Prediction of ACE-I Inhibitory Peptides Derived from Chickpea (Cicer arietinum L.): In Silico Assessments Using Simulated Enzymatic Hydrolysis, Molecular Docking and ADMET Evaluation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Sequences
2.2. Analysis of ACE-I Inhibitory Peptides from Chickpea (Cicer arietinum L.) Proteins
2.3. In Silico Proteolysis Analysis and Virtual Screening of ACE-I Inhibitory Peptides
2.4. Molecular Docking of Predicted ACE-Inhibitory Peptides from Chickpea Proteins on the ACE-I Binding Site
2.5. ADMET Prediction Analysis
3. Results and Discussion
3.1. ACE-I Inhibitory Peptides Prediction from Chickpea Seed Legumin and Provicilin
3.2. Molecular Interaction of Chickpea (Cicer arietinum L.) ACE-I Inhibitory Peptides and ACE-I
3.3. ADMET Analyses Showed That CP Are Bioavailable and Non-Toxic
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Toto, R.D. Defining Hypertension: Role of New Trials and Guidelines. Clin. J. Am. Soc. Nephrol. 2018, 13, 1578–1580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardiovascular Diseases (CVDs). Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 2 March 2022).
- Chobanian, A.V. Guidelines for the Management of Hypertension. Med. Clin. N. Am. 2017, 101, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Colafella, K.M.M.; Bovée, D.M.; Danser, A.J. The Renin-Angiotensin-Aldosterone System and Its Therapeutic Targets. Exp. Eye Res. 2019, 186, 107680. [Google Scholar] [CrossRef] [PubMed]
- Real Hernandez, L.M.; Gonzalez de Mejia, E. Enzymatic Production, Bioactivity, and Bitterness of Chickpea (Cicer arietinum) Peptides. Compr. Rev. Food Sci. 2019, 18, 1913–1946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.-W.; Alli, I. In Silico Assessment: Suggested Homology of Chickpea (Cicer arietinum L.) Legumin and Prediction of ACE-Inhibitory Peptides from Chickpea Proteins Using BLAST and BIOPEP Analyses. Food Res. Int. 2012, 49, 477–486. [Google Scholar] [CrossRef]
- Pedroche, J.; Yust, M.M.; Girón-Calle, J.; Alaiz, M.; Millán, F.; Vioque, J. Utilisation of Chickpea Protein Isolates for Production of Peptides with Angiotensin I-Converting Enzyme (ACE)-Inhibitory Activity. J. Sci. Food Agric. 2002, 82, 960–965. [Google Scholar] [CrossRef]
- Barbana, C.; Boye, J.I. Angiotensin I-Converting Enzyme Inhibitory Activity of Chickpea and Pea Protein Hydrolysates. Food Res. Int. 2010, 43, 1642–1649. [Google Scholar]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [Green Version]
- Dziuba, J.; Iwaniak, A.; Minkiewicz, P. Computer-Aided Characteristics of Proteins as Potential Precursors of Bioactive Peptides. Polimery 2003, 48, 50–53. [Google Scholar] [CrossRef] [Green Version]
