Lactic Acid Bacteria Isolated from Fermented Doughs in Spain Produce Dextrans and Riboflavin
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation
2.2. DNA Extraction and Random Amplified Polymorphic DNA (RAPD) Pattern Analysis
2.3. Characterization of rrs and pheS Genes
2.4. Detection of EPS Production on Solid Media and by Transmission Electron Microscopy (TEM)
2.5. Production, Purification, and Quantification of Dextran
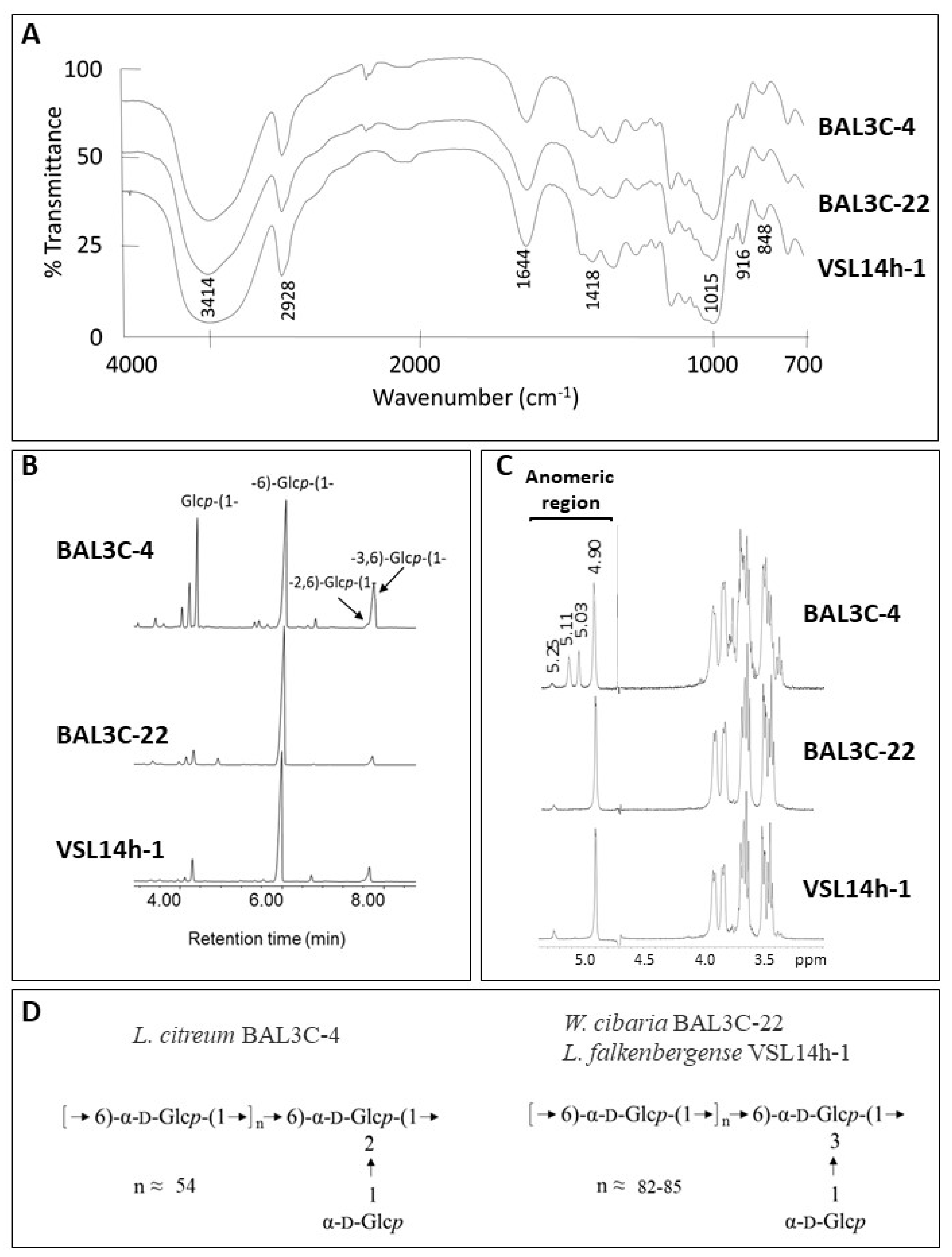
2.6. Characterization of the EPS
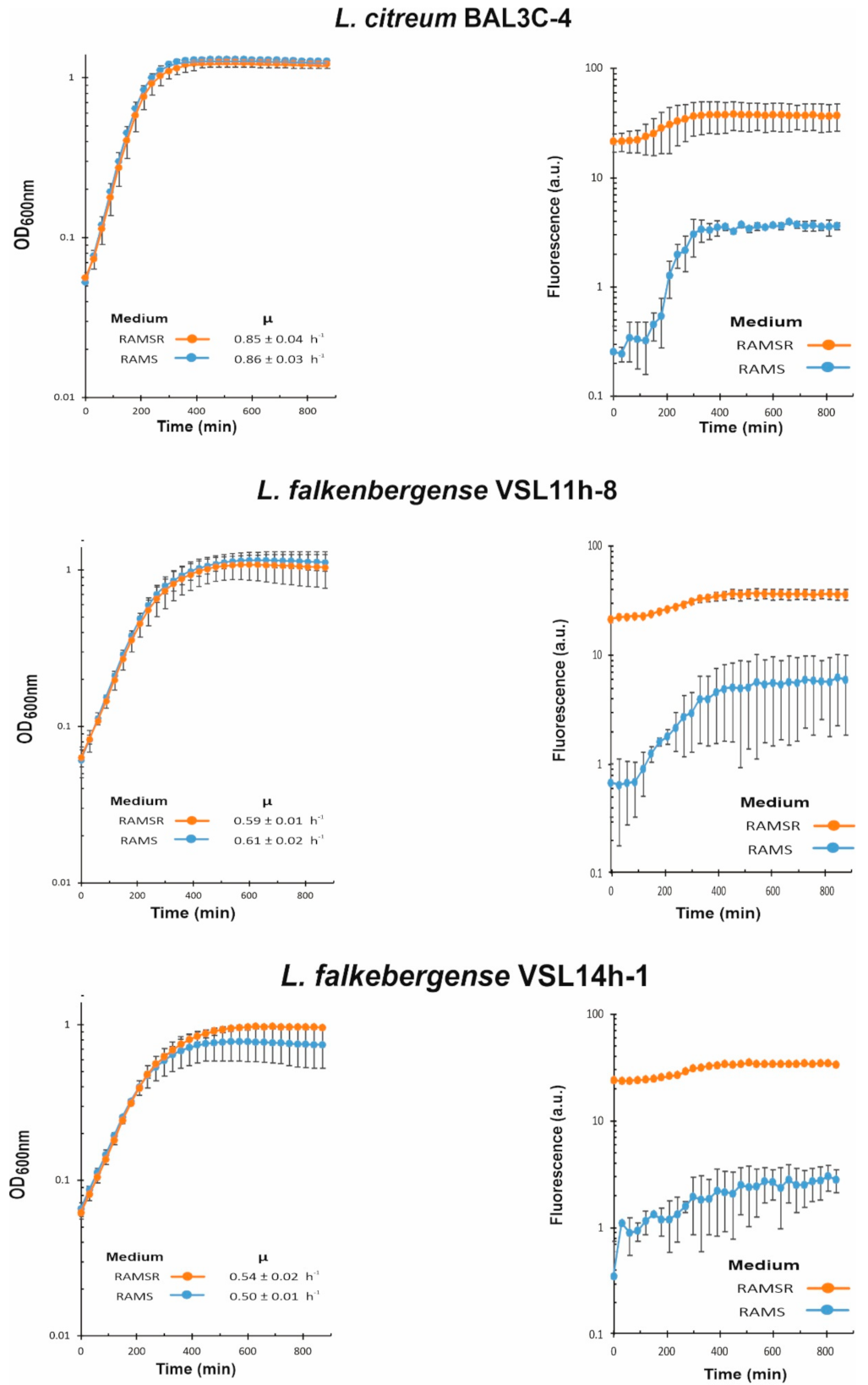
2.7. Analysis of Bacterial Growth and Riboflavin Production and Quantification
2.8. Statistical Analysis
3. Results and Discussion
3.1. Isolated Strains
3.2. RAPD Fingerprinting
3.3. Analysis of the rrs and pheS Gene Sequences
3.4. Detection of EPS Production by LAB Strains
3.5. Influence of Carbon Source on LAB Growth
3.6. Analysis of Riboflavin Production by LAB during Growth
3.7. Production and Purification of the EPS
3.8. Characterization of the EPS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Mora-Villalobos, J.A.; Montero-Zamora, J.; Barboza, N.; Rojas-Garbanzo, C.; Usaga, J.; Redondo-Solano, M.; Schroedter, L.; Olszewska-Widdrat, A.; Lopez-Gomez, J.P. Multi-product lactic acid bacteria fermentations: A review. Fermentation 2020, 6, 23. [Google Scholar] [CrossRef]
- Ercolini, D.; Pontonio, E.; De Filippis, F.; Minervini, F.; La Storia, A.; Gobbetti, M.; Di Cagno, R. Microbial ecology dynamics during rye and wheat sourdough preparation. Appl. Environ. Microbiol. 2013, 79, 7827–7836. [Google Scholar] [CrossRef] [PubMed]
- Bessmeltseva, M.; Viiard, E.; Simm, J.; Paalme, T.; Sarand, I. Evolution of bacterial consortia in spontaneously started rye sourdoughs during two months of daily propagation. PLoS ONE 2014, 9, e95449. [Google Scholar] [CrossRef] [PubMed]
- Nataraj, B.H.; Ali, S.A.; Behare, P.V.; Yadav, H. Postbiotics-parabiotics: The new horizons in microbial biotherapy and functional foods. Microb. Cell Fact. 2020, 19, 168. [Google Scholar] [CrossRef]
- Katina, K.; Maina, N.H.; Juvonen, R.; Flander, L.; Johansson, L.; Virkki, L.; Tenkanen, M.; Laitila, A. In situ production and analysis of Weissella confusa dextran in wheat sourdough. Food Microbiol. 2009, 26, 734–743. [Google Scholar] [CrossRef]
- Galle, S.; Schwab, C.; Arendt, E.; Gänzle, M. Exopolysaccharide-forming Weissella strains as starter cultures for sorghum and wheat sourdoughs. J. Agric. Food Chem. 2010, 58, 5834–5841. [Google Scholar] [CrossRef]
