Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry Combined with Chemometrics for Protein Profiling and Classification of Boiled and Extruded Quinoa from Conventional and Organic Crops
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Samples
2.3. Extrusion Process
2.4. Boiling Process
2.5. Sample Preparation
2.6. MALDI-TOF-MS
2.7. Data Analysis
2.7.1. MALDIquant Data Processing
2.7.2. Multivariate Data Analysis
3. Results and Discussion
3.1. MALDI-TOF-MS Analysis
3.2. Multivariate Data Analysis
3.2.1. Discrimination of Conventional and Organic Quinoa
3.2.2. Discrimination of Raw and Processed Quinoa
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aloisi, I.; Parrotta, L.; Ruiz, K.B.; Landi, C.; Bini, L.; Cai, G.; Biondi, S.; Del Duca, S. New Insight into Quinoa Seed Quality under Salinity: Changes in Proteomic and Amino Acid Profiles, Phenolic Content, and Antioxidant Activity of Protein Extracts. Front. Plant Sci. 2016, 7, 183977. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Xue, S.; Sun, Q.; Shi, J.; Zhang, D.; Wang, D.; Wei, J. Research Progress of Quinoa Seeds (Chenopodium quinoa Wild.): Nutritional Components, Technological Treatment, and Application. Foods 2023, 12, 2087. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Walia, S.; Kumar, R. Functional Composition, Physiological Effect and Agronomy of Future Food Quinoa (Chenopodium Quinoa Willd.): A Review. J. Food Compos. Anal. 2023, 118, 105192. [Google Scholar] [CrossRef]
- Angeli, V.; Silva, P.M.; Massuela, D.C.; Khan, M.W.; Hamar, A.; Khajehei, F.; Graeff-Hönninger, S.; Piatti, C. Quinoa (Chenopodium quinoa Willd.): An Overview of the Potentials of the “Golden Grain” and Socio-Economic and Environmental Aspects of Its Cultivation and Marketization. Foods 2020, 9, 216. [Google Scholar] [CrossRef] [PubMed]
- Niro, S.; D’Agostino, A.; Fratianni, A.; Cinquanta, L.; Panfili, G. Gluten-Free Alternative Grains: Nutritional Evaluation and Bioactive Compounds. Foods 2019, 8, 208. [Google Scholar] [CrossRef] [PubMed]
- Mhada, M.; Metougui, M.L.; El Hazzam, K.; El Kacimi, K.; Yasri, A. Variations of Saponins, Minerals and Total Phenolic Compounds Due to Processing and Cooking of Quinoa (Chenopodium quinoa Willd.) Seeds. Foods 2020, 9, 660. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.I.; Farooq, M.; Syed, Q.A.; Ishaq, A.; Al-Ghamdi, A.A.; Hatamleh, A.A. Botany, Nutritional Value, Phytochemical Composition and Biological Activities of Quinoa. Plants 2021, 10, 2258. [Google Scholar] [CrossRef] [PubMed]
- Ceyhun Sezgin, A.; Sanlier, N. A New Generation Plant for the Conventional Cuisine: Quinoa (Chenopodium quinoa Willd.). Trends Food Sci. Technol. 2019, 86, 51–58. [Google Scholar] [CrossRef]
- El-Serafy, R.S.; El-Sheshtawy, A.-N.A.; Abd El-Razek, U.A.; Abd El-Hakim, A.F.; Hasham, M.M.A.; Sami, R.; Khojah, E.; Al-Mushhin, A.A.M. Growth, Yield, Quality, and Phytochemical Behavior of Three Cultivars of Quinoa in Response to Moringa and Azolla Extracts under Organic Farming Conditions. Agronomy 2021, 11, 2186. [Google Scholar] [CrossRef]
- Gomiero, T. Food Quality Assessment in Organic vs. Conventional Agricultural Produce: Findings and Issues. Appl. Soil Ecol. 2018, 123, 714–728. [Google Scholar] [CrossRef]
- Cancino-Espinoza, E.; Vázquez-Rowe, I.; Quispe, I. Organic Quinoa (Chenopodium quinoa L.) Production in Peru: Environmental Hotspots and Food Security Considerations Using Life Cycle Assessment. Sci. Total Environ. 2018, 637–638, 221–232. [Google Scholar] [CrossRef] [PubMed]
- FAO; CIRAD. State of the Art Report on Quinoa around the World in 2013; Bazile, D., Bertero, D., Nieto, C., Eds.; FAO: Rome, Italy, 2015; ISBN 978-92-5-108558-5. [Google Scholar]
- Huang, R.; Huang, K.; Guan, X.; Li, S.; Cao, H.; Zhang, Y.; Lao, X.; Bao, Y.; Wang, J. Effect of Defatting and Extruding Treatment on the Physicochemical and Storage Properties of Quinoa (Chenopodium quinoa Wild) Flour. LWT 2021, 147, 111612. [Google Scholar] [CrossRef]
- Naozuka, J.; Oliveira, P.V. Cooking Effects on Iron and Proteins Content of Beans (Phaseolus vulgaris L.) by GF AAS and MALDI-TOF MS. J. Braz. Chem. Soc. 2012, 23, 156–162. [Google Scholar]
- Scanlin, L.; Lewis, K.A. Quinoa as a Sustainable Protein Source: Production, Nutrition, and Processing. In Sustainable Protein Sources; Nadathur, S.R., Wanasundara, J.P.D., Scanlin, L., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 223–238. ISBN 9780128027769. [Google Scholar]
- Poza-Viejo, L.; Redondo-Nieto, M.; Matías, J.; Granado-Rodríguez, S.; Maestro-Gaitán, I.; Cruz, V.; Olmos, E.; Bolaños, L.; Reguera, M. Shotgun Proteomics of Quinoa Seeds Reveals Chitinases Enrichment under Rainfed Conditions. Sci. Rep. 2023, 13, 4951. [Google Scholar] [CrossRef] [PubMed]
- Di Silvestre, D.; Passignani, G.; Rossi, R.; Ciuffo, M.; Turina, M.; Vigani, G.; Mauri, P.L. Presence of a Mitovirus Is Associated with Alteration of the Mitochondrial Proteome, as Revealed by Protein–Protein Interaction (PPI) and Co-Expression Network Models in Chenopodium Quinoa Plants. Biology 2022, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Derbali, W.; Manaa, A.; Spengler, B.; Goussi, R.; Abideen, Z.; Ghezellou, P.; Abdelly, C.; Forreiter, C.; Koyro, H.W. Comparative Proteomic Approach to Study the Salinity Effect on the Growth of Two Contrasting Quinoa Genotypes. Plant Physiol. Biochem. 2021, 163, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Rasouli, F.; Kiani-Pouya, A.; Shabala, L.; Li, L.; Tahir, A.; Yu, M.; Hedrich, R.; Chen, Z.; Wilson, R.; Zhang, H.; et al. Salinity Effects on Guard Cell Proteome in Chenopodium Quinoa. Int. J. Mol. Sci. 2021, 22, 428. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, Q.; Kamruzzaman, M. Portable NIR Spectroscopy and PLS Based Variable Selection for Adulteration Detection in Quinoa Flour. Food Control 2022, 138, 108970. [Google Scholar] [CrossRef]
