, 181, 106177. [Google Scholar] [CrossRef] [PubMed]
- Shivani, T.M.; Sathiavelu, M. Probiotic evaluation, adherence capability and safety assessment of Lactococcus lactis strain isolated from an important herb “Murraya koenigii". Sci. Rep. 2024, 14, 15565. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh Behbahani, B.; Noshad, M.; Falah, F. Inhibition of Escherichia coli adhesion to human intestinal Caco-2 cells by probiotic candidate Lactobacillus plantarum strain L15. Microb. Pathog. 2019, 136, 103677. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Halami, P.M.; Tamang, J.P. Genome Analysis of Lactobacillus plantarum Isolated From Some Indian Fermented Foods for Bacteriocin Production and Probiotic Marker Genes. Front. Microbiol. 2020, 11, 40. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef]
- Liu, Y.; Tempelaars, M.H.; Boeren, S.; Alexeeva, S.; Smid, E.J.; Abee, T. Extracellular vesicle formation in Lactococcus lactis is stimulated by prophage-encoded holin-lysin system. Microb. Biotechnol. 2022, 15, 1281–1295. [Google Scholar] [CrossRef]
- Wu, J.-J.; Zhou, Q.-Y.; Liu, D.-M.; Xiong, J.; Liang, M.-H.; Tang, J.; Xu, Y.-Q. Evaluation of the safety and probiotic properties of Lactobacillus gasseri LGZ1029 based on whole genome analysis. LWT 2023, 184, 114759. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, D.; Jia, X.; Liang, M.; Lu, Y.; Liu, J. Whole genome sequencing of Lactobacillus plantarum DMDL 9010 and its effect on growth phenotype under nitrite stress. LWT 2021, 149, 111778. [Google Scholar] [CrossRef]
- Liu, D.M.; Huang, Y.Y.; Liang, M.-H. Analysis of the probiotic characteristics and adaptability of Lactiplantibacillus plantarum DMDL 9010 to gastrointestinal environment by complete genome sequencing and corresponding phenotypes. LWT 2022, 158, 113129. [Google Scholar] [CrossRef]
- Lu, J.; Mao, Y.; Ma, T.; Liu, X.; Cheng, X.; Bai, Y.; Li, S. Screening and genome analysis of lactic acid bacteria with high exopolysaccharide production and good probiotic properties. Food Biosci. 2023, 56, 103211. [Google Scholar] [CrossRef]
- Ardèvol, A.; Rovira, C. Reaction Mechanisms in Carbohydrate-Active Enzymes: Glycoside Hydrolases and Glycosyltransferases. Insights from ab Initio Quantum Mechanics/Molecular Mechanics Dynamic Simulations. J. Am. Chem. Soc. 2015, 137, 7528–7547. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Zhang, X.; Sui, S.; Ye, F.; Dai, J. Exploring and applying the substrate promiscuity of a C-glycosyltransferase in the chemo-enzymatic synthesis of bioactive C-glycosides. Nat. Commun. 2020, 11, 5162. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, Y.; Sun, M.; Zhang, H.; Mu, G.; Tuo, Y. Physiological function analysis of Lactobacillus plantarum Y44 based on genotypic and phenotypic characteristics. J. Dairy Sci. 2020, 103, 5916–5930. [Google Scholar] [CrossRef] [PubMed]