- Wolter, A.; Hager, A.S.; Zannini, E.; Galle, S.; Gänzle, M.G.; Waters, D.M.; Arendt, E.K. Evaluation of exopolysaccharide producing Weissella cibaria MG1 strain for the production of sourdough from various flours. Food Microbiol. 2014, 37, 44–50. [Google Scholar] [CrossRef]
- Lynch, K.M.; Coffey, A.; Arendt, E.K. Exopolysaccharide producing lactic acid bacteria: Their techno-functional role and potential application in gluten-free bread products. Food Res. Int. 2018, 110, 52–61. [Google Scholar] [CrossRef]
- Boyaci Gunduz, C.P.; Gaglio, R.; Franciosi, E.; Settanni, L.; Erten, H. Molecular analysis of the dominant lactic acid bacteria of chickpea liquid starters and doughs and propagation of chickpea sourdoughs with selected Weissella confusa. Food Microbiol. 2020, 91, 103490. [Google Scholar] [CrossRef]
- Bounaix, M.S.; Robert, H.; Gabriel, V.; Morel, S.; Remaud-Siméon, M.; Gabriel, B.; Fontagné-Faucher, C. Characterization of dextran-producing Weissella strains isolated from sourdoughs and evidence of constitutive dextransucrase expression. FEMS Microbiol. Lett. 2010, 311, 18–26. [Google Scholar] [CrossRef]
- Zarour, K.; Llamas-Arriba, M.G.; Prieto, A.; Ruas-Madiedo, P.; Dueñas, M.T.; Fernández de Palencia, P.; Aznar, R.; Kihal, M.; López, P. Rheology and bioactivity of high molecular weight dextrans synthesised by lactic acid bacteria. Carbohydr. Polym. 2017, 174, 646–657. [Google Scholar] [CrossRef]
- Nácher-Vázquez, M.; Ballesteros, N.; Canales, Á.; Rodríguez Saint-Jean, S.; Pérez-Prieto, S.I.; Prieto, A.; Aznar, R.; López, P. Dextrans produced by lactic acid bacteria exhibit antiviral and immunomodulatory activity against salmonid viruses. Carbohydr. Polym. 2015, 124, 292–301. [Google Scholar] [CrossRef]
- Păcularu-Burada, B.; Georgescu, L.A.; Vasile, M.A.; Rocha, J.M.; Bahrim, G.E. Selection of wild lactic acid bacteria strains as promoters of postbiotics in gluten-free sourdoughs. Microorganisms 2020, 8, 643. [Google Scholar] [CrossRef]
- Galli, V.; Venturi, M.; Coda, R.; Maina, N.H.; Granchi, L. Isolation and characterization of indigenous Weissella confusa for in situ bacterial exopolysaccharides (EPS) production in chickpea sourdough. Food Res. Int. 2020, 138, 109785. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Laiño, J.E.; Juarez del Valle, M.; Vannini, V.; Savoy de Giori, G.; Sesma, F.; Taranto, M.P. B-group vitamins production by probiotic lactic acid bacteria. In Biotechnology of Lactic Acid Bacteria: Novel Applications; Mozzi, F.R., Raya, R., Vignolo, G., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 279–296. [Google Scholar]
