Screening and Probiotic Potential Evaluation of Bacteriocin-Producing Lactiplantibacillus plantarum In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Screening of Bacteriocin-Producing L. plantarum
2.3. Antibacterial Activity against Common Intestinal Pathogens
2.4. Acid and Bile Salt Tolerance
2.5. Gastric and Intestinal Juice Tolerance
2.6. Hydrophobicity Assay
2.7. Self-Aggregation Assay
2.8. Cytotoxic Activity Determination
2.9. Anti-Inflammatory Activity Detection
2.10. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.11. Statistical Analysis
3. Results and Discussion
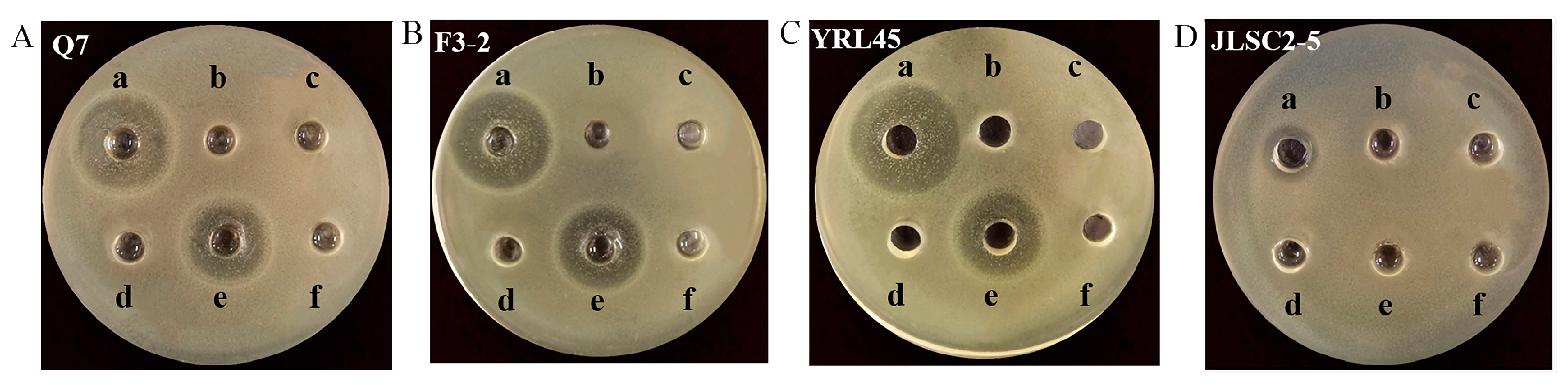
3.1. Screening of Bacteriocin-Producing L. plantarum
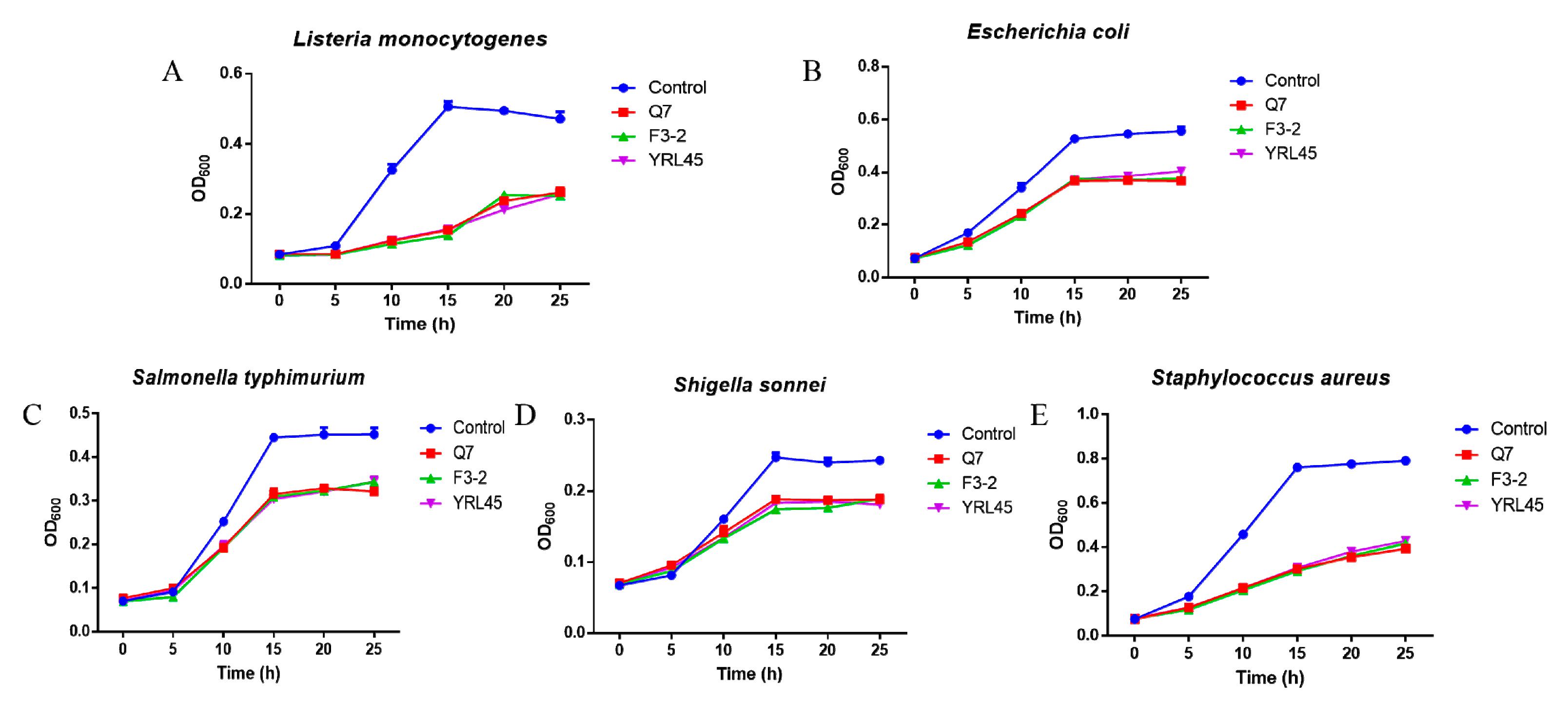
3.2. Antibacterial Activity of Bacteriocin-Producing L. plantarum against Common Intestinal Pathogens
3.3. Tolerance of Bacteriocin-Producing L. plantarum to Acid and Bile Salt
3.4. Tolerance of Bacteriocin-Producing L. plantarum to Gastric and Intestinal Juice
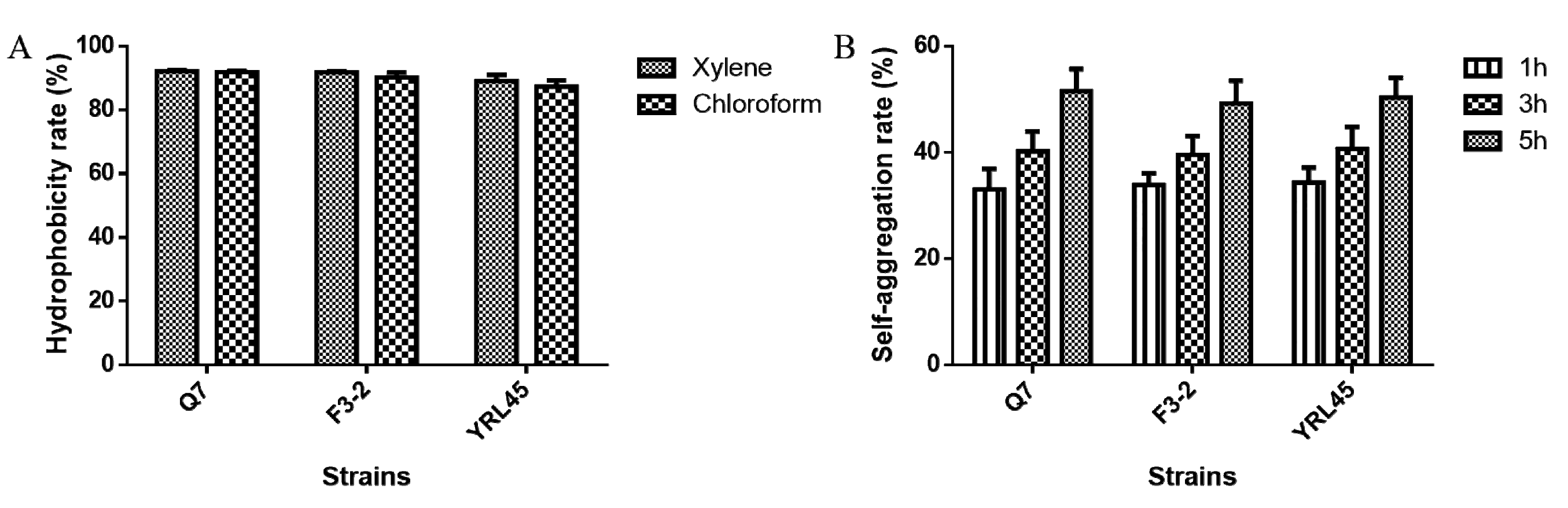
3.5. Hydrophobicity and Self-Aggregation Ability of Bacteriocin-Producing L. plantarum
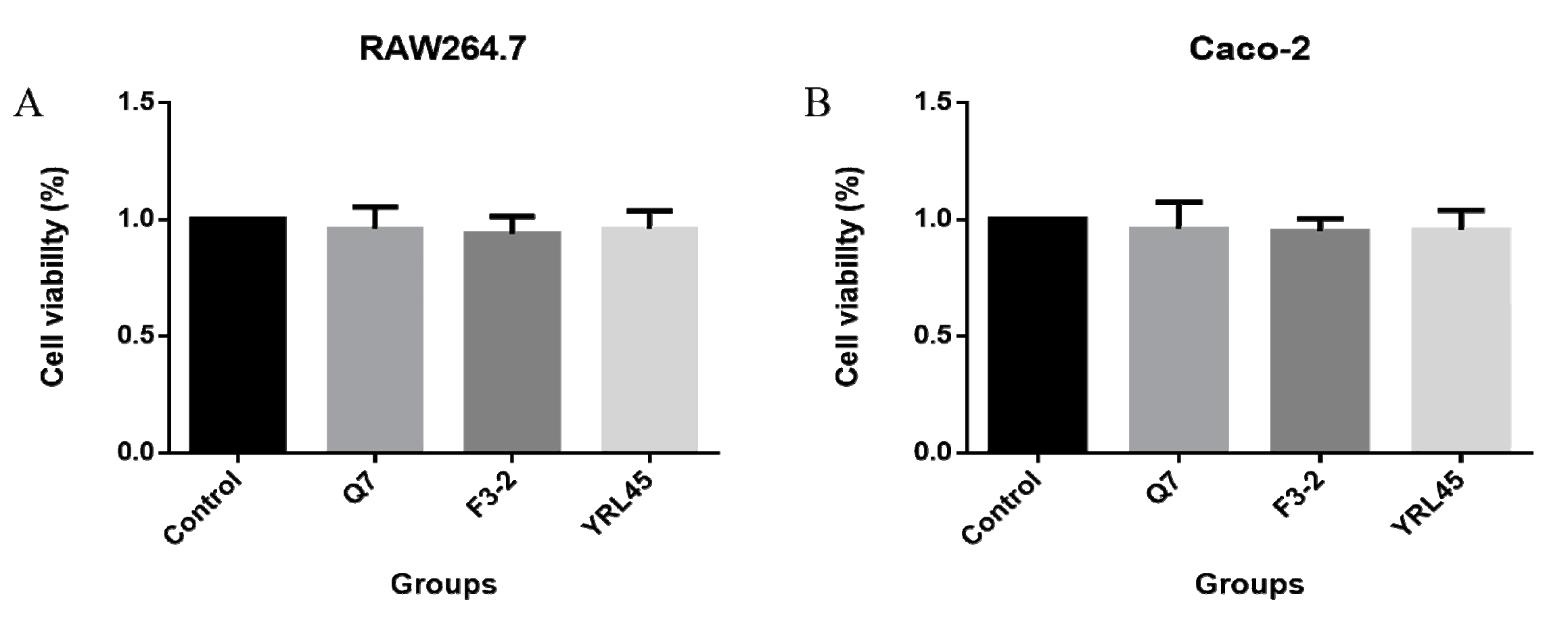
3.6. Cytotoxicity of Bacteriocin-Producing L. plantarum
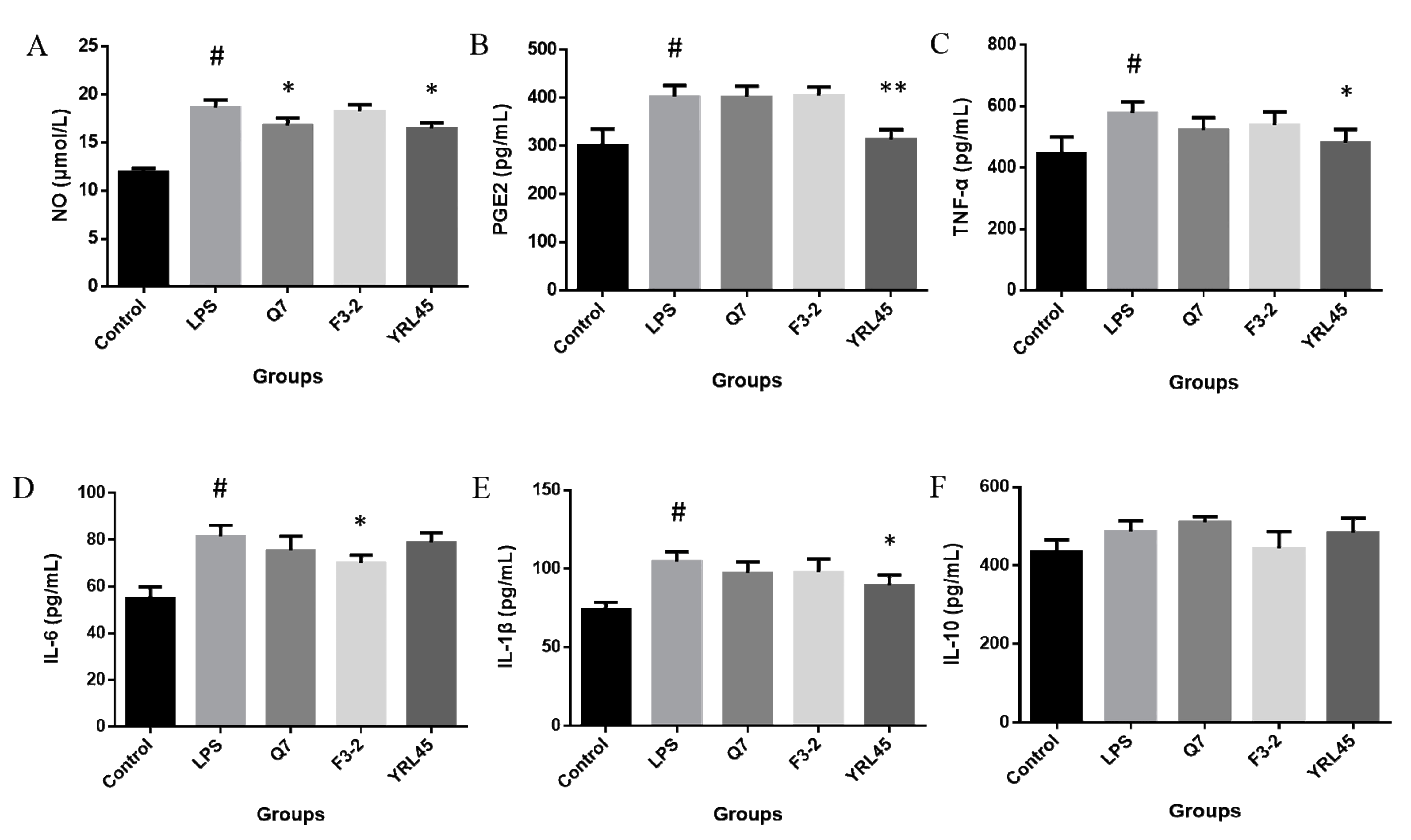
3.7. Anti-Inflammatory Activity of Bacteriocin-Producing L. plantarum
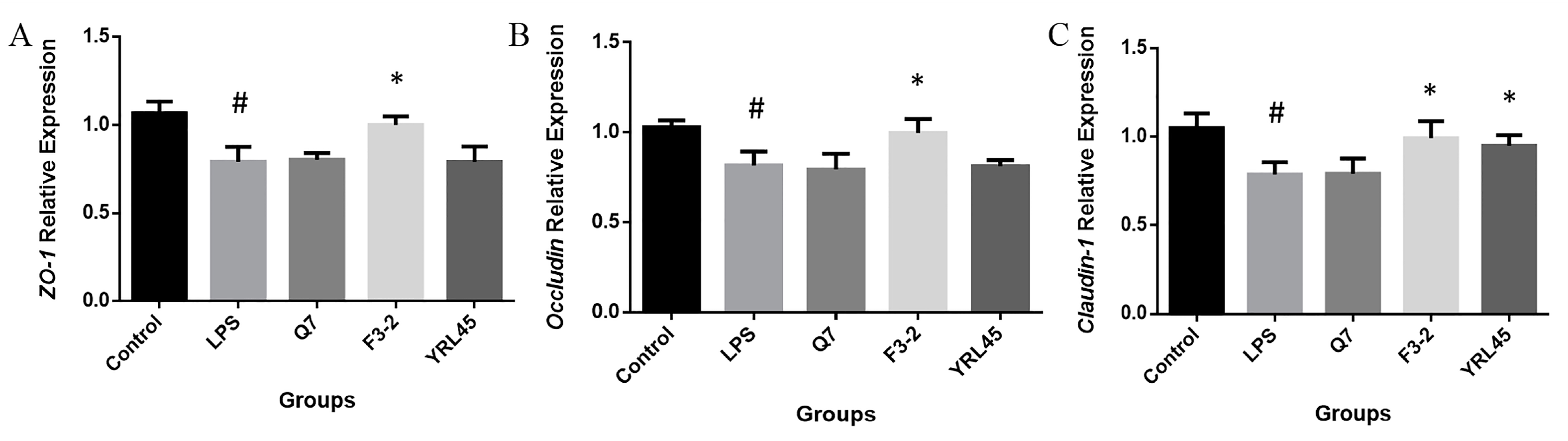
3.8. Tight Junction Intestinal Barrier Function of Bacteriocin-Producing L. plantarum
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peng, M.; Tabashsum, Z.; Anderson, M.; Truong, A.; Houser, A.K.; Padilla, J.; Akmel, A.; Bhatti, J.; Rahaman, S.O.; Biswas, D. Effectiveness of probiotics, prebiotics, and prebiotic-like components in common functional foods. Compr. Rev. Food Sci. F. 2020, 19, 1908–1933. [Google Scholar] [CrossRef] [PubMed]