- Shotts, M.L.; Plans Pujolras, M.; Rossell, C.; Rodriguez-Saona, L. Authentication of Indigenous Flours (Quinoa, Amaranth and Kañiwa) from the Andean Region Using a Portable ATR-Infrared Device in Combination with Pattern Recognition Analysis. J. Cereal Sci. 2018, 82, 65–72. [Google Scholar] [CrossRef]
- Rodríguez, S.D.; Rolandelli, G.; Buera, M.P. Detection of Quinoa Flour Adulteration by Means of FT-MIR Spectroscopy Combined with Chemometric Methods. Food Chem. 2019, 274, 392–401. [Google Scholar] [CrossRef]
- Xue, S.S.; Tan, J.; Xie, J.Y.; Li, M.F. Rapid, Simultaneous and Non-Destructive Determination of Maize Flour and Soybean Flour Adulterated in Quinoa Flour by Front-Face Synchronous Fluorescence Spectroscopy. Food Control 2021, 130, 108329. [Google Scholar] [CrossRef]
- Kelis Cardoso, V.G.; Poppi, R.J. Cleaner and Faster Method to Detect Adulteration in Cassava Starch Using Raman Spectroscopy and One-Class Support Vector Machine. Food Control 2021, 125, 107917. [Google Scholar] [CrossRef]
- Ellis, D.I.; Brewster, V.L.; Dunn, W.B.; Allwood, J.W.; Golovanov, A.P.; Goodacre, R. Fingerprinting Food: Current Technologies for the Detection of Food Adulteration and Contamination. Chem. Soc. Rev. 2012, 41, 5706–5727. [Google Scholar] [CrossRef] [PubMed]
- Bansal, S.; Singh, A.; Mangal, M.; Mangal, A.K.; Kumar, S. Food Adulteration: Sources, Health Risks, and Detection Methods. Crit. Rev. Food Sci. Nutr. 2017, 57, 1174–1189. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xing, B.; Guo, Y.; Wang, S.; Guo, H.; Qin, P.; Hou, C.; Ren, G. Rapid, Accurate and Simply-Operated Determination of Laboratory-Made Adulteration of Quinoa Flour with Rice Flour and Wheat Flour by Headspace Gas Chromatography-Ion Mobility Spectrometry. LWT 2022, 167, 113814. [Google Scholar] [CrossRef]
- Galindo-Luján, R.; Pont, L.; Sanz-Nebot, V.; Benavente, F. Classification of Quinoa Varieties Based on Protein Fingerprinting by Capillary Electrophoresis with Ultraviolet Absorption Diode Array Detection and Advanced Chemometrics. Food Chem. 2021, 341, 128207. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Luján, R.; Caballero-Alcazar, N.; Pont, L.; Sanz-Nebot, V.; Benavente, F. Fingerprinting of Quinoa Grain Protein Extracts by Liquid Chromatography with Spectrophotometric Detection for Chemometrics Discrimination. LWT 2023, 187, 115289. [Google Scholar] [CrossRef]
- Galindo-Luján, R.; Pont, L.; Minic, Z.; Berezovski, M.V.; Sanz-Nebot, V.; Benavente, F. Characterization and Differentiation of Quinoa Seed Proteomes by Label-Free Mass Spectrometry-Based Shotgun Proteomics. Food Chem. 2021, 363, 130250. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Luján, R.; Pont, L.; Sanz-Nebot, V.; Benavente, F. Protein Profiling and Classification of Commercial Quinoa Grains by MALDI-TOF-MS and Chemometrics. Food Chem. 2023, 398, 133895. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Luján, R.; Pont, L.; Sanz-Nebot, V.; Benavente, F. A Proteomics Data Mining Strategy for the Identification of Quinoa Grain Proteins with Potential Immunonutritional Bioactivities. Foods 2023, 12, 390. [Google Scholar] [CrossRef]
- R Development Core Team: R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing. Available online: http://www.r-project.org/ (accessed on 1 January 2024).
- Gibb, S. MALDIquantForeign: Import/Export Routines for MALDIquant. 2014; pp. 1–7. Available online: https://cran.r-project.org/package=MALDIquantForeign (accessed on 1 January 2024).
- Gibb, S.; Strimmer, K. Mass Spectrometry Analysis Using MALDIquant. In Statistical Analysis of Proteomics, Metabolomics, and Lipidomics Data Using Mass Spectrometry; Springer: Cham, Switzerland, 2017; pp. 101–124. [Google Scholar]
- Purohit, P.V.; Rocke, D.M. Discriminant Models for High-Throughput Proteomics Mass Spectrometer Data. Proteomics 2003, 3, 1699–1703. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1639–1643. [Google Scholar] [CrossRef]
- Ryan, C.G.; Clayton, E.; Griffin, W.L.; Sie, S.H.; Cousens, D.R. SNIP, a Statistics-Sensitive Background Treatment for the Quantitative Analysis of PIXE Spectra in Geoscience Applications. Nucl. Instrum. Methods Phys. Res. B 1988, 34, 396–402. [Google Scholar] [CrossRef]
- Borgaonkar, S.P.; Hocker, H.; Shin, H.; Markey, M.K. Comparison of Normalization Methods for the Identification of Biomarkers Using MALDI-TOF and SELDI-TOF Mass Spectra. OMICS 2010, 14, 115–126. [Google Scholar] [CrossRef]
- Cleveland, W.S. Robust Locally Weighted Regression and Smoothing Scatterplots. J. Am. Stat. Assoc. 1979, 74, 829–836. [Google Scholar] [CrossRef]
- Friedman, J.H. A Variable Span Smoother. Laboratory for Computational Statistics, Stanford University Technical Report No. 5. J. Am. Stat. Assoc. 1984, 5, 1–32. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial Least Squares for Discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Ballabio, D.; Consonni, V. Classification Tools in Chemistry. Part 1: Linear Models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-Regression: A Basic Tool of Chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Worley, B.; Halouska, S.; Powers, R. Utilities for Quantifying Separation in PCA/PLS-DA Scores Plots. Anal. Biochem. 2013, 433, 102–104. [Google Scholar] [CrossRef]
- Mehmood, T.; Liland, K.H.; Snipen, L.; Sæbø, S. A Review of Variable Selection Methods in Partial Least Squares Regression. Chemom. Intell. Lab. Syst. 2012, 118, 62–69. [Google Scholar] [CrossRef]
- Galindo-Luján, R.; Pont, L.; Minic, Z.; Berezovski, M.V.; Quispe, F.; Sanz-Nebot, V.; Benavente, F. Comprehensive Characterization of Raw and Processed Quinoa from Conventional and Organic Farming by Label-Free Shotgun Proteomics. 2024. Manuscript submitted for publication. Available online: https://ssrn.com/abstract=4774018 (accessed on 27 March 2024).