- Gu, Q.; Liu, P. Biosynthesis of vitamins by probiotic bacteria. In Probiotics and Prebiotics in Human Nutrition and Health; Rao, V., Rao, L., Eds.; InTech: London, UK, 2016; pp. 135–148. [Google Scholar]
- Titcomb, T.J.; Tanumihardjo, S.A. Global concerns with B vitamin statuses: Biofortification, fortification, hidden hunger, interactions, and toxicity. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1968–1984. [Google Scholar] [CrossRef]
- Rollán, G.C.; Gerez, C.L.; Leblanc, J.G. Lactic fermentation as a strategy to improve the nutritional and functional values of pseudocereals. Front. Nutr. 2019, 6, 98. [Google Scholar] [CrossRef]
- FAO/WHO. FAO/WHO Vitamin and Mineral Requirements in Human Nutrition, 2nd ed.; FAO/WHO: Geneva, Switzerland, 2004. [Google Scholar]
- Ventura, M.; Turroni, F.; Zomer, A.; Foroni, E.; Giubellini, V.; Bottacini, F.; Canchava, C.; Claesson, M.J.; He, F.; Mantzourani, M.; et al. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 2009, 5, e1000785. [Google Scholar] [CrossRef]
- Papagianni, M. Metabolic engineering of lactic acid bacteria for the production of industrially important compounds. Comput. Struct. Biotechnol. J. 2012, 3, e201210003. [Google Scholar] [CrossRef]
- Juarez del Valle, M.; Laiño, J.E.; Savoy de Giori, G.; LeBlanc, J.G. Riboflavin producing lactic acid bacteria as a biotechnological strategy to obtain bio-enriched soymilk. Food Res. Int. 2014, 62, 1015–1019. [Google Scholar] [CrossRef]
- Capozzi, V.; Russo, P.; Dueñas, M.T.; López, P.; Spano, G. Lactic acid bacteria producing B-group vitamins: A great potential for functional cereals products. Appl. Microbiol. Biotechnol. 2012, 96, 1383–1394. [Google Scholar] [CrossRef]
- Ewe, J.-A.; Wan-Abdullah, W.-N.; Liong, M.-T. Viability and growth characteristics of Lactobacillus in soymilk supplemented with B-vitamins. Int. J. Food Sci. Nutr. 2010, 61, 87–107. [Google Scholar] [CrossRef]
- Russo, P.; Capozzi, V.; Arena, M.P.; Spadaccino, G.; Dueñas, M.T.; López, P.; Fiocco, D.; Spano, G. Riboflavin-overproducing strains of Lactobacillus fermentum for riboflavin-enriched bread. Appl. Microbiol. Biotechnol. 2014, 98, 3691–3700. [Google Scholar] [CrossRef]