- Koskenniemi, K.; Laakso, K.; Koponen, J.; Kankainen, M.; Greco, D.; Auvinen, P.; Savijoki, K.; Nyman, T.A.; Surakka, A.; Salusjärvi, T.; et al. Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol. Cell. Proteom. 2011, 10, S1–S18. [Google Scholar] [CrossRef]
- Li, J.; Mu, G.; Tuo, Y. Phenotypic Traits and Probiotic Functions of Lactiplantibacillus plantarum Y42 in Planktonic and Biofilm Forms. Foods 2023, 12, 1516. [Google Scholar] [CrossRef]
- Bove, P.; Capozzi, V.; Garofalo, C.; Rieu, A.; Spano, G.; Fiocco, D. Inactivation of the ftsH gene of Lactobacillus plantarum WCFS1: Effects on growth, stress tolerance, cell surface properties and biofilm formation. Microbiol. Res. 2012, 167, 187–193. [Google Scholar] [CrossRef]
- Zschiedrich, C.P.; Keidel, V.; Szurmant, H. Molecular Mechanisms of Two-Component Signal Transduction. J. Mol. Biol. 2016, 428, 3752–3775. [Google Scholar] [CrossRef]
- Zou, F.; Huo, Y.; Gao, W.; Dai, M.; Zhao, G.; Zhang, S. Physicochemical, microbiological and sensory characterization of yogurt fermented by Weissella confusa SW1 and traditional starters. LWT 2024, 201, 116229. [Google Scholar] [CrossRef]
- Jantama, S.S.; Pichayajittipong, P.; Maneewong, R.Y.; Cheng, K.-C.; Jantama, K. Transcriptional analysis of oxidative-tolerant and temperature-sensitive genes of Bifidobacterium animalis BF052 during freeze-drying process and development of its soymilk-synbiotic product containing banana and jicama powders. Food Res. Int. 2025, 221, 117600. [Google Scholar] [CrossRef]
- Bergagnini, I.; Hmoud, H.; Nocerino, A.; Ivanina, E. S1804 Gut Friend or Foe: A Case of Lactobacillus Rhamnosus Endocarditis from Probiotic Use. Am. J. Gastroenterol. 2020, 115, S933. [Google Scholar] [CrossRef]
- Li, T.; Wang, Z.; Guo, J.; de la Fuente-Nunez, C.; Wang, J.; Han, B.; Tao, H.; Liu, J.; Wang, X. Bacterial resistance to antibacterial agents: Mechanisms, control strategies, and implications for global health. Sci. Total Environ. 2023, 860, 160461. [Google Scholar] [CrossRef] [PubMed]
- Méhi, O.; Bogos, B.; Csörgő, B.; Pál, F.; Nyerges, A.; Papp, B.; Pál, C. Perturbation of iron homeostasis promotes the evolution of antibiotic resistance. Mol. Biol. Evol. 2014, 31, 2793–2804. [Google Scholar] [CrossRef] [PubMed]
- Gueimonde, M.; Sánchez, B.; de los Reyes-Gavilán, C.G.; Margolles, A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013, 4, 202. [Google Scholar] [CrossRef]
- Anisimova, E.A.; Yarullina, D.R. Antibiotic Resistance of LACTOBACILLUS Strains. Curr. Microbiol. 2019, 76, 1407–1416. [Google Scholar] [CrossRef]
- Katla, A.-K.; Kruse, H.; Johnsen, G.; Herikstad, H. Antimicrobial susceptibility of starter culture bacteria used in Norwegian dairy products. Int. J. Food Microbiol. 2001, 67, 147–152. [Google Scholar] [CrossRef]
- Okoye, C.O.; Dong, K.; Wang, Y.; Gao, L.; Li, X.; Wu, Y.; Jiang, J. Comparative genomics reveals the organic acid biosynthesis metabolic pathways among five lactic acid bacterial species isolated from fermented vegetables. New Biotechnol. 2022, 70, 73–83. [Google Scholar] [CrossRef]