- Soladoye, O.P.; Juárez, M.L.; Aalhus, J.L.; Shand, P.; Estévez, M. Protein Oxidation in Processed Meat: Mechanisms and Potential Implications on Human Health. Compr. Rev. Food Sci. Food Saf. 2015, 14, 106–122. [Google Scholar] [CrossRef] [PubMed]
- Santé-Lhoutellier, V.; Astruc, T.; Marinova, P.; Greve, E.; Gatellier, P. Effect of Meat Cooking on Physicochemical State and in Vitro Digestibility of Myofibrillar Proteins. J. Agric. Food Chem. 2008, 56, 1488–1494. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Fu, T.; Feng, Y.; Zhang, S.; Wang, C.; Zhang, D. Effect of Heat Treatment on Solubility, Surface Hydrophobicity and Structure of Rice Bran Albumin and Globulin. Qual. Assur. Saf. Crops Foods 2019, 11, 499–509. [Google Scholar] [CrossRef]
- Xiao, R.; Li, L.; Ma, Y. A Label-Free Proteomic Approach Differentiates between Conventional and Organic Rice. J. Food Compos. Anal. 2019, 80, 51–61. [Google Scholar] [CrossRef]
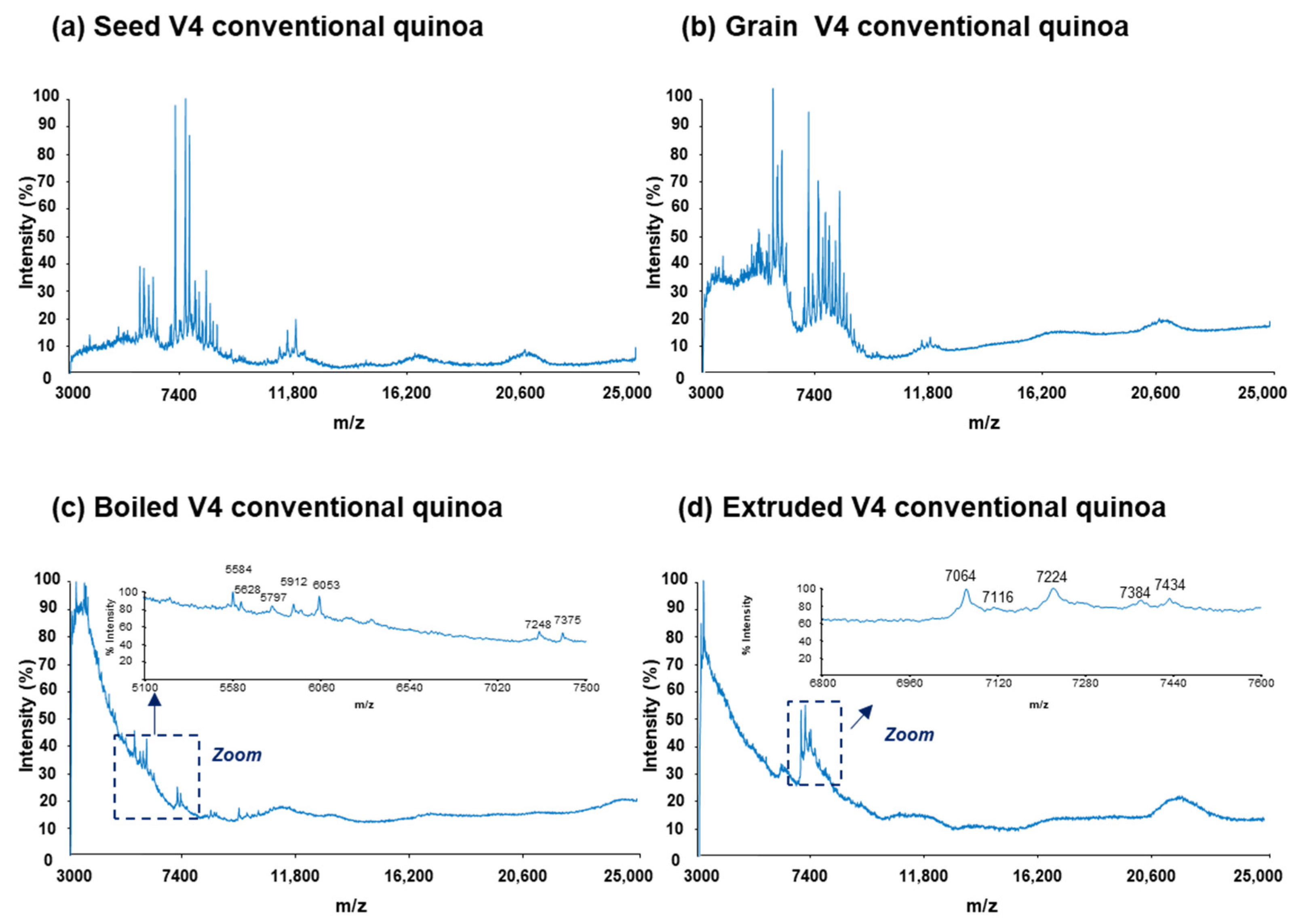
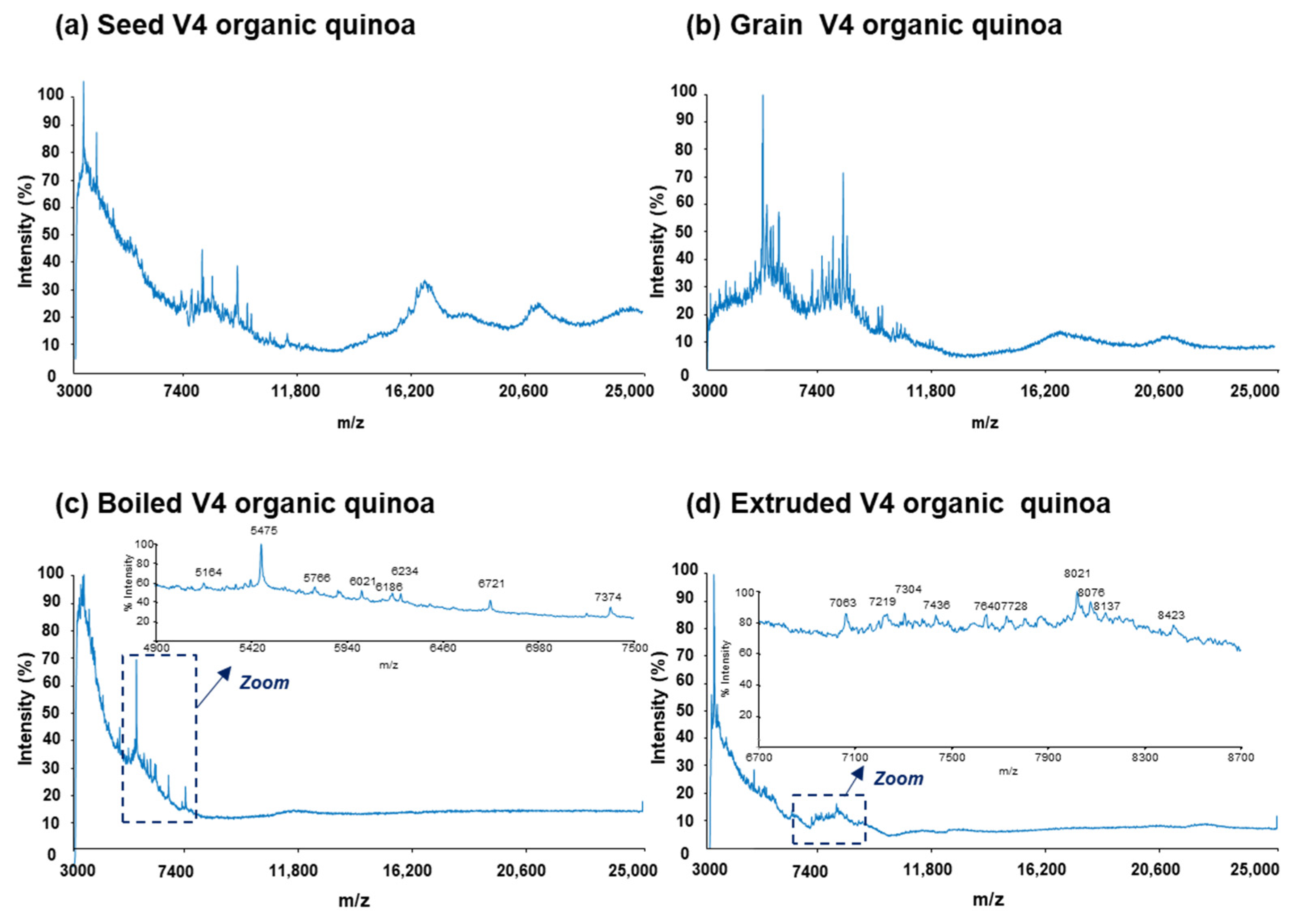

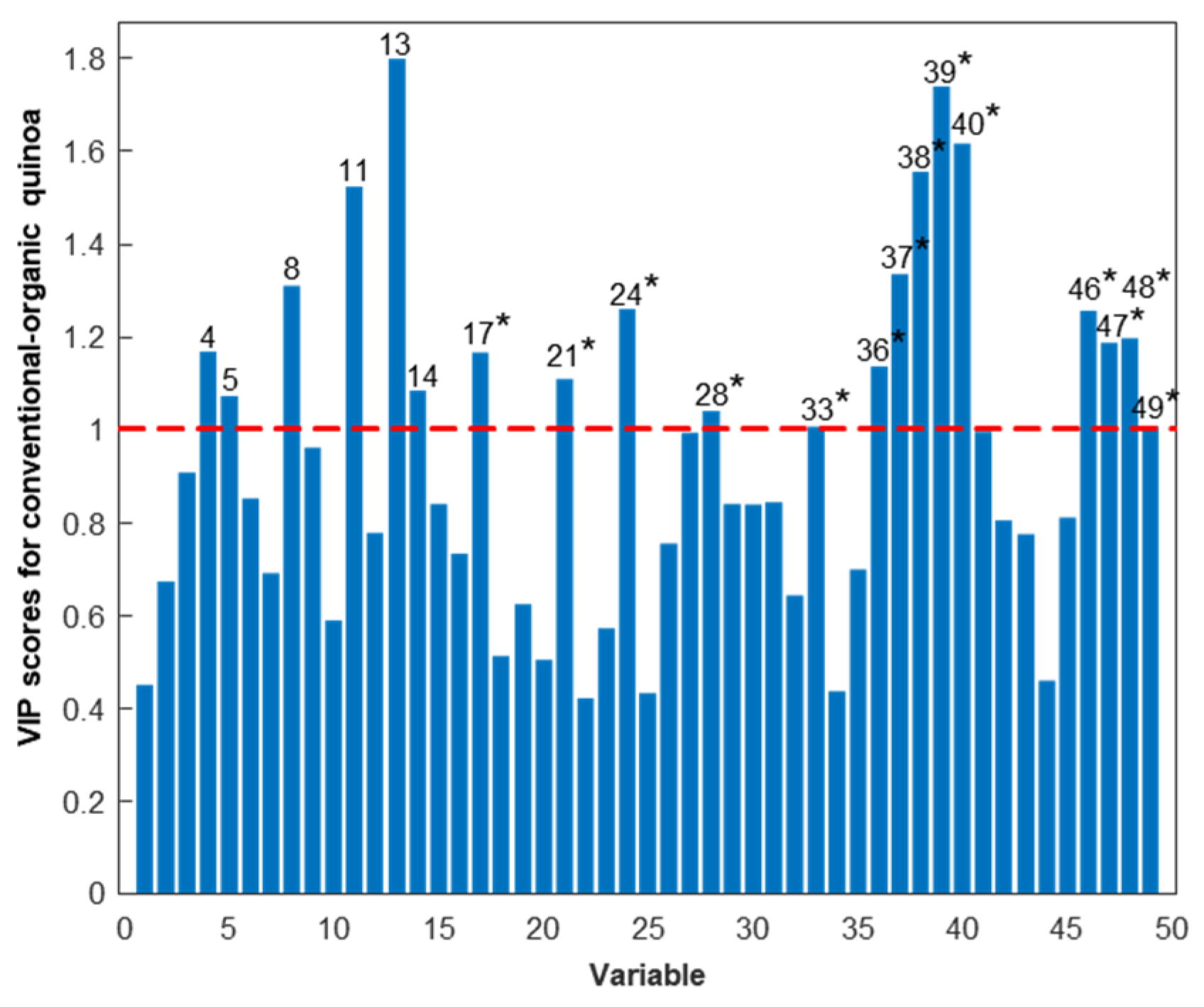
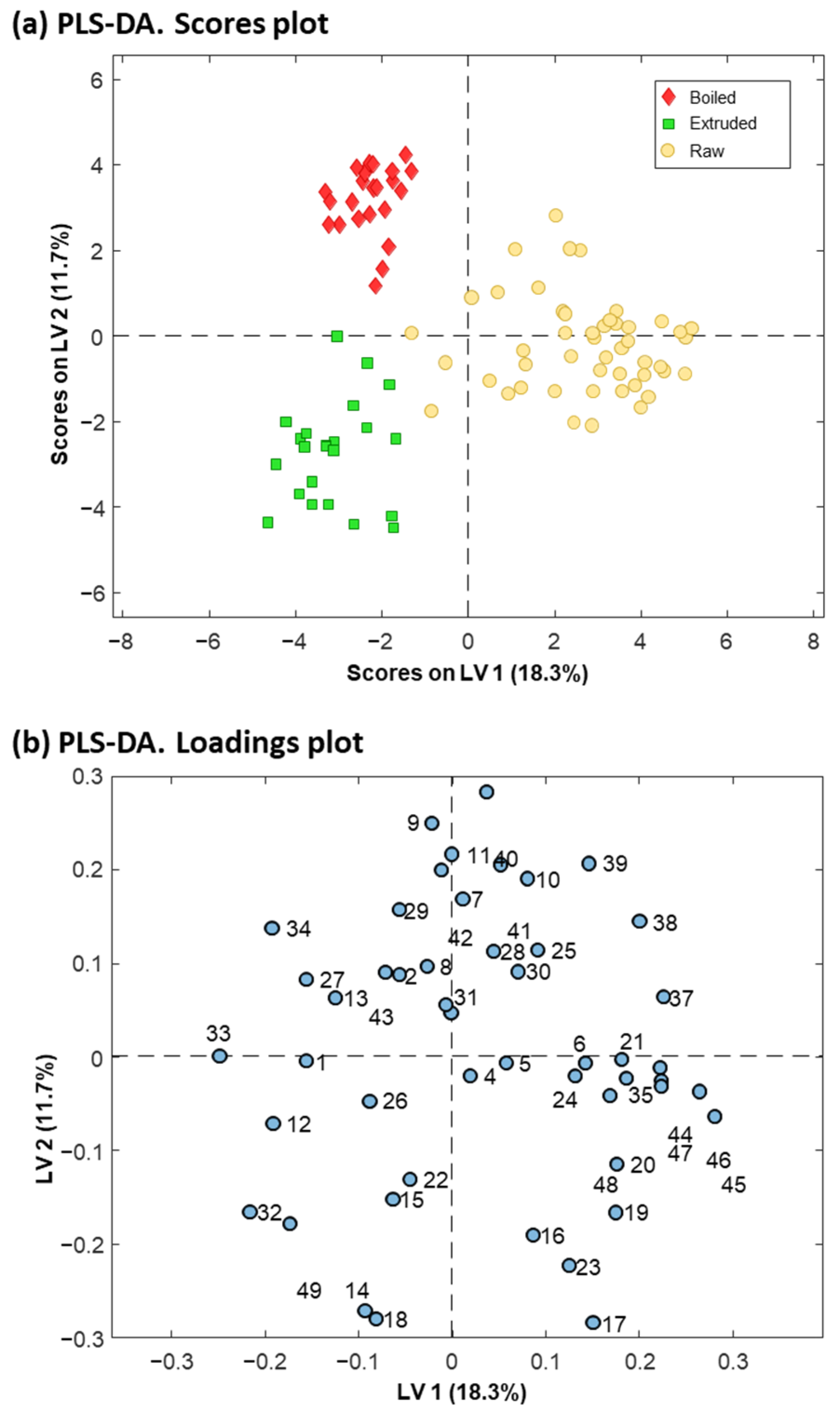
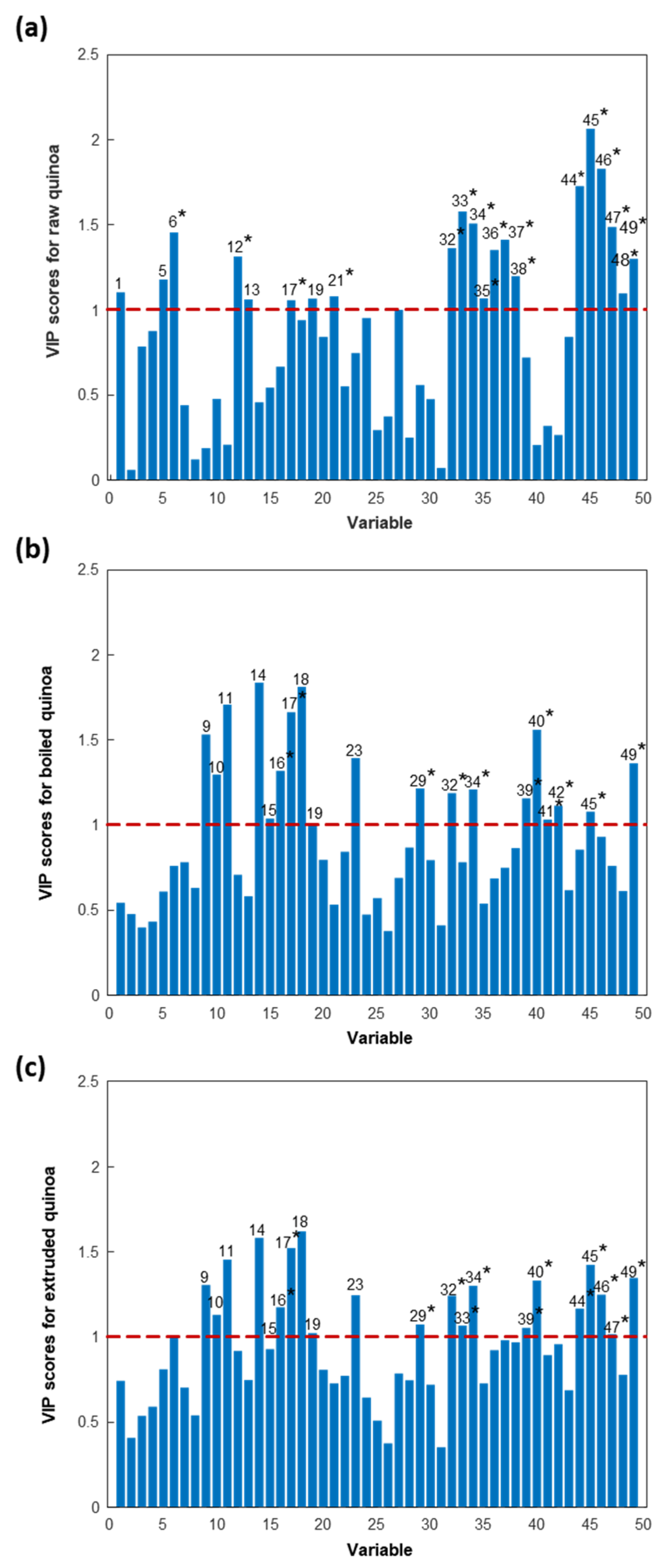
| Multivariate Data Analysis Protein Variables a | Tentative Identifications b | |||||||
|---|---|---|---|---|---|---|---|---|
| Protein | Experimental Mr c | PLS-DA VIP Scores d | Theoretical Mr | Accession Number (ID) e and Protein Name | ||||
| Farming | Processing | |||||||
| Craw f | Oraw f | Raw | Boiled | Extruded | ||||
| 1 | 4231 | 0.45 | 0.45 | 1.10 | 0.54 | 0.74 | - | |
| 2 | 4445 | 0.67 | 0.67 | 0.06 | 0.48 | 0.41 | - | |
| 3 | 4650 | 0.91 | 0.91 | 0.78 | 0.40 | 0.53 | - | |
| 4 | 4951 | 1.17 | 1.17 | 0.87 | 0.43 | 0.59 | - | |
| 5 | 5180 | 1.07 | 1.07 | 1.18 | 0.61 | 0.81 | - | |
| 6 | 5304 | 0.85 | 0.85 | 1.45 * | 0.76 | 1.00 | 5307 | XP_021736943.1 wound-induced basic protein-like |
| 7 | 5460 | 0.69 | 0.69 | 0.44 | 0.78 | 0.70 | - | |
| 8 | 5587 | 1.31 | 1.31 | 0.12 | 0.63 | 0.54 | - | |
| 9 | 5767 | 0.96 | 0.96 | 0.19 | 1.53 | 1.30 | - | |
| 10 | 5934 | 0.59 | 0.59 | 0.47 | 1.29 | 1.13 | - | |
| 11 | 6184 | 1.52 | 1.52 | 0.20 | 1.70 | 1.45 | - | |
| 12 | 6391 | 0.78 | 0.78 | 1.31 * | 0.70 | 0.91 | 6413 | XP_021764391.1 40S ribosomal protein S29 |
| 13 | 6818 | 1.80 | 1.80 | 1.06 | 0.58 | 0.75 | - | |
| 14 | 7063 | 1.08 | 1.08 | 0.46 | 1.84 | 1.58 | - | |
| 15 | 7435 | 0.84 | 0.84 | 0.54 | 1.04 | 0.93 | ||
| 16 | 7730 | 0.73 | 0.73 | 0.67 | 1.32 * | 1.17 * | 7747 | XP_021714409.1 uncharacterized protein LOC110682385 |
| 17 | 7983 | 1.17 * | 1.17 * | 1.06 * | 1.66 * | 1.52 * | 7974 | XP_021773921.1 metallothionein-like protein 4B |
| 18 | 8221 | 0.51 | 0.51 | 0.94 | 1.81 | 1.62 | - | |
| 19 | 8465 | 0.62 | 0.62 | 1.06 | 1.01 | 1.02 | - | |
| 20 | 8635 | 0.50 | 0.50 | 0.84 | 0.79 | 0.81 | - | |