- Thakur, K.; Lule, V.K.; Rajni, C.S.; Kumar, N.; Mandal, S.; Anand, S.; Kumari, V.; Tomar, S.K. Riboflavin-producing probiotic Lactobacilli as a biotechnological strategy to obtain riboflavin-enriched fermented food. J. Pure Appl. Microbiol. 2016, 10, 161–166. [Google Scholar]
- Yépez, A.; Russo, P.; Spano, G.; Khomenko, I.; Biasioli, F.; Capozzi, V.; Aznar, R. In situ riboflavin fortification of different kefir-like cereal-based beverages using selected Andean LAB strains. Food Microbiol. 2019, 77, 61–68. [Google Scholar] [CrossRef]
- Russo, P.; Valeria de Chiara, M.L.; Capozzi, V.; Pia Arena, M.; Amodio, M.L.; Rascon, A.; Dueñas, M.T.; López, P.; Spano, G. Lactobacillus plantarum strains for multifunctional oat-based foods. LWT Food Sci. Technol. 2016, 68, 288–294. [Google Scholar] [CrossRef]
- Rodrigo-Torres, L.; Yépez, A.; Aznar, R.; Arahal, D.R. Genomic insights into five strains of Lactobacillus plantarum with biotechnological potential isolated from chicha, a traditional maize-based fermented beverage from northwestern Argentina. Front. Microbiol. 2019, 10, 2232. [Google Scholar] [CrossRef]
- Chiva, R.; Celador-Lera, L.; Uña, J.A.; Jiménez-López, A.; Espinosa-Alcantud, M.; Mateos-Horganero, E.; Vega, S.; Santos, M.Á.; Velázquez, E.; Tamame, M. Yeast biodiversity in fermented doughs and raw cereal matrices and the study of technological traits of selected strains isolated in Spain. Microorganisms 2021, 9, 47. [Google Scholar] [CrossRef]
- Rivas, R.; Peix, A.; Mateos, P.F.; Trujillo, M.E.; Martínez-Molina, E.; Velázquez, E. Biodiversity of populations of phosphate solubilizing rhizobia that nodulates chickpea in different Spanish soils. Plant Soil 2006, 287, 23–33. [Google Scholar] [CrossRef]
- Carro, L.; Spröer, C.; Alonso, P.; Trujillo, M.E. Diversity of Micromonospora strains isolated from nitrogen fixing nodules and rhizosphere of Pisum sativum analyzed by multilocus sequence analysis. Syst. Appl. Microbiol. 2012, 35, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Naser, S.M.; Thompson, F.L.; Hoste, B.; Gevers, D.; Dawyndt, P.; Vancanneyt, M.; Swings, J. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology 2005, 151, 2141–2150. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions Through Comparative Studies of Nucleotides Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2021, 33, 1870–1874. [Google Scholar] [CrossRef]