- Natesh, R.; Schwager, S.L.; Sturrock, E.D.; Acharya, K.R. Crystal Structure of the Human Angiotensin-Converting Enzyme–Lisinopril Complex. Nature 2003, 421, 551–554. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Lu, X.; Sun, Q.; Gao, J.; Ma, L.; Huang, J. Novel ACE Inhibitory Peptides Derived from Simulated Gastrointestinal Digestion in Vitro of Sesame (Sesamum indicum L.) Protein and Molecular Docking Study. Int. J. Mol. Sci. 2020, 21, 1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapovalov, M.V.; Dunbrack, R.L., Jr. A Smoothed Backbone-Dependent Rotamer Library for Proteins Derived from Adaptive Kernel Density Estimates and Regressions. Structure 2011, 19, 844–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Q.; Jia, J.; Yan, H.; Du, J.; Gui, Z. A Novel Angiotensin-I Converting Enzyme (ACE) Inhibitory Peptide from Gastrointestinal Protease Hydrolysate of Silkworm Pupa (Bombyx mori) Protein: Biochemical Characterization and Molecular Docking Study. Peptides 2015, 68, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An Integrated Online Platform for Accurate and Comprehensive Predictions of ADMET Properties. Nucleic. Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef] [PubMed]
- Iwaniak, A.; Dziuba, J. Animal and Plant Proteins as Precursors of Peptides with ACE Inhibitory Activity—An in Silico Strategy of Protein Evaluation. Food Technol. Biotechnol. 2009, 31, 441. [Google Scholar]
- Fernández-Lucas, J.; Castañeda, D.; Hormigo, D. New Trends for a Classical Enzyme: Papain, a Biotechnological Success Story in the Food Industry. Trends Food Sci. Technol. 2017, 68, 91–101. [Google Scholar] [CrossRef]
- Tacias-Pascacio, V.G.; Morellon-Sterling, R.; Siar, E.-H.; Tavano, O.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R. Use of Alcalase in the Production of Bioactive Peptides: A Review. Int. J. Biol. Macromol. 2020, 165, 2143–2196. [Google Scholar] [CrossRef]
- Fan, H.; Liao, W.; Wu, J. Molecular Interactions, Bioavailability, and Cellular Mechanisms of Angiotensin-Converting Enzyme Inhibitory Peptides. J. Food Biochem. 2019, 43, e12572. [Google Scholar] [CrossRef] [Green Version]
- Karaś, M. Influence of Physiological and Chemical Factors on the Absorption of Bioactive Peptides. Int. J. Food Sci. Technol. 2019, 54, 1486–1496. [Google Scholar] [CrossRef]
- Zarei, M.; Abidin, N.B.Z.; Auwal, S.M.; Chay, S.Y.; Abdul Haiyee, Z.; Md Sikin, A.; Saari, N. Angiotensin Converting Enzyme (ACE)-Peptide Interactions: Inhibition Kinetics, in Silico Molecular Docking and Stability Study of Three Novel Peptides Generated from Palm Kernel Cake Proteins. Biomolecules 2019, 9, 569. [Google Scholar] [CrossRef] [Green Version]