- WHO/FAO|Probiotics in Food. Health and Nutritional Properties and Guidelines for Evaluation. Available online: https://www.fao.org/3/a0512e/a0512e.pdf (accessed on 10 May 2022).
- Dobson, A.; Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocin production: A probiotic trait? Appl. Environ. Microb. 2012, 78, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egan, K.; Field, D.; Rea, M.C.; Ross, R.P.; Hill, C.; Cotter, P.D. Bacteriocins: Novel solutions to age old spore-related problems? Front. Microbiol. 2016, 7, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, C.C.; Silva, S.P.; Ribeiro, S.C. Application of bacteriocins and protective cultures in dairy food preservation. Front. Microbiol. 2018, 9, 594. [Google Scholar] [CrossRef]
- Field, D.; Ross, R.P.; Hill, C. Developing bacteriocins of lactic acid bacteria into next generation biopreservatives. Curr. Opin. Food Sci. 2018, 20, 1–6. [Google Scholar] [CrossRef]
- Cagno, R.D.; Angelis, M.D.; Calasso, M.; Vincentini, O.; Vernocchi, P.; Ndagijimana, M.; Vincenzi, M.D.; Dessì, M.R.; Guerzoni, M.E.; Gobbetti, M. Quorum sensing in sourdough Lactobacillus plantarum DC400: Induction of plantaricin A (PlnA) under co-cultivation with other lactic acid bacteria and effect of PlnA on bacterial and Caco-2 cells. Proteomics 2010, 10, 2175–2190. [Google Scholar] [CrossRef] [Green Version]
- Guinane, C.M.; Lawton, E.M.; O’Connor, P.M.; O’Sullivan, Ó.; Hill, C.; Ross, R.P.; Cotter, P.D. The bacteriocin bactofencin A subtly modulates gut microbial populations. Anaerobe 2016, 40, 41–49. [Google Scholar] [CrossRef]
- Bai, L.; Kumar, S.; Verma, S.; Seshadri, S. Bacteriocin PJ4 from probiotic lactobacillus reduced adipokine and inflammasome in high fat diet induced obesity. Biotech 2020, 10, 355. [Google Scholar] [CrossRef]