- Pérez-Ramos, A.; Mohedano, M.L.; Pardo, M.Á.; López, P. β-glucan-producing Pediococcus parvulus 2.6: Test of probiotic and immunomodulatory properties in zebrafish models. Front. Microbiol. 2018, 9, 1684. [Google Scholar] [CrossRef]
- Dueñas-Chasco, M.T.; Rodríguez-Carvajal, M.A.; Tejero-Mateo, P.; Espartero, J.L.; Irastorza-Iribas, A.; Gil-Serrano, A.M. Structural analysis of the exopolysaccharides produced by Lactobacillus spp. G-77. Carbohydr. Res. 1998, 307, 125–133. [Google Scholar] [CrossRef]
- Llamas-arriba, M.G.; Puertas, A.I.; Prieto, A.; López, P.; Cobos, M.; Miranda, J.I.; Marieta, C.; Ruas-madiedo, P.; Dueñas, M.T. Characterization of dextrans produced by Lactobacillus mali CUPV271 and Leuconostoc carnosum CUPV. Food Hydrocoll. 2019, 89, 613–622. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Notararigo, S.; Nácher-Vázquez, M.; Ibarburu, I.; Werning, M.L.; Fernández de Palencia, P.; Dueñas, M.T.; Aznar, R.; López, P.; Prieto, A. Comparative analysis of production and purification of homo- and hetero-polysaccharides produced by lactic acid bacteria. Carbohydr. Polym. 2013, 94, 57–64. [Google Scholar] [CrossRef]
- Mohedano, M.L.; Hernández-Recio, S.; Yépez, A.; Requena, T.; Martínez-Cuesta, M.C.; Peláez, C.; Russo, P.; LeBlanc, J.G.; Spano, G.; Aznar, R.; et al. Real-time detection of riboflavin production by Lactobacillus plantarum strains and tracking of their gastrointestinal survival and functionality in vitro and in vivo using mCherry labeling. Front. Microbiol. 2019, 10, 1748. [Google Scholar] [CrossRef]
- Widdel, F. Theory and Measurement of Bacterial Growth. In Grundpraktikum Mikrobiologie; Bremen University: Bremen, Germany, 2010; pp. 1–11. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Cirrincione, S.; Breuer, Y.; Mangiapane, E.; Mazzoli, R.; Pessione, E. ’Ropy’ phenotype, exopolysaccharides and metabolism: Study on food isolated potential probiotics LAB. Microbiol. Res. 2018, 214, 137–145. [Google Scholar] [CrossRef]
- Palla, M.; Cristani, C.; Giovannetti, M.; Agnolucci, M. Large genetic intraspecific diversity of autochthonous lactic acid bacteria and yeasts isolated from PDO tuscan bread sourdough. Appl. Sci. 2020, 10, 1043. [Google Scholar] [CrossRef]
- Montemurro, M.; Celano, G.; De Angelis, M.; Gobbetti, M.; Rizzello, C.G.; Pontonio, E. Selection of non-Lactobacillus strains to be used as starters for sourdough fermentation. Food Microbiol. 2020, 90, 103491. [Google Scholar] [CrossRef]