- Álvarez-Martínez, F.J.; Barrajón-Catalán, E.; Herranz-López, M.; Micol, V. Antibacterial plant compounds, extracts and essential oils: An updated review on their effects and putative mechanisms of action. Phytomedicine 2021, 90, 153626. [Google Scholar] [CrossRef]
- Perricone, M.; Arace, E.; Corbo, M.R.; Sinigaglia, M.; Bevilacqua, A. Bioactivity of essential oils: A review on their interaction with food components. Front. Microbiol. 2015, 6, 76. [Google Scholar] [CrossRef]
- Saini, R.K.; Nile, S.H.; Keum, Y.-S. Folates: Chemistry, analysis, occurrence, biofortification and bioavailability. Food Res. Int. 2016, 89, 1–13. [Google Scholar] [CrossRef]
- Liu, C.-J.; Wu, B.; Zhang, S.-Y.; Li, Q.-K.; Zeng, X.-Q.; Yang, E.; Luo, Y.-Y.; Li, X.-R. Transcriptomic analysis of de novo folate biosynthetic genes in Lactobacillus plantarum strain 4_3 in fermented soybean. Food Funct. 2019, 10, 2426–2438. [Google Scholar] [CrossRef]
- An, F.; Wu, J.; Feng, Y.; Pan, G.; Ma, Y.; Jiang, J.; Yang, X.; Xue, R.; Wu, R.; Zhao, M. A systematic review on the flavor of soy-based fermented foods: Core fermentation microbiome, multisensory flavor substances, key enzymes, and metabolic pathways. Compr. Rev. Food Sci. Food Saf. 2023, 22, 2773–2801. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, L.; Cai, J.; Yang, T.; Li, J.; Shu, G. Characterization of fermented pomegranate juice: ACE inhibitory activity under in vitro digestion, antioxidant capacity, phenolics composition, chemical properties and sensory evaluation. Food Sci. Biotechnol. 2024, 33, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Spano, G.; Russo, P.; Lonvaud-Funel, A.; Lucas, P.; Alexandre, H.; Grandvalet, C.; Coton, E.; Coton, M.; Barnavon, L.; Bach, B.; et al. Biogenic amines in fermented foods. Eur. J. Clin. Nutr. 2010, 64, S95–S100. [Google Scholar] [CrossRef] [PubMed]
- Shiling, L.; Caihong, J.; Xinglian, X.; Chengjian, X.; Kaixiong, L.; Ruihua, S. Improved screening procedure for biogenic amine production by lactic acid bacteria and Enterobacteria. Czech J. Food Sci. 2015, 33, 19–26. [Google Scholar] [CrossRef]
- Capozzi, V.; Russo, P.; Ladero, V.; Fernandez, M.; Fiocco, D.; Alvarez, M.A.; Grieco, F.; Spano, G. Biogenic Amines Degradation by Lactobacillus plantarum: Toward a Potential Application in Wine. Front. Microbiol. 2012, 3, 122. [Google Scholar] [CrossRef]
- Zhang, Q.; Lin, S.; Nie, X. Reduction of biogenic amine accumulation in silver carp sausage by an amine-negative Lactobacillus plantarum. Food Control 2013, 32, 496–500. [Google Scholar] [CrossRef]
- Li, H.; Huang, J.; Wang, Y.; Wang, X.; Ren, Y.; Yue, T.; Wang, Z.; Gao, Z. Study on the nutritional characteristics and antioxidant activity of dealcoholized sequentially fermented apple juice with Saccharomyces cerevisiae and Lactobacillus plantarum fermentation. Food Chem. 2021, 363, 130351. [Google Scholar] [CrossRef]