| 21 | 8840 | 1.11 * | 1.11 * | 1.08 * | 0.53 | 0.73 | 8806 | XP_021718956.1 protein DELETION OF SUV3 SUPPRESSOR 1(I)-like |
| 8814 | XP_021729007.1 uncharacterized protein LOC110696048 | |||||||
| 8823 | XP_021762602.1 defensin-like protein | |||||||
| 22 | 9054 | 0.42 | 0.42 | 0.55 | 0.84 | 0.77 | 9076 | XP_021768105.1 late seed maturation protein P8B6-like |
| 23 | 9314 | 0.57 | 0.57 | 0.74 | 1.39 | 1.24 | - | |
| 24 | 9695 | 1.26 * | 1.26 * | 0.95 | 0.47 | 0.64 | 9692 | XP_021747091.1 sm-like protein LSM5 |
| 25 | 10,106 | 0.43 | 0.43 | 0.29 | 0.57 | 0.51 | - | |
| 26 | 10,703 | 0.75 | 0.75 | 0.37 | 0.37 | 0.37 | 10,689 | XP_021714810.1 uncharacterized protein LOC110682782 |
| 10,724 | XP_021756490.1 sm-like protein LSM8 | |||||||
| 10,736 | XP_021756753.1 60S ribosomal protein L37-3 | |||||||
| 10,750 | XP_021768220.1 sm-like protein LSM7 | |||||||
| 27 | 10,947 | 0.99 | 0.99 | 1.00 | 0.69 | 0.79 | 10,920 | XP_021761138.1 mitochondrial import inner membrane translocase subunit Tim9 |
| 10,928 | XP_021746329.1 probable steroid-binding protein 3 | |||||||
| 10,989 | XP_021761862.1 peamaclein-like | |||||||
| 28 | 11,274 | 1.04 * | 1.04 * | 0.25 | 0.86 | 0.75 | 11,270 | XP_021771595.1 sm-like protein LSM3A |
| 11,308 | XP_021716413.1 60S acidic ribosomal protein P2-4-like | |||||||
| 29 | 11,489 | 0.84 | 0.84 | 0.56 | 1.21 * | 1.07 * | 11,449 | XP_021727941.1 NADH dehydrogenase |
| 11,458 | XP_021766637.1 non-specific lipid-transfer protein-like | |||||||
| 11,517 | XP_021754863.1 thioredoxin M-type, chloroplastic-like isoform X2 | |||||||
| 30 | 11,779 | 0.84 | 0.84 | 0.47 | 0.79 | 0.72 | 11,723 | XP_021772578.1 RNA polymerase II transcriptional coactivator KIWI-like isoform X1 |
| 11,772 | XP_021755694.1 uncharacterized protein LOC110720913 | |||||||
| 11,797 | XP_021763483.1 small ubiquitin-related modifier 1-like | |||||||
| 31 | 12,043 | 0.84 | 0.84 | 0.07 | 0.41 | 0.35 | 11,992 | YP_009380236.1 ribosomal protein S18 (chloroplast) |
| 12,050 | XP_021776279.1 peptidyl-prolyl cis-trans isomerase FKBP12-like | |||||||
| 12,055 | XP_021765385.1 NADH dehydrogenase | |||||||
| 12,064 | XP_021774292.1 huntingtin-interacting protein K-like | |||||||
| 32 | 12,362 | 0.64 | 0.64 | 1.36 * | 1.19 * | 1.24 * | 12,301 | XP_021760438.1 gibberellin-regulated protein 9-like |
| 12,315 | XP_021716755.1 uncharacterized protein At2g27730, mitochondrial-like | |||||||
| 12,332 | XP_021765334.1 V-type proton ATPase subunit G 1-like | |||||||
| 12,375 | XP_021720641.1 60S ribosomal protein L30 | |||||||
| 12,407 | XP_021761775.1 uncharacterized protein LOC110726608 | |||||||
| 12,413 | XP_021773050.1 60S ribosomal protein L36-2-like | |||||||
| 12,420 | XP_021738644.1 40S ribosomal protein S25-like | |||||||
| 33 | 12,801 | 1.01 * | 1.01 * | 1.58 * | 0.78 | 1.06 * | 12,827 | XP_021769120.1 60S ribosomal protein L35a-3 |