- Yang, H.; Liu, T.; Zhang, G.; He, G. Intraspecific diversity and fermentative properties of Saccharomyces cerevisiae from Chinese traditional sourdough. LWT 2020, 124, 109195. [Google Scholar] [CrossRef]
- Syrokou, M.K.; Themeli, C.; Paramithiotis, S.; Mataragas, M.; Bosnea, L.; Argyri, A.A.; Chorianopoulos, N.G.; Skandamis, P.N.; Drosinos, E.H. Microbial Ecology of Greek Wheat Sourdoughs, Identified by a Culture-Dependent and a Culture-Independent Approach. Foods 2020, 9, 1603. [Google Scholar] [CrossRef]
- Vandamme, P.; De Bruyne, K.; Pot, B. Phylogenetics and Systematics. In Lactic acid Bacteria: Biodiversity and Taxonomy; Holzapfel, W.H., Wood, B.J.B., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2014; pp. 31–39. [Google Scholar]
- Oshiro, M.; Momoda, R.; Tanaka, M.; Zendo, T.; Nakayama, J. Dense tracking of the dynamics of the microbial community and chemicals constituents in spontaneous wheat sourdough during two months of backslopping. J. Biosci. Bioeng. 2019, 128, 170–176. [Google Scholar] [CrossRef]
- Di Cagno, R.; Pontonio, E.; Buchin, S.; De Angelis, M.; Lattanzi, A.; Valerio, F.; Gobbetti, M.; Calasso, M. Diversity of the lactic acid bacterium and yeast microbiota in the switch from firm- to liquid-sourdough fermentation. Appl. Environ. Microbiol. 2014, 80, 3161–3172. [Google Scholar] [CrossRef]
- Sánchez-juanes, F.; Teixeira-martín, V.; González-buitrago, J.M.; Velázquez, E.; Flores-félix, J.D. Identification of species and subspecies of lactic acid bacteria present in spanish cheeses type “Torta” by MALDI-TOF MS and pheS gene analyses. Microorganisms 2020, 8, 301. [Google Scholar] [CrossRef]
- Bounaix, M.S.; Gabriel, V.; Robert, H.; Morel, S.; Remaud-Siméon, M.; Gabriel, B.; Fontagné-Faucher, C. Characterization of glucan-producing Leuconostoc strains isolated from sourdough. Int. J. Food Microbiol. 2010, 144, 1–9. [Google Scholar] [CrossRef]
- Manini, F.; Casiraghi, M.C.; Poutanen, K.; Brasca, M.; Erba, D.; Plumed-Ferrer, C. Characterization of lactic acid bacteria isolated from wheat bran sourdough. LWT Food Sci. Technol. 2016, 66, 275–283. [Google Scholar] [CrossRef]
- Liu, A.; Jia, Y.; Zhao, L.; Gao, Y.; Liu, G.; Chen, Y.; Zhao, G.; Xu, L.; Shen, L.; Liu, Y.; et al. Diversity of isolated lactic acid bacteria in Ya’an sourdoughs and evaluation of their exopolysaccharide production characteristics. LWT 2018, 95, 17–22. [Google Scholar] [CrossRef]