- Yang, S.; Hou, M.; Tan, W.; Chen, Y.; Li, H.; Song, J.; Wang, X.; Ren, J.; Gao, Z. Lactic acid bacteria sequential fermentation improves viable counts and quality of fermented apple juice via generating two logarithmic phases. Food Chem. 2025, 464, 141635. [Google Scholar] [CrossRef]
- Song, H.S.; Lee, S.H.; Ahn, S.W.; Kim, J.Y.; Rhee, J.-K.; Roh, S.W. Effects of the main ingredients of the fermented food, kimchi, on bacterial composition and metabolite profile. Food Res. Int. 2021, 149, 110668. [Google Scholar] [CrossRef]
- Song, H.S.; Whon, T.W.; Kim, J.; Lee, S.H.; Kim, J.Y.; Kim, Y.B.; Choi, H.-J.; Rhee, J.-K.; Roh, S.W. Microbial niches in raw ingredients determine microbial community assembly during kimchi fermentation. Food Chem. 2020, 318, 126481. [Google Scholar] [CrossRef] [PubMed]
- Palmnäs-Bédard, M.; de Santa Izabel, A.; Dicksved, J.; Landberg, R. Characterization of the Bacterial Composition of 47 Fermented Foods in Sweden. Foods 2023, 12, 3827. [Google Scholar] [CrossRef] [PubMed]
- López-Sánchez, R.; Hernández-Oaxaca, D.; Escobar-Zepeda, A.; Ramos Cerrillo, B.; López-Munguía, A.; Segovia, L. Analysing the dynamics of the bacterial community in pozol, a Mexican fermented corn dough. Microbiology 2023, 169, 001355. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, Y.; Wen, Z.; Liu, W.; Meng, L.; Huang, H. Oscillospira—A candidate for the next-generation probiotics. Gut Microbes 2021, 13, 1987783. [Google Scholar] [CrossRef]
- Shang, Q.; Liu, S.; Liu, H.; Mahfuz, S.; Piao, X. Impact of sugar beet pulp and wheat bran on serum biochemical profile, inflammatory responses and gut microbiota in sows during late gestation and lactation. J. Anim. Sci. Biotechnol. 2021, 12, 54. [Google Scholar] [CrossRef]
- Imran, M.; Rauf, A.; Abu-Izneid, T.; Nadeem, M.; Shariati, M.A.; Khan, I.A.; Imran, A.; Orhan, I.E.; Rizwan, M.; Atif, M.; et al. Luteolin, a flavonoid, as an anticancer agent: A review. Biomed. Pharmacother. 2019, 112, 108612. [Google Scholar] [CrossRef]
- Lien, E.J.; Ren, S.; Bui, H.-H.; Wang, R. Quantitative structure-activity relationship analysis of phenolic antioxidants. Free Radic. Biol. Med. 1999, 26, 285–294. [Google Scholar] [CrossRef]
- Bi, F.; Qin, Y.; Chen, D.; Kan, J.; Liu, J. Development of active packaging films based on chitosan and nano-encapsulated luteolin. Int. J. Biol. Macromol. 2021, 182, 545–553. [Google Scholar] [CrossRef]
- Punia Bangar, S.; Kajla, P.; Chaudhary, V.; Sharma, N.; Ozogul, F. Luteolin: A flavone with myriads of bioactivities and food applications. Food Biosci. 2023, 52, 102366. [Google Scholar] [CrossRef]
- Liu, J.Y.; Zheng, R.Q.; Wang, Y.; Liu, Y.H.; Jiang, S.; Wang, X.-Z.; He, K.; Pan, X.; Zhou, T.; Li, T.; et al. The Endogenous Metabolite Glycerophosphocholine Promotes Longevity and Fitness in Caenorhabditis elegans. Metabolites 2022, 12, 177. [Google Scholar] [CrossRef]