- Małaczewska, J.; Kaczorek-Łukowska, E.; Wójcik, R.; Rękawek, W.; Siwicki, A.K. In vitro immunomodulatory effect of nisin on porcine leucocytes. J. Anim. Physiol. Anim. Nutr. 2019, 103, 882–893. [Google Scholar] [CrossRef]
- Wang, S.; Ye, Q.; Wang, K.; Zeng, X.; Huang, S.; Yu, H.; Ge, Q.; Qi, D.; Qiao, S. Enhancement of macrophage function by the antimicrobial peptide sublancin protects mice from methicillin-resistant Staphylococcus aureus. J. Immunol. Res. 2019, 2019, 3979352. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.; Teng, K.; Liu, Y.; Cao, Y.; Wang, T.; Ma, C.; Zhang, J.; Zhong, J. Bacteriocins: Potential for human health. Oxid. Med. Cell. Longev. 2021, 2021, 5518825. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sheng, Y.; Pan, Q.; Xue, Y.; Yu, L.; Tian, F.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Identification of the key physiological characteristics of Lactobacillus plantarum strains for ulcerative colitis alleviation. Food Funct. 2020, 11, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Chang, H.C.; Kim, H.Y. Complete genome sequence of Lactobacillus plantarum EM, a putative probiotic strain with the cholesterol-lowering effect and antimicrobial activity. Curr. Microbiol. 2020, 77, 1871–1882. [Google Scholar] [CrossRef] [PubMed]
- Maldonado-Barragán, A.; Caballero-Guerrero, B.; Lucena-Padrós, H.; Ruiz-Barba, J.L. Induction of bacteriocin production by coculture is widespread among plantaricin-producing Lactobacillus plantarum strains with different regulatory operons. Food Microbiol. 2013, 33, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Cho, G.S.; Huch, M.; Hanak, A.; Holzapfel, W.H.; Franz, C.M. Genetic analysis of the plantaricin EFI locus of Lactobacillus plantarum PCS20 reveals an unusual plantaricin E gene sequence as a result of mutation. Int. J. Food Microbiol. 2010, 141, S117–S124. [Google Scholar] [CrossRef]