| 12,849 | XP_021757241.1 nodulin-related protein 1-like | |||||||
| 12,855 | XP_021718430.1 nodulin-related protein 1-like | |||||||
| 34 | 13,220 | 0.43 | 0.43 | 1.50 * | 1.21 * | 1.30 * | 13,205 | XP_021758336.1 thioredoxin H-type 1-like |
| 13,231 | XP_021759897.1 thioredoxin H-type 1-like | |||||||
| 35 | 16,215 | 0.70 | 0.70 | 1.06 * | 0.54 | 0.72 | 16,134 | XP_021716351.1 ferredoxin, root R-B2-like |
| 16,149 | XP_021747601.1 uncharacterized protein LOC110713466 | |||||||
| 16,165 | XP_021730244.1 outer envelope pore protein 16-2, chloroplastic-like isoform X2 | |||||||
| 16,200 | XP_021717733.1 high mobility group B protein 3-like | |||||||
| 16,215 | XP_021716749.1 ferredoxin, root R-B2-like | |||||||
| 16,216 | XP_021754488.1 high mobility group B protein 3-like | |||||||
| 16,239 | XP_021762815.1 uncharacterized protein At5g48480-like | |||||||
| 16,250 | XP_021766528.1 40S ribosomal protein S14-2 | |||||||
| 16,289 | XP_021721762.1 oleosin 1-like | |||||||
| 36 | 16,514 | 1.13 * | 1.13 * | 1.35 * | 0.68 | 0.92 | 16,458 | XP_021733518.1 uncharacterized protein At5g48480-like |
| 16,464 | XP_021761922.1 uncharacterized protein LOC110726743 | |||||||
| 16,469 | XP_021746531.1 60S ribosomal protein L27a-3-like | |||||||
| 16,474 | XP_021769235.1 glycine cleavage system H protein 2, mitochondrial-like | |||||||
| 16,524 | XP_021768671.1 60S ribosomal protein L27a-3-like | |||||||
| 16,568 | XP_021762909.1 uncharacterized protein LOC110727639 | |||||||
| 16,570 | XP_021732568.1 uncharacterized protein LOC110699354 | |||||||
| 37 | 16,693 | 1.34 * | 1.34 * | 1.41 * | 0.75 | 0.98 | 16,616 | XP_021755504.1 2S albumin-like |
| 16,624 | XP_021751394.1 60S ribosomal protein L26-1 | |||||||
| 16,625 | XP_021730224.1 probable calcium-binding protein CML13 | |||||||
| 16,636 | XP_021760375.1 eukaryotic translation initiation factor 1A | |||||||
| 16,651 | XP_021731588.1 glycine-rich RNA-binding, abscisic acid-inducible protein-like | |||||||
| 16,685 | XP_021735190.1 ubiquitin-conjugating enzyme E2 variant 1D-like | |||||||
| 16,693 | XP_021774210.1 60S ribosomal protein L28-1-like | |||||||
| 16,702 | XP_021717270.1 blue copper protein-like isoform X2 | |||||||
| 16,742 | XP_021720407.1 17.4 kDa class III heat shock protein-like | |||||||
| 16,758 | XP_021766054.1 uncharacterized protein LOC110730552 | |||||||
| 38 | 16,897 | 1.56 * | 1.56 * | 1.20 * | 0.86 | 0.97 | 16,833 | XP_021733717.1 40S ribosomal protein S16-like |
| 16,834 | XP_021776507.1 calmodulin-7-like | |||||||
| 16,860 | XP_021754554.1 calmodulin | |||||||
| 16,877 | XP_021749775.1 peptidyl-prolyl cis-trans isomerase FKBP15-1-like | |||||||
| 16,884 | XP_021716580.1 17.4 kDa class III heat shock protein-like | |||||||
| 16,933 | XP_021735458.1 probable prefoldin subunit 5 | |||||||