- Syme, C.; Czajkowski, S.; Shin, J.; Abrahamowicz, M.; Leonard, G.; Perron, M.; Richer, L.; Veillette, S.; Gaudet, D.; Strug, L.; et al. Glycerophosphocholine Metabolites and Cardiovascular Disease Risk Factors in Adolescents. Circulation 2016, 134, 21. [Google Scholar] [CrossRef]
- Wu, J.; Gao, J.; Lin, J.; Cui, C.; Li, L.; He, S.; Brennan, C. Preparation and Taste Characteristics of Kokumi N-Lactoyl Phenylalanine in the Presence of Phenylalanine and Lactate. J. Agric. Food Chem. 2022, 70, 5396–5407. [Google Scholar] [CrossRef]
- Li, V.L.; He, Y.; Contrepois, K.; Liu, H.; Kim, J.T.; Wiggenhorn, A.L.; Tanzo, J.T.; Tung, A.S.-H.; Lyu, X.; Zushin, P.-J.H.; et al. An exercise-inducible metabolite that suppresses feeding and obesity. Nature 2022, 606, 785–790. [Google Scholar] [CrossRef]

| Items | Simulated Gastric Fluid (logCFU/mL) | Survival (%) | Simulated Intestinal Fluid (logCFU/mL) | Survival (%) | Artificial Gastrointestinal Fluid (logCFU/mL) | Survival (%) | ||
|---|---|---|---|---|---|---|---|---|
| 0 h | 3 h | 0 h | 3 h | 6 h | ||||
| Z15 | 7.05 ± 0.10 | 6.17 ± 0.17 b | 87.45 ± 1.60 b | 8.02 ± 0.20 | 7.83 ± 0.14 a | 97.60 ± 0.72 a | 6.02 ± 0.12 | 85.43 ± 0.64 |
| Z45 | 7.25 ± 0.14 | 7.06 ± 0.03 a | 97.37 ± 1.56 a | 8.08 ± 0.06 | 7.28 ± 0.04 b | 90.14 ± 0.17 b | 6.08 ± 0.03 | 83.87 ± 1.83 |
| VFDB ID | Description | Gene | Virulence Factor |
|---|---|---|---|
| VFG000964 (gb|WP_010922799) | UTP-glucose-1-phosphate uridylyltransferase | HasC | Immune modulation |
| VFG002190 (gb|WP_002362225) | undecaprenyl diphosphate synthase | Cpsa/Upps | |
| VFG048830 (gb|WP_014907233) | NADP-dependent phosphogluconate dehydrogenase | GndA | |
| VFG046465 (gb|WP_003028672) | elongation factor Tu | TufA | Adherence |
| VFG012095 (gb|WP_003435012) | chaperonin GroEL | GroEL | |
| VFG037100 (gb|WP_010980745) | trifunctional thioredoxin/methionine sulfoxide reductase A/B protein | Msra | Stress |
| VFG000077 (gb|NP_465991) | ATP-dependent Clp protease proteolytic subunit | ClpP | |
| VFG000080 (gb|NP_464522) | ATP-dependent protease | ClpE |
| Genes Detected in Z45 | ID Number | Predicted Function |
|---|---|---|
| ATP synthase subunit a | 00997 | Acid tolerance |
| ATP synthase subunit b | 00999 | |
| ATP synthase subunit c | 00998 | |
| ATP synthase subunit alpha | 01001 | |
| ATP synthase subunit gamma | 01002 | |
| ATP synthase subunit beta | 01003 | |
| ATP synthase subunit epsilon | 01004 | |
| ATP synthase subunit delta | 01000 | |
| Na+/H+ antiporter napA | 00663 | |
| Na+/H+ antiporter nhaC | 00168/02830 | |
| Phosphotransferase system cellobiose-specific EIIB component (celA) | 00673/01861 | |
| L-lactate dehydrogenase | 01212 | |
| L-lactate permease | 01495 | |
| ATP-dependent Clp protease ATP-binding subunit ClpX | 01242 | |
| ATP-dependent Clp protease proteolytic subunit ClpP | 01908 | |
| ATP-dependent Clp protease ATP-binding subunit ClpE | 02036 | |
| ATP-dependent Clp protease ATP-binding subunit ClpC | 02257 | |
| Glucose-6-phosphate isomerase | 00895 | |
| Pyruvate kinase | 01430 | |
| Glucosamine-6-phosphate deaminase | 00197 | Acid/Bile tolerance |