- Corsetti, A.; Lavermicocca, P.; Morea, M.; Baruzzi, F.; Tosti, N.; Gobbetti, M. Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of Southern Italy. Int. J. Food Microbiol. 2001, 64, 95–104. [Google Scholar] [CrossRef]
- Yu, J.; Wang, W.H.; Menghe, B.L.G.; Jiri, M.T.; Wang, H.M.; Liu, W.J.; Bao, Q.H.; Lu, Q.; Zhang, J.C.; Wang, F.; et al. Diversity of lactic acid bacteria associated with traditional fermented dairy products in Mongolia. J. Dairy Sci. 2011, 94, 3229–3241. [Google Scholar] [CrossRef]
- Laguerre, S.; Amari, M.; Vuillemin, M.; Robert, H.; Loux, V.; Klopp, C.; Morel, S.; Gabriel, B.; Remaud-Siméon, M.; Gabriel, V.; et al. Genome sequences of three Leuconostoc citreum strains, LBAE C10, LBAE C11, and LBAE E16, Isolated from wheat sourdoughs. J. Bacteriol. 2012, 194, 1610–1611. [Google Scholar] [CrossRef][Green Version]
- Lattanzi, A.; Minervini, F.; Di Cagno, R.; Diviccaro, A.; Antonielli, L.; Cardinali, G.; Cappelle, S.; De Angelis, M.; Gobbetti, M. The lactic acid bacteria and yeast microbiota of eighteen sourdoughs used for the manufacture of traditional Italian sweet leavened baked goods. Int. J. Food Microbiol. 2013, 163, 71–79. [Google Scholar] [CrossRef]
- Zotta, T.; Ricciardi, A.; Parente, E. Enzymatic activities of lactic acid bacteria isolated from Cornetto di Matera sourdoughs. Int. J. Food Microbiol. 2007, 115, 165–172. [Google Scholar] [CrossRef]
- Moroni, A.V.; Arendt, E.K.; Bello, F.D. Biodiversity of lactic acid bacteria and yeasts in spontaneously-fermented buckwheat and teff sourdoughs. Food Microbiol. 2011, 28, 497–502. [Google Scholar] [CrossRef]
- Wu, Y.; Gu, C.T. Leuconostoc falkenbergense sp. Nov., isolated from a lactic culture, fermentating string beans and traditional yogurt. Int. J. Syst. Evol. Microbiol. 2021, 71, 004602. [Google Scholar] [CrossRef]
- Besrour-Aouam, N.; Fhoula, I.; Hernández-Alcántara, A.M.; Mohedano, M.L.; Najjari, A.; Prieto, A.; Ruas-Madiedo, P.; López, P.; Ouzari, H.I. The role of dextran production in the metabolic context of Leuconostoc and Weissella Tunisian strains. Carbohydr. Polym. 2021, 253, 117254. [Google Scholar] [CrossRef]
- Falasconi, I.; Fontana, A.; Patrone, V.; Rebecchi, A.; Duserm Garrido, G.; Principato, L.; Callegari, M.L.; Spigno, G.; Morelli, L. Genome-assisted characterization of Lactobacillus fermentum, Weissella cibaria and Weissella confusa strains isolated from sorghum as starters for sourdough fermentation. Microorganisms 2020, 8, 1388. [Google Scholar] [CrossRef]