| 16,942 | XP_021731073.1 thiosulfate sulfurtransferase 16, chloroplastic-like isoform X2 | |||||||
| 16,946 | XP_021743153.1 uncharacterized protein LOC110709246 | |||||||
| 16,962 | XP_021758167.1 transcription initiation factor TFIID subunit 15b-like | |||||||
| 39 | 17,101 | 1.74 * | 1.74 * | 0.72 | 1.16 * | 1.05 * | 17,026 | XP_021751891.1 NADH dehydrogenase |
| 17,040 | XP_021771944.1 DNA-directed RNA polymerases II, IV and V subunit 8B-like | |||||||
| 17,048 | XP_021739940.1 uncharacterized protein LOC110706342 | |||||||
| 17,111 | XP_021765383.1 40S ribosomal protein S13-like | |||||||
| 17,129 | XP_021766190.1 uncharacterized protein LOC110730679 | |||||||
| 17,131 | XP_021740721.1 MLP-like protein 423 | |||||||
| 17,143 | XP_021769150.1 17.8 kDa class I heat shock protein-like | |||||||
| 40 | 17,326 | 1.62 * | 1.62 * | 0.20 | 1.56 * | 1.33 * | 17,290 | XP_021770408.1 outer envelope pore protein 16-3, chloroplastic/mitochondrial-like |
| 17,301 | XP_021729636.1 NADH dehydrogenase | |||||||
| 17,330 | XP_021717756.1 uncharacterized protein LOC110685525 | |||||||
| 17,340 | XP_021747441.1 eukaryotic translation initiation factor 5A-4-like | |||||||
| 17,350 | XP_021764293.1 40S ribosomal protein S15-4-like | |||||||
| 17,355 | YP_009380273.1 ribosomal protein S7 (chloroplast) | |||||||
| 17,366 | XP_021747435.1 eukaryotic translation initiation factor 5A-like | |||||||
| 17,376 | XP_021720177.1 ubiquitin-NEDD8-like protein RUB2 | |||||||
| 17,385 | XP_021748235.1 60S ribosomal protein L23A | |||||||
| 41 | 17,617 | 1.00* | 1.00* | 0.32 | 1.03 * | 0.89 | 17,532 | XP_021765685.1 glycine cleavage system H protein, mitochondrial |
| 17,543 | XP_021768154.1 glycine cleavage system H protein, mitochondrial-like | |||||||
| 17,560 | XP_021731505.1 oleosin 1-like | |||||||
| 17,562 | XP_021736891.1 peroxiredoxin-2B-like | |||||||
| 17,572 | XP_021732018.1 peroxiredoxin-2B-like | |||||||
| 17,592 | XP_021756471.1 putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 3 | |||||||
| 17,604 | XP_021735589.1 nascent polypeptide-associated complex subunit beta-like | |||||||
| 17,622 | XP_021733122.1 protein mago nashi homolog 2 | |||||||
| 17,652 | XP_021743932.1 histidine-containing phosphotransfer protein 1-like | |||||||
| 17,665 | XP_021753630.1 uncharacterized protein LOC110719020 | |||||||
| 17,675 | XP_021759953.1 nascent polypeptide-associated complex subunit beta-like | |||||||
| 17,700 | XP_021745442.1 40S ribosomal protein S11-3 | |||||||
| 42 | 17,879 | 0.80 | 0.80 | 0.26 | 1.11 * | 0.96 | 17,803 | XP_021769395.1 40S ribosomal protein S11-like |
| 17,855 | XP_021773311.1 60S ribosomal protein L12-1 | |||||||
| 17,916 | XP_021748317.1 desiccation protectant protein Lea14 homolog | |||||||
| 17,939 | XP_021749487.1 MLP-like protein 43 | |||||||
| 17,969 | XP_021715429.1 universal stress protein PHOS34-like | |||||||