- Bu, Y.; Yang, H.; Li, J.; Liu, Y.; Liu, T.; Gong, P.; Zhang, L.; Wang, S.; Yi, H. Comparative Metabolomics Analyses of plantaricin Q7 production by Lactobacillus plantarum Q7. J. Agric. Food Chem. 2021, 69, 10741–10748. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, Q.; Lu, F.; Bie, X.; Zhao, H.; Lu, Z.; Lu, Y. A novel class IIb bacteriocin-plantaricin EmF effectively inhibits Listeria monocytogenes and extends the shelf life of beef in combination with chitosan. J. Agric. Food Chem. 2022, 70, 2187–2196. [Google Scholar] [CrossRef]
- Bu, Y.; Liu, Y.; Li, J.; Liu, T.; Gong, P.; Zhang, L.; Wang, Y.; Yi, H. Analyses of plantaricin Q7 synthesis by Lactobacillus plantarum Q7 based on comparative transcriptomics. Food Control. 2021, 124, 107909. [Google Scholar] [CrossRef]
- Reuben, R.C.; Roy, P.C.; Sarkar, S.L.; Alam, A.R.U.; Jahid, I.K. Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J. Dairy Sci. 2020, 103, 1223–1237. [Google Scholar] [CrossRef]
- Wang, G.; Yu, Y.; Garcia-Gutierrez, E.; Jin, X.; He, Y.; Wang, L.; Tian, P.; Liu, Z.; Zhao, J.; Zhang, H.; et al. Lactobacillus acidophilus JCM 1132 strain and its mutant with different bacteriocin-producing behaviour have various in situ effects on the gut microbiota of healthy mice. Microorganisms 2019, 8, 49. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Lv, Y.; Zhang, Z.; Yi, H.; Liu, T.; Li, R.; Yu, Z.; Zhang, L. Study on intestinal survival and cholesterol metabolism of probiotics. LWT-Food Sci. Technol. 2020, 124, 109132. [Google Scholar] [CrossRef]