| Cyclopropane-fatty-acyl-phospholipid synthase (CfA) | 00407/01598 | |
| Manganese-dependent inorganic pyrophosphatase (ppaC) | 01478 | |
| Alkaline shock protein | 02335 | |
| Putative universal stress protein | 00498/00590/00707/00787/01022/01557/01593/01983/02081 | |
| ABC-type transporter ATP-binding protein EcsA | 01641/01812 | |
| General stress protein (YugL) | 01117 | |
| Heat-inducible transcriptional repressor (HrcA) | 01330 | Heat shock defense |
| Molecular chaperone Hsp31 and glyoxalase (HchA) | 00068 | |
| Chaperone protein (DnaJ) | 01333 | |
| Chaperone protein (DnaK) | 01332 | |
| Chaperone protein (ClpB) | 01425 | |
| Molecular chaperone (GrpE) | 01331 | |
| Cold shock protein (Csp) | 02145 | |
| Cold shock protein (CspL) | 00029 | |
| Cold shock-like protein CspLA | 02280 | |
| ATP-dependent RNA helicase DeaD | 02143 | |
| DEAD-box ATP-dependent RNA helicase CshB | 01075 | |
| S-ribosylhomocysteine lyase luxS | 02472 | Biofilm formation |
| Catabolite control protein A ccpA | 00146/01095 | |
| Catabolite control protein B ccpB | 00521/00820 | |
| Family DNA-binding protein ComEA | 01227 | |
| DNA internalization-related competence protein ComEC | 01229 | |
| Biofilm regulatory protein A (BrpA) | 00256 | |
| Two-component system WalR/WalK regulatory protein (YycL) | 00035 | |
| Hydrogen peroxide-inducible genes activator OxyR | 00543/00755/02182 | Protection against peroxids |
| Organic hydroperoxide resistance transcriptional regulator (OhrR) | 02368 | |
| Peroxide operon regulator (PerR) | 02758 | |
| Putative oxidoreductase/MSMEI_2347 | 00113/00655/02869 | Protection against Hydroxyl radicals |
| Putative oxidoreductase | 00119/00615/01269 | |
| Putative oxidoreductase (YghA) | 00466 | |
| Putative oxidoreductase (YhhX) | 00816 | |
| Putative oxidoreductase (YjmC) | 02208 | |
| Putative oxidoreductase (YceM) | 02435 | |
| Putative FAD-linked oxidoreductase | 00257 | |
| Quinone oxidoreductase (QorB) | 02748/02751 | |
| Oxidoreductase YdhF | 02817 | |
| Thiol peroxidase Tpx | 01042 | Oxidative stress |
| Thioredoxin TrxA | 00205/01082/02904 | |
| Thioredoxin-like protein YtpP | 00798 | |
| Thioredoxin reductase TrxB | 02483 | |
| Hsp33 family molecular chaperone HslO | 02676 | |
| I-methionine (R)-S-oxide reductase MsrC | 01037 | |
| Peptide-methionine (S)-S-oxide reductase MsrA | 01354/01480/01969 | |
| Peptide-methionine (R)-S-oxide reductase MsrB | 01479 | |
| NADH peroxidase NpR | 00858/01822 | |
| NADH oxidase NoX | 01387/02478/02484/02914 | |
| Elongation factor Tu (TuF) | 01240 | Adhesion and aggregation |
| Pyruvate dehydrogenase El component beta (pdhB) | 01209 | |
| Enolase (eno) | 01408/01410/02456 | |
| Glyceraldehyde-3-phosphate dehydrogenase GapA | 02459 | |
| Triosephosphate isomerase TpiA | 02457 |
| Antimicrobial Classes | Antimicrobial Agents | Disk Dose (μg) | Inhibition Zone Diameters/mm (IZD) a |
|---|---|---|---|
| β-lactams antibiotics | Penicillin | 10 | S |