| 43 | 18,311 | 0.77 | 0.77 | 0.84 | 0.62 | 0.69 | 18,221 | XP_021738830.1 oleosin 16 kDa |
| 18,224 | XP_021737967.1 MFP1 attachment factor 1-like | |||||||
| 18,238 | XP_021765145.1 60S ribosomal protein L24-like | |||||||
| 18,240 | XP_021753128.1 peptidyl-prolyl cis-trans isomerase 1-like | |||||||
| 18,252 | XP_021763237.1 pathogenesis-related protein STH-21-like | |||||||
| 18,254 | XP_021775867.1 peptidyl-prolyl cis-trans isomerase 1 | |||||||
| 18,258 | XP_021769094.1 18.3 kDa class I heat shock protein-like | |||||||
| 18,271 | XP_021730326.1 universal stress protein PHOS32 | |||||||
| 18,276 | XP_021744114.1 17.3 kDa class II heat shock protein-like | |||||||
| 18,317 | XP_021752091.1 probable NADH dehydrogenase | |||||||
| 18,348 | XP_021738936.1 17.3 kDa class II heat shock protein-like | |||||||
| 18,348 | XP_021732306.1 pathogenesis-related protein STH-21-like | |||||||
| 18,348 | XP_021725562.1 deoxyuridine 5-triphosphate nucleotidohydrolase | |||||||
| 18,366 | XP_021774711.1 50S ribosomal protein L18, chloroplastic | |||||||
| 44 | 20,349 | 0.46 | 0.46 | 1.72 * | 0.85 | 1.16 * | 20,301 | XP_021763546.1 30S ribosomal protein 3, chloroplastic |
| 20,406 | XP_021734303.1 HMG-Y-related protein A-like | |||||||
| 45 | 20,556 | 0.81 | 0.81 | 2.06 * | 1.08 * | 1.42 * | 20,466 | XP_021727144.1 21 kDa seed protein-like |
| 20,499 | XP_021763320.1 photosystem II reaction center Psb28 protein-like | |||||||
| 20,522 | XP_021744010.1 succinate dehydrogenase assembly factor 2, mitochondrial-like | |||||||
| 20,523 | XP_021729294.1 uncharacterized protein LOC110696308 | |||||||
| 20,557 | XP_021766022.1 PLAT domain-containing protein 3-like | |||||||
| 20,565 | XP_021741243.1 putative H/ACA ribonucleoprotein complex subunit 1-like protein 1 | |||||||
| 20,592 | XP_021769990.1 ADP-ribosylation factor 1-like | |||||||
| 20,619 | XP_021752903.1 thioredoxin-like protein CITRX, chloroplastic | |||||||
| 46 | 20,780 | 1.25 * | 1.25 * | 1.83 * | 0.93 | 1.25 * | 20,736 | XP_021773813.1 adenylate kinase isoenzyme 6 homolog |
| 20,739 | XP_021740322.1 protein CutA, chloroplastic-like | |||||||
| 20,778 | XP_021761077.1 peroxiredoxin-2F, mitochondrial-like isoform X1 | |||||||
| 20,799 | XP_021753718.1 60S ribosomal protein L11-1 | |||||||
| 20,801 | XP_021772257.1 HMG-Y-related protein A-like | |||||||
| 20,844 | XP_021763208.1 60S ribosomal protein L18-3-like | |||||||
| 20,848 | XP_021738998.1 protein OPI10 homolog | |||||||
| 20,852 | XP_021763370.1 monothiol glutaredoxin-S10-like | |||||||
| 47 | 21,075 | 1.19 * | 1.19 * | 1.48 * | 0.76 | 1.01 * | 21,027 | XP_021750037.1 uncharacterized protein LOC110715738 |
| 21,031 | XP_021730777.1 thioredoxin O2, mitochondrial-like isoform X2 | |||||||
| 21,055 | XP_021766443.1 lactoylglutathione lyase isoform X2 | |||||||