- Rohit, A.; Sathisha, K.; Aparna, H.S. A Variant Peptide of Buffalo Colostrum β-Lactoglobulin Inhibits Angiotensin I-Converting Enzyme Activity. Eur. J. Med. Chem. 2012, 53, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Alashi, A.M.; Young, J.F.; Therkildsen, M.; Aluko, R.E. Enzyme Inhibition Kinetics and Molecular Interactions of Patatin Peptides with Angiotensin I-Converting Enzyme and Renin. Int. J. Biol. Macromol. 2017, 101, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Du, J.; Jia, J.; Kuang, C. Production of ACE Inhibitory Peptides from Sweet Sorghum Grain Protein Using Alcalase: Hydrolysis Kinetic, Purification and Molecular Docking Study. Food Chem. 2016, 199, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Natesh, R.; Schwager, S.L.; Evans, H.R.; Sturrock, E.D.; Acharya, K.R. Structural Details on the Binding of Antihypertensive Drugs Captopril and Enalaprilat to Human Testicular Angiotensin I-Converting Enzyme. Biochemistry 2004, 43, 8718–8724. [Google Scholar] [CrossRef] [PubMed]
- Vercruysse, L.; Van Camp, J.; Morel, N.; Rougé, P.; Herregods, G.; Smagghe, G. Ala-Val-Phe and Val-Phe: ACE Inhibitory Peptides Derived from Insect Protein with Antihypertensive Activity in Spontaneously Hypertensive Rats. Peptides 2010, 31, 482–488. [Google Scholar] [CrossRef]
- Wang, Z.-L.; Zhang, S.-S.; Wei, W.; Feng, F.-Q.; Shan, W.-G. A Novel Angiotensin I Converting Enzyme Inhibitory Peptide from the Milk Casein: Virtual Screening and Docking Studies. Agric. Sci. China 2011, 10, 463–467. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Armayor, G.M.; Lopez, L.M. Lisinopril: A New Angiotensin-Converting Enzyme Inhibitor. Drug Intell. Clin. Pharm. 1988, 22, 365–372. [Google Scholar] [CrossRef]
- Beermann, B. Pharmacokinetics of Lisinopril. Am. J. Med. 1988, 85, 25–30. [Google Scholar] [CrossRef]
- Iwaniak, A.; Minkiewicz, P.; Pliszka, M.; Mogut, D.; Darewicz, M. Characteristics of Biopeptides Released in Silico from Collagens Using Quantitative Parameters. Foods 2020, 9, 965. [Google Scholar] [CrossRef]
- Duchin, K.L.; Singhvi, S.M.; Willard, D.A.; Migdalof, B.H.; McKinstry, D.N. Captopril Kinetics. Clin. Pharmacol. Ther. 1982, 31, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Duchin, K.; McKinstry, D.; Cohen, A.; Migdalof, B. Pharmacokinetics of Captopril in Healthy Subjects and in Patients with Cardiovascular Diseases. Clin. Pharmacokinet. 1988, 14, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Torres, G.; Ontiveros, N.; Lopez-Teros, V.; Ibarra-Diarte, J.A.; Reyes-Moreno, C.; Cuevas-Rodríguez, E.O.; Cabrera-Chávez, F. Amaranth Protein Hydrolysates Efficiently Reduce Systolic Blood Pressure in Spontaneously Hypertensive Rats. Molecules 2017, 22, 1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ontiveros, N.; López-Teros, V.; de Jesús Vergara-Jiménez, M.; Islas-Rubio, A.R.; Cárdenas-Torres, F.I.; Cuevas-Rodríguez, E.-O.; Reyes-Moreno, C.; Granda-Restrepo, D.M.; Lopera-Cardona, S.; Ramírez-Torres, G.I.; et al. Amaranth-Hydrolyzate Enriched Cookies Reduce the Systolic Blood Pressure in Spontaneously Hypertensive Rats. J. Funct. Foods 2020, 64, 103613. [Google Scholar] [CrossRef]