- Chen, T.; Wang, L.; Li, Q.; Long, Y.; Lin, Y.; Yin, J.; Zeng, Y.; Huang, L.; Yao, T.; Abbasi, M.N.; et al. Functional probiotics of lactic acid bacteria from Hu sheep milk. BMC Microbiol. 2020, 20, 228. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Tian, L.; Huang, X.; Liu, Z.; Jia, K.; Wei, H.; Tao, X. Characterization and antitumor activity of novel exopolysaccharide APS of Lactobacillus plantarum WLPL09 from human breast milk. Int. J. Biol. Macromol. 2020, 163, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Qiu, Z.; Tian, F.; Yu, L.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Effect of bacteriocin-producing Pediococcus acidilactici strains on the immune system and intestinal flora of normal mice. Food Sci. Hum. Well. 2022, 11, 238–246. [Google Scholar] [CrossRef]
- Kaewiad, K.; Kaewnopparat, S.; Kaewnopparat, N. In vitro comparison of probiotic properties of Lactobacillus fermentum SK54 isolated from new born baby with Lactobacillus rhamnosus GG ATCC 53103. Adv. Mat. Res. 2015, 1060, 215–218. [Google Scholar] [CrossRef]
- Davoodabadi, A.; Dallal, M.M.S.; Foroushani, A.R.; Douraghi, M.; Yazdi, M.K.S.; Harati, F.A. Antibacterial activity of Lactobacillus spp. isolated from the feces of healthy infants against enteropathogenic bacteria. Anaerobe 2015, 34, 53–58. [Google Scholar] [CrossRef]
- Saini, K.; Tomar, S.K. In vitro evaluation of probiotic potential of Lactobacillus cultures of human origin capable of selenium bioaccumulation. LWT-Food Sci. Technol. 2017, 84, 497–504. [Google Scholar] [CrossRef]
- Yin, X.; Heeney, D.; Srisengfa, Y.; Golomb, B.; Griffey, S.; Marco, M. Bacteriocin biosynthesis contributes to the anti-inflammatory capacities of probiotic Lactobacillus plantarum. Benef. Microbes 2018, 9, 333–344. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, X.; Hao, Y.; Ding, J.; Shen, J.; Xue, Z.; Qi, W.; Li, Z.; Song, Y.; Zhang, T.; et al. Protective effects of a novel probiotic strain, Lactococcus lactis ML2018, in colitis: In vivo and in vitro evidence. Food Funct. 2019, 10, 1132–1145. [Google Scholar] [CrossRef]