| Ampicillin | 10 | I | |
| Imipenem | 10 | S | |
| Aminoglycosides antibiotics | Gentamicin | 10 | R |
| Tetracyclines | Minocycline | 30 | S |
| Doxycycline | 30 | S | |
| Chloramphenicol | 30 | S |
| Items | Appearance | Odor | Acidity | Hardness | Viscosity | Elasticity | Overall Score |
|---|---|---|---|---|---|---|---|
| CK | 10.75 ± 1.86 a | 13.92 ± 1.62 a | 21.42 ± 2.54 a | 7.33 ± 0.89 a | 7.50 ± 0.80 | 12.17 ± 1.11 | 73.08 ± 4.29 a |
| Z15 | 11.92 ± 1.00 b | 14.83 ± 2.48 ab | 21.50 ± 2.32 a | 7.50 ± 0.80 a | 7.75 ± 1.06 | 12.75 ± 0.97 | 76.25 ± 4.63 b |
| Z45 | 13.42 ± 0.90 c | 16.25 ± 1.29 b | 23.67 ± 1.30 b | 8.42 ± 0.79 b | 8.33 ± 0.89 | 13.08 ± 1.08 | 83.17 ± 3.04 c |
| M | 11.92 ± 1.00 b | 15.08 ± 0.67 ab | 22.33 ± 1.23 ab | 7.92 ± 0.67 ab | 7.83 ± 0.58 | 12.58 ± 1.00 | 77.67 ± 2.50 b |
| Items | Content | |||
|---|---|---|---|---|
| CK | Z15 | Z45 | M | |
| pH | 3.87 ± 0.05 a | 3.80 ± 0.04 ab | 3.70 ± 0.09 b | 3.83 ± 0.03 a |
| TA (g/L) | 10.33 ± 0.50 a | 11.70 ± 0.56 b | 12.37 ± 0.55 b | 11.53 ± 0.50 b |
| Viable count (log CFU/mL) | 7.57 ± 0.25 a | 7.99 ± 0.18 b | 8.15 ± 0.13 b | 8.03 ± 0.21 b |
| Putrescine (mg/kg) | 17.05 ± 1.88 a | 12.04 ± 1.70 b | 4.12 ± 0.63 d | 8.23 ± 0.21 c |
| Tryptamine (mg/kg) | 17.87 ± 1.11 a | 16.51 ± 1.31 a | 2.99 ± 0.22 c | 11.11 ± 1.17 b |
| Phenylethylamine (mg/kg) | 5.31 ± 0.54 a | 0.48 ± 0.07 b | 0.24 ± 0.04 b | 0.42 ± 0.07 b |
| Cadaverine (mg/kg) | 8.55 ± 1.04 a | 0.51 ± 0.05 b | 0.45 ± 0.08 b | 0.45 ± 0.07 b |
| Histamine (mg/kg) | 3.94 ± 0.36 a | 3.80 ± 0.31 a | 0.26 ± 0.08 c | 0.26 ± 0.08 b |
| Tyramine (mg/kg) | 4.73 ± 0.59 a | 4.25 ± 0.90 a | 0.45 ± 0.09 c | 2.70 ± 0.46 b |
| Spermidine (mg/kg) | 0.28 ± 0.07 | 0.26 ± 0.05 | 0.16 ± 0.04 | 2.73 ± 0.32 |
| Spermine (mg/kg) | 0.13 ± 0.05 ab | 0.21 ± 0.05 a | 0.08 ± 0.03 b | 0.16 ± 0.05 ab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, M.; Zhang, Y.; Wu, Y.; Liang, S.; Li, G. Lactiplantibacillus plantarum Z45 from Sour Soup Improves Flavor and Safety of Fermented Corn: Insights from Genomic and Metabolomic Approaches. Foods 2025, 14, 3803. https://doi.org/10.3390/foods14213803
Zhao M, Zhang Y, Wu Y, Liang S, Li G. Lactiplantibacillus plantarum Z45 from Sour Soup Improves Flavor and Safety of Fermented Corn: Insights from Genomic and Metabolomic Approaches. Foods. 2025; 14(21):3803. https://doi.org/10.3390/foods14213803
Chicago/Turabian StyleZhao, Mengdi, Yuanyuan Zhang, Yi Wu, Shuang Liang, and Guangyu Li. 2025. "Lactiplantibacillus plantarum Z45 from Sour Soup Improves Flavor and Safety of Fermented Corn: Insights from Genomic and Metabolomic Approaches" Foods 14, no. 21: 3803. https://doi.org/10.3390/foods14213803
APA StyleZhao, M., Zhang, Y., Wu, Y., Liang, S., & Li, G. (2025). Lactiplantibacillus plantarum Z45 from Sour Soup Improves Flavor and Safety of Fermented Corn: Insights from Genomic and Metabolomic Approaches. Foods, 14(21), 3803. https://doi.org/10.3390/foods14213803