- Wu, J.; Aluko, R.E.; Nakai, S. Structural Requirements of Angiotensin I-Converting Enzyme Inhibitory Peptides: Quantitative Structure- Activity Relationship Study of Di-and Tripeptides. J. Agric. Food Chem. 2006, 54, 732–738. [Google Scholar] [CrossRef]
- Suetsuna, K.; Chen, J.-R. Identification of Antihypertensive Peptides from Peptic Digest of Two Microalgae, Chlorella Vulgaris and Spirulina Platensis. Mar. Biotechnol. 2001, 3, 305–309. [Google Scholar] [CrossRef]

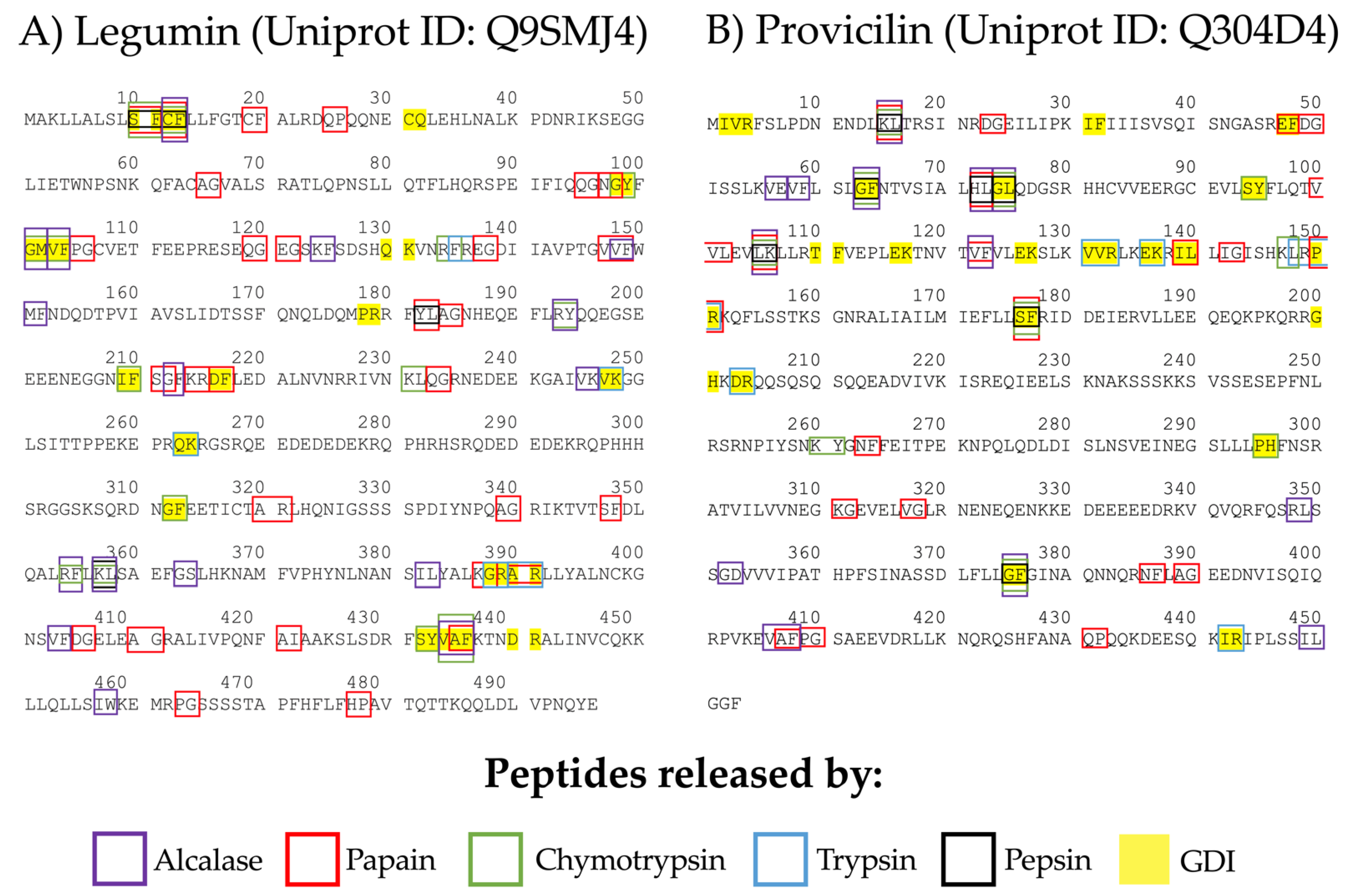

| Protein | A | B | In Silico Hydrolysis | ||
|---|---|---|---|---|---|
| Proteases | AE | BE | |||
| Legumin | 0.4335 | 0.0201 | Pepsin | 0.0101 | 0.00112 |
| Trypsin | 0.0101 | 0.00018 | |||
| Chymotrypsin | 0.0282 | 0.00168 | |||
| Gastrointestinal digestion | 0.0343 | 0.00206 | |||
| Papain | 0.0605 | 0.00302 | |||
| Alcalase | 0.0323 | 0.00273 | |||
| Provicilin | 0.3642 | 0.0110 | Pepsin | 0.0155 | 0.00011 |
| Trypsin | 0.0132 | 0.00058 | |||
| Chymotrypsin | 0.0221 | 0.00035 | |||
| Gastrointestinal digestion | 0.0419 | 0.00024 | |||
| Papain | 0.0442 | 0.00139 | |||
| Alcalase | 0.0287 | 0.00067 | |||
| Peptide/Ligand | BIOPEP ID | Binding Energy (Kcal/Mol) | Protein | Location | Released by | EC50 (µM/L) | PubChem/Satpdb ID |
|---|---|---|---|---|---|---|---|
| Lisinopril | −8.6 | 5362119 | |||||
| VVF | 9044 | −9.2 | Legumin | [147–149] | Papain | 35.45 | 7014911 |
| VAF | 8126 | −8.6 | Legumin | [434–436] | GID; Chymotrypsin (A) | 35.8 | satpdb14951 |
| [406–408] [434–436] | Alcalase | ||||||
| IW | 7544 | −8.5 | Legumin | [457,458] | Alcalase | 4.7 | 7019084 |
| RY | 3380 | −8.4 | Legumin | [193,194] | Chymotrypsin (A); Alcalase | 10.5 | 7021456 |
| RF | 3489 | −8.2 | Legumin | [134,135] | Chymotrypsin (A) | 93 | 150964 |
| [354,355] | Chymotrypsin (A); Pepsin (pH 1.3);Alcalase | ||||||
| IVR | 7502 | −8.0 | Provicilin | [2–4] | GID | 0.81 | 25217595 |
| YL | 3350 | −7.9 | Legumin | [182–183] | Papain; GID | 122 | 87071 |
| VF | 3384 | −7.8 | Legumin | [103,104] | Chymotrypsin (A); GID; Alcalase | 9.2 | 6993120 |
| [148,149] [403,404] | Alcalase | ||||||
| Provicilin | [122,123] | Papain; Alcalase | |||||
| [58,59] | Alcalase | ||||||
| SF | 7685 | −7.7 | Provicilin | [176,177] | Papain; Chymotrypsin (A), Pepsin (pH 1.3); GID | 130.2 | 7009597 |
| Legumin | [10,11] | Papain; Chymotrypsin (A), Pepsin (pH 1.3); GID | |||||
| [347,348] | Papain | ||||||
| AF | 7583 | −7.5 | Provicilin | [407,408] | Papain | 190 | 6992394 |
| Legumin | [435,436] | Papain | |||||
| KF | 7692 | −7.4 | Legumin | [124,125] | Alcalase | 28.3 | 151410 |
| CF | 7751 | −7.3 | Legumin | [12,13] | Papain; Chymotrypsin (A), Pepsin (pH 1.3); GID; Alcalase | 1.96 | 25051327 |