- Sheibanie, A.F.; Yen, J.H.; Khayrullina, T.; Emig, F.; Zhang, M.; Tuma, R.; Ganea, D. The proinflammatory effect of prostaglandin E2 in experimental inflammatory bowel disease is mediated through the IL-23→IL-17 axis. J. Immunol. 2007, 178, 8138–8147. [Google Scholar] [CrossRef] [Green Version]
- Rojas, J.M.; Avia, M.; Martín, V.; Sevilla, N. IL-10: A multifunctional cytokine in viral infections. J. Immunol. Res. 2017, 2017, 6104054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, F.; Teng, K.; Liu, Y.; Wang, T.; Xia, T.; Yun, F.; Zhong, J. Nisin Z attenuates lipopolysaccharide-induced mastitis by inhibiting the ERK1/2 and p38 mitogen-activated protein kinase signaling pathways. J. Dairy Sci. 2022, 105, 3530–3543. [Google Scholar] [CrossRef] [PubMed]
- Mouritzen, M.V.; Andrea, A.; Qvist, K.; Poulsen, S.S.; Jenssen, H. Immunomodulatory potential of Nisin A with application in wound healing. Wound. Repair. Regen. 2019, 27, 650–660. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Ding, X.; Shang, L.; Zeng, X.; Liu, H.; Li, N.; Huang, S.; Wang, Y.; Wang, G.; Cai, S.; et al. Protective ability of biogenic antimicrobial peptide microcin J25 against enterotoxigenic Escherichia coli-induced intestinal epithelial dysfunction and inflammatory responses IPEC-J2 cells. Front. Cell. Infect. Microbiol. 2018, 8, 242. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Mariscal, L.; Betanzos, A.; Nava, P.; Jaramillo, B.E. Tight junction proteins. Prog. Biophys. Mol. Biol. 2003, 81, 1–44. [Google Scholar] [CrossRef]
- Blackwood, B.P.; Yuan, C.Y.; Wood, D.R.; Nicolas, J.D.; Grothaus, J.S.; Hunter, C.J. Probiotic Lactobacillus species strengthen intestinal barrier function and tight junction integrity in experimental necrotizing enterocolitis. J. Prob. Health 2017, 5, 1000159. [Google Scholar] [CrossRef]