| 21,077 | XP_021756715.1 uncharacterized protein LOC110721825 | |||||||
| 21,107 | XP_021727997.1 50S ribosomal protein L27, chloroplastic | |||||||
| 21,121 | XP_021733985.1 glycine-rich RNA-binding protein 3, mitochondrial-like | |||||||
| 21,149 | XP_021736893.1 probable inactive nicotinamidase At3g16190 | |||||||
| 21,170 | XP_021763161.1 uncharacterized protein Os08g0359500-like | |||||||
| 21,172 | XP_021732021.1 probable inactive nicotinamidase At3g16190 | |||||||
| 48 | 21,343 | 1.20 * | 1.20 * | 1.09 * | 0.61 | 0.78 | 21,241 | XP_021720070.1 ankyrin repeat and SAM domain-containing protein 6-like isoform X2 |
| 21,268 | XP_021771518.1 uncharacterized protein LOC110735639 | |||||||
| 21,294 | XP_021764214.1 cyclic phosphodiesterase-like | |||||||
| 21,326 | XP_021763910.1 60S ribosomal protein L18a-2 | |||||||
| 21,364 | XP_021754795.1 50S ribosomal protein L24, chloroplastic-like | |||||||
| 21,376 | XP_021718085.1 60S ribosomal protein L18a | |||||||
| 21,433 | XP_021730369.1 probable prefoldin subunit 3 | |||||||
| 49 | 22,079 | 1.01 * | 1.01 * | 1.30 * | 1.36* | 1.34 * | 21,984 | XP_021772119.1 RNA-binding protein Y14-like |
| 22,018 | XP_021763572.1 40S ribosomal protein S7-like | |||||||
| 22,034 | XP_021761714.1 histone H1-like | |||||||
| 22,040 | YP_009380239.1 ClpP (chloroplast) | |||||||
| 22,088 | XP_021766393.1 50S ribosomal protein L9, chloroplastic-like | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galindo-Luján, R.; Pont, L.; Quispe, F.; Sanz-Nebot, V.; Benavente, F. Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry Combined with Chemometrics for Protein Profiling and Classification of Boiled and Extruded Quinoa from Conventional and Organic Crops. Foods 2024, 13, 1906. https://doi.org/10.3390/foods13121906
Galindo-Luján R, Pont L, Quispe F, Sanz-Nebot V, Benavente F. Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry Combined with Chemometrics for Protein Profiling and Classification of Boiled and Extruded Quinoa from Conventional and Organic Crops. Foods. 2024; 13(12):1906. https://doi.org/10.3390/foods13121906
Chicago/Turabian StyleGalindo-Luján, Rocío, Laura Pont, Fredy Quispe, Victoria Sanz-Nebot, and Fernando Benavente. 2024. "Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry Combined with Chemometrics for Protein Profiling and Classification of Boiled and Extruded Quinoa from Conventional and Organic Crops" Foods 13, no. 12: 1906. https://doi.org/10.3390/foods13121906
APA StyleGalindo-Luján, R., Pont, L., Quispe, F., Sanz-Nebot, V., & Benavente, F. (2024). Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry Combined with Chemometrics for Protein Profiling and Classification of Boiled and Extruded Quinoa from Conventional and Organic Crops. Foods, 13(12), 1906. https://doi.org/10.3390/foods13121906