| [19,20] | Papain | ||||||
| PR | 3537 | −7.2 | Provicilin | [150,151] | Papain; Trypsin; GID | 4.1 | 151004 |
| Legumin | [178,179] | GID | |||||
| TF | 8185 | −7.1 | Provicilin | [110,111] | GID | 18 | 7010580 |
| DR | 10091 | −7.0 | Provicilin | [203,204] | Trypsin; GID | 110.5 | 16122509 |
| Legumin | [440,441] | GID | |||||
| LR | 9213 | −6.8 | Provicilin | [148,149] | Trypsin | 158 | 152914 |
| IL | 9079 | −6.5 | Provicilin | [139,140] | Papain; GID | 54.95 | 7019083 |
| [449,450] | Alcalase | ||||||
| Legumin | [382,383] | Alcalase | |||||
| AR | 7742 | −6.4 | Legumin | [390,391] | GID; Trypsin; Papain | 95.5 | 446132 |
| [320,321] | Papain | ||||||
| DG | 7681 | −5.8 | Provicilin | [23,24] | Papain | 190.1410 | 151148 |
| [49,50] | Papain | ||||||
| [405,406] | Papain | ||||||
| VK | 7558 | −5.7 | Legumin | [247,248] | Trypsin; GID | 13 | 168058 |
| [245,246] | Alcalase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arámburo-Gálvez, J.G.; Arvizu-Flores, A.A.; Cárdenas-Torres, F.I.; Cabrera-Chávez, F.; Ramírez-Torres, G.I.; Flores-Mendoza, L.K.; Gastelum-Acosta, P.E.; Figueroa-Salcido, O.G.; Ontiveros, N. Prediction of ACE-I Inhibitory Peptides Derived from Chickpea (Cicer arietinum L.): In Silico Assessments Using Simulated Enzymatic Hydrolysis, Molecular Docking and ADMET Evaluation. Foods 2022, 11, 1576. https://doi.org/10.3390/foods11111576
Arámburo-Gálvez JG, Arvizu-Flores AA, Cárdenas-Torres FI, Cabrera-Chávez F, Ramírez-Torres GI, Flores-Mendoza LK, Gastelum-Acosta PE, Figueroa-Salcido OG, Ontiveros N. Prediction of ACE-I Inhibitory Peptides Derived from Chickpea (Cicer arietinum L.): In Silico Assessments Using Simulated Enzymatic Hydrolysis, Molecular Docking and ADMET Evaluation. Foods. 2022; 11(11):1576. https://doi.org/10.3390/foods11111576
Chicago/Turabian StyleArámburo-Gálvez, Jesús Gilberto, Aldo Alejandro Arvizu-Flores, Feliznando Isidro Cárdenas-Torres, Francisco Cabrera-Chávez, Giovanni I. Ramírez-Torres, Lilian Karem Flores-Mendoza, Pedro Erick Gastelum-Acosta, Oscar Gerardo Figueroa-Salcido, and Noé Ontiveros. 2022. "Prediction of ACE-I Inhibitory Peptides Derived from Chickpea (Cicer arietinum L.): In Silico Assessments Using Simulated Enzymatic Hydrolysis, Molecular Docking and ADMET Evaluation" Foods 11, no. 11: 1576. https://doi.org/10.3390/foods11111576
APA StyleArámburo-Gálvez, J. G., Arvizu-Flores, A. A., Cárdenas-Torres, F. I., Cabrera-Chávez, F., Ramírez-Torres, G. I., Flores-Mendoza, L. K., Gastelum-Acosta, P. E., Figueroa-Salcido, O. G., & Ontiveros, N. (2022). Prediction of ACE-I Inhibitory Peptides Derived from Chickpea (Cicer arietinum L.): In Silico Assessments Using Simulated Enzymatic Hydrolysis, Molecular Docking and ADMET Evaluation. Foods, 11(11), 1576. https://doi.org/10.3390/foods11111576