- Heeney, D.D.; Zhai, Z.; Bendiks, Z.; Barouei, J.; Martinic, A.; Slupsky, C.; Marco, M.L. Lactobacillus plantarum bacteriocin is associated with intestinal and systemic improvements in diet-induced obese mice and maintains epithelial barrier integrity in vitro. Gut Microbes 2019, 10, 382–397. [Google Scholar] [CrossRef] [Green Version]
| Genes | Primer Sequences (5′-3′) |
|---|---|
| GAPDH | F: AACGGATTTGGTCGTATTG |
| R: GCTCCTGGAAGATGGTGAT | |
| ZO-1 | F: CGGGACTGTTGGTATTGGCTAGA |
| R: GGCCAGGGCCATAGTAAAGTTTG | |
| Occludin | F: GGGCATTGCTCATCCTGAAG |
| R: GCCTGTAAGGAGGTGGACTT | |
| Claudin-1 | F: GCGCGATATTTCTTCTTGCAGG |
| R: TTCGTACCTGGCATTGACTGG |
| Strains | Inhibition Zone Diameter (mm) | |
|---|---|---|
| Acid Not Excluded | Acid Excluded | |
| Q7 | 26.85 ± 0.40 | 26.94 ± 0.56 |
| F3-2 | 27.08 ± 0.51 | 26.98 ± 0.41 |
| YRL45 | 26.77 ± 0.22 | 26.89 ± 0.29 |
| JLSC2-5 | 13.89 ± 0.39 | 13.87 ± 0.38 |
| CXG-2 | 13.30 ± 0.33 | 7.80 |
| HZPS-2 | 13.00 ± 0.65 | 7.80 |
| JLSC1-2 | 12.97 ± 0.29 | 7.80 |
| CXG-8 | 12.91 ± 0.41 | 7.80 |
| CXG-6 | 12.49 ± 0.50 | 7.80 |
| NDF1000S-4 | 12.41 ± 0.41 | 7.80 |
| JLSC2-4 | 12.40 ± 0.43 | 7.80 |
| JNS-3 | 12.00 ± 0.33 | 7.80 |
| SXLJ-2 | 11.84 ± 0.26 | 7.80 |
| HZZC-2 | 11.53 ± 0.47 | 7.80 |
| HCG4-2 | 11.47 ± 0.33 | 7.80 |
| SHG-1 | 10.87 ± 0.51 | 7.80 |
| CXG-4 | 10.77 ± 0.40 | 7.80 |
| HZPS-1 | 10.14 ± 0.54 | 7.80 |
| Strains | Inhibition Zone Diameter (mm) | Titer (AU/mL) | |
|---|---|---|---|
| Without Catalase | With Catalase | With or Without Catalase | |
| Q7 | 27.03 ± 0.23 a | 26.96 ± 0.40 a | 400 |
| F3-2 | 26.95 ± 0.36 a | 26.99 ± 0.39 a | 400 |
| YRL45 | 26.95 ± 0.50 a | 26.89 ± 0.34 a | 400 |
| JLSC2-5 | 13.93 ± 0.36 a | 13.90 ± 0.25 a | 50 |
| Strains | Acid (3 h of Incubation) | Bile Salt (5 h of Incubation) | ||
|---|---|---|---|---|
| Viable Counts (lg CFU/mL) | Survival Rate (%) | Viable Counts (lg CFU/mL) | Survival Rate (%) | |
| Q7 | 8.71 ± 0.06 | 93.43 ± 0.16 | 7.44 ± 0.04 | 80.05 ± 0.84 |
| F3-2 | 8.68 ± 0.04 | 92.82 ± 0.97 | 7.12 ± 0.03 | 76.37 ± 0.24 |
| YRL45 | 8.99 ± 0.07 | 96.40 ± 0.63 | 7.29 ± 0.03 | 78.08 ± 0.28 |
| Strains | Gastric Juice (3 h of Incubation) | Intestinal Juice (8 h of Incubation) | ||
|---|---|---|---|---|
| Viable Counts (lg CFU/mL) | Survival Rate (%) | Viable Counts (lg CFU/mL) | Survival Rate (%) | |
| Q7 | 8.75 ± 0.05 | 93.97 ± 1.04 | 8.64 ± 0.04 | 99.02 ± 0.89 |
| F3-2 | 8.67 ± 0.01 | 93.04 ± 0.52 | 8.48 ± 0.07 | 98.77 ± 0.55 |
| YRL45 | 8.65 ± 0.06 | 92.91 ± 1.18 | 8.52 ± 0.04 | 98.96 ± 0.95 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bu, Y.; Liu, Y.; Liu, Y.; Wang, S.; Liu, Q.; Hao, H.; Yi, H. Screening and Probiotic Potential Evaluation of Bacteriocin-Producing Lactiplantibacillus plantarum In Vitro. Foods 2022, 11, 1575. https://doi.org/10.3390/foods11111575
Bu Y, Liu Y, Liu Y, Wang S, Liu Q, Hao H, Yi H. Screening and Probiotic Potential Evaluation of Bacteriocin-Producing Lactiplantibacillus plantarum In Vitro. Foods. 2022; 11(11):1575. https://doi.org/10.3390/foods11111575
Chicago/Turabian StyleBu, Yushan, Yisuo Liu, Yinxue Liu, Shaolei Wang, Qiqi Liu, Haining Hao, and Huaxi Yi. 2022. "Screening and Probiotic Potential Evaluation of Bacteriocin-Producing Lactiplantibacillus plantarum In Vitro" Foods 11, no. 11: 1575. https://doi.org/10.3390/foods11111575
APA StyleBu, Y., Liu, Y., Liu, Y., Wang, S., Liu, Q., Hao, H., & Yi, H. (2022). Screening and Probiotic Potential Evaluation of Bacteriocin-Producing Lactiplantibacillus plantarum In Vitro. Foods, 11(11), 1575. https://doi.org/10.3390/foods11111575