- Son, J.; Jang, S.H.; Cha, J.W.; Jeong, K.J. Development of CRISPR interference (CRISPRi) platform for metabolic engineering of Leuconostoc citreum and its application for engineering riboflavin biosynthesis. Int. J. Mol. Sci. 2020, 21, 5614. [Google Scholar] [CrossRef]
- Han, J.; Hang, F.; Guo, B.; Liu, Z.; You, C.; Wu, Z. Dextran synthesized by Leuconostoc mesenteroides BD1710 in tomato juice supplemented with sucrose. Carbohydr. Polym. 2014, 112, 556–562. [Google Scholar] [CrossRef]
- Domingos-Lopes, M.F.P.; Lamosa, P.; Stanton, C.; Ross, R.P.; Silva, C.C.G. Isolation and characterization of an exopolysaccharide-producing Leuconostoc citreum strain from artisanal cheese. Lett. Appl. Microbiol. 2018, 67, 570–578. [Google Scholar] [CrossRef]
- Kothari, D.; Das, D.; Patel, S.; Goyal, A. Dextran and food application. In Polysaccharides: Bioactivity and Biotechnology; Gopal Ramawat, K., Mérillon, J.-M., Eds.; Springer International Publishing: Cham, Switzerland, 2014; ISBN 978-3-319-03751-6. [Google Scholar]
- Wang, Y.; Maina, N.H.; Coda, R.; Katina, K. Challenges and opportunities for wheat alternative grains in breadmaking: Ex-situ- versus in-situ-produced dextran. Trends Food Sci. Technol. 2021, 113, 232–244. [Google Scholar] [CrossRef]
- Salazar, N.; Ruas-Madiedo, P.; Prieto, A.; Calle, L.P.; de los Reyes-Gavilán, C.G. Characterization of exopolysaccharides produced by Bifidobacterium longum NB667 and its cholate-resistant derivative strain IPLA B667dCo. J. Agric. Food Chem. 2012, 60, 1028–1035. [Google Scholar] [CrossRef]
- Rosca, I.; Petrovici, A.R.; Peptanariu, D.; Nicolescu, A.; Dodi, G.; Avadanei, M.; Ivanov, I.C.; Bostanaru, A.C.; Mares, M.; Ciolacu, D. Biosynthesis of dextran by Weissella confusa and its In vitro functional characteristics. Int. J. Biol. Macromol. 2018, 107, 1765–1772. [Google Scholar] [CrossRef]
- Siddiqui, N.N.; Aman, A.; Silipo, A.; Alil Ul Qader, S.; Molinaro, A. Structural analysis and characterization of dextran produced by wild and mutant strains of Leuconostoc mesenteroides. Carbohydr. Polym. 2014, 99, 331–338. [Google Scholar] [CrossRef]
- Maina, N.H.; Tenkanen, M.; Maaheimo, H.; Juvonen, R.; Virkki, L. NMR spectroscopic analysis of exopolysaccharides produced by Leuconostoc citreum and Weissella confusa. Carbohydr. Res. 2008, 343, 1446–1455. [Google Scholar] [CrossRef]
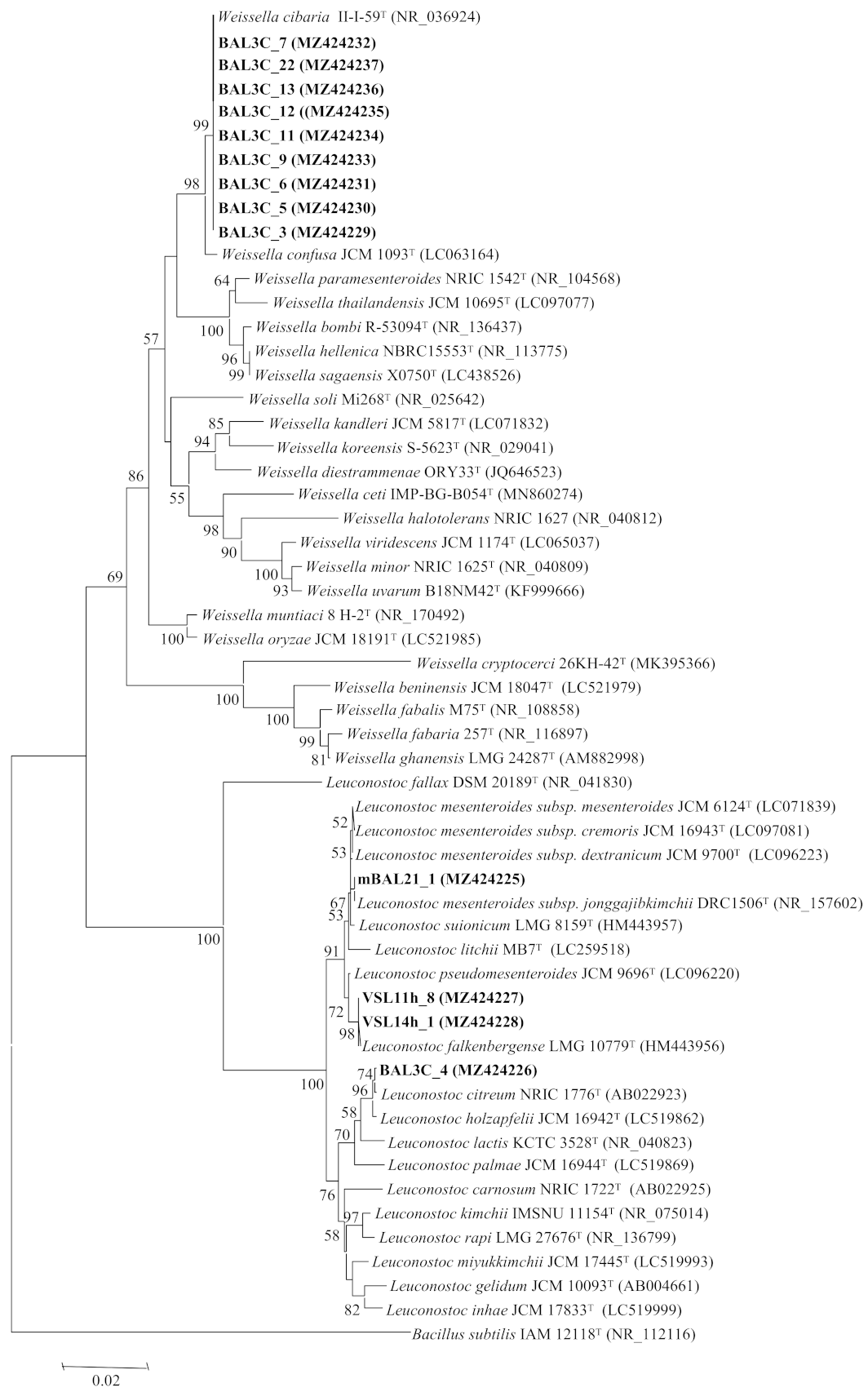
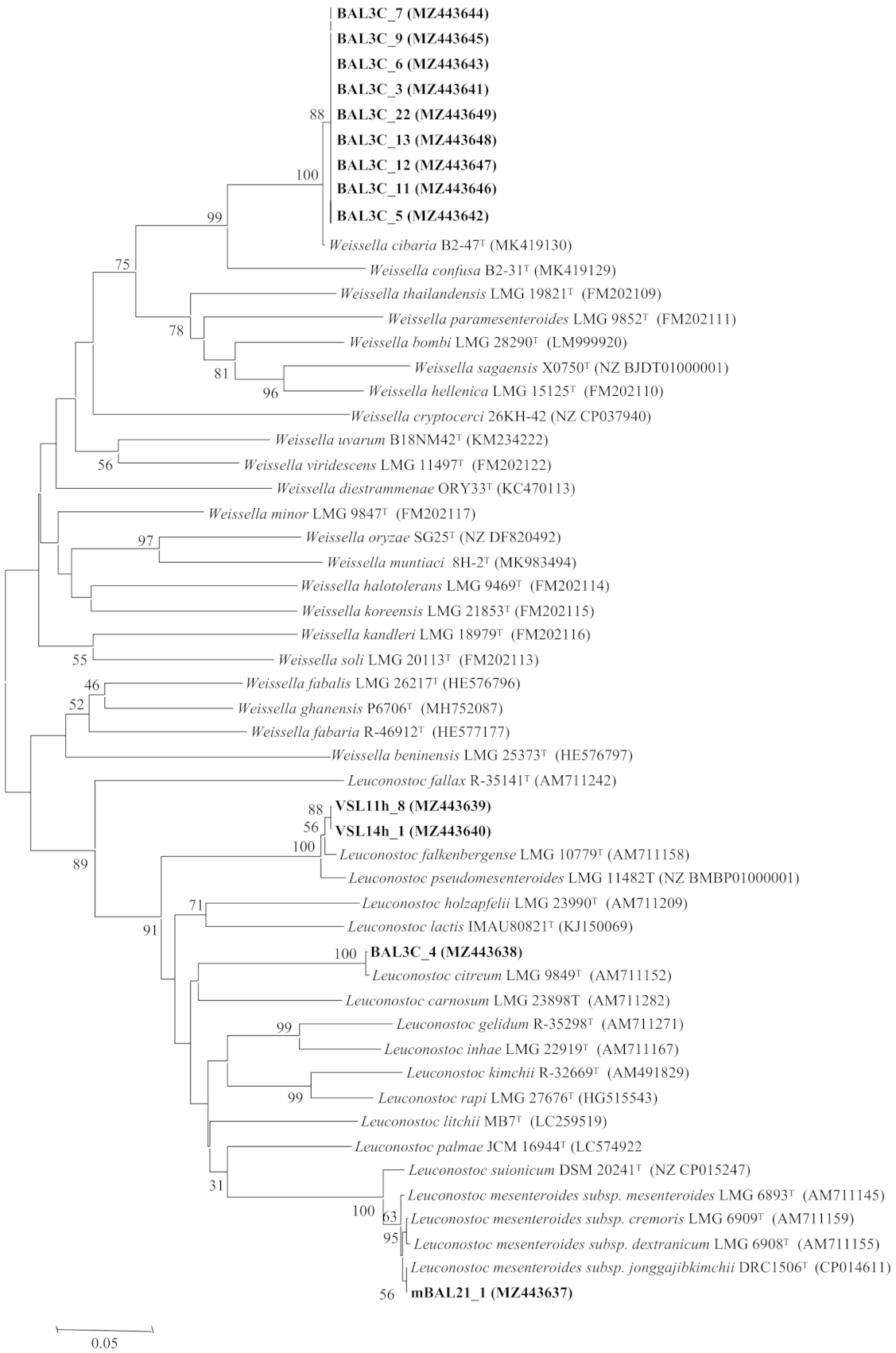
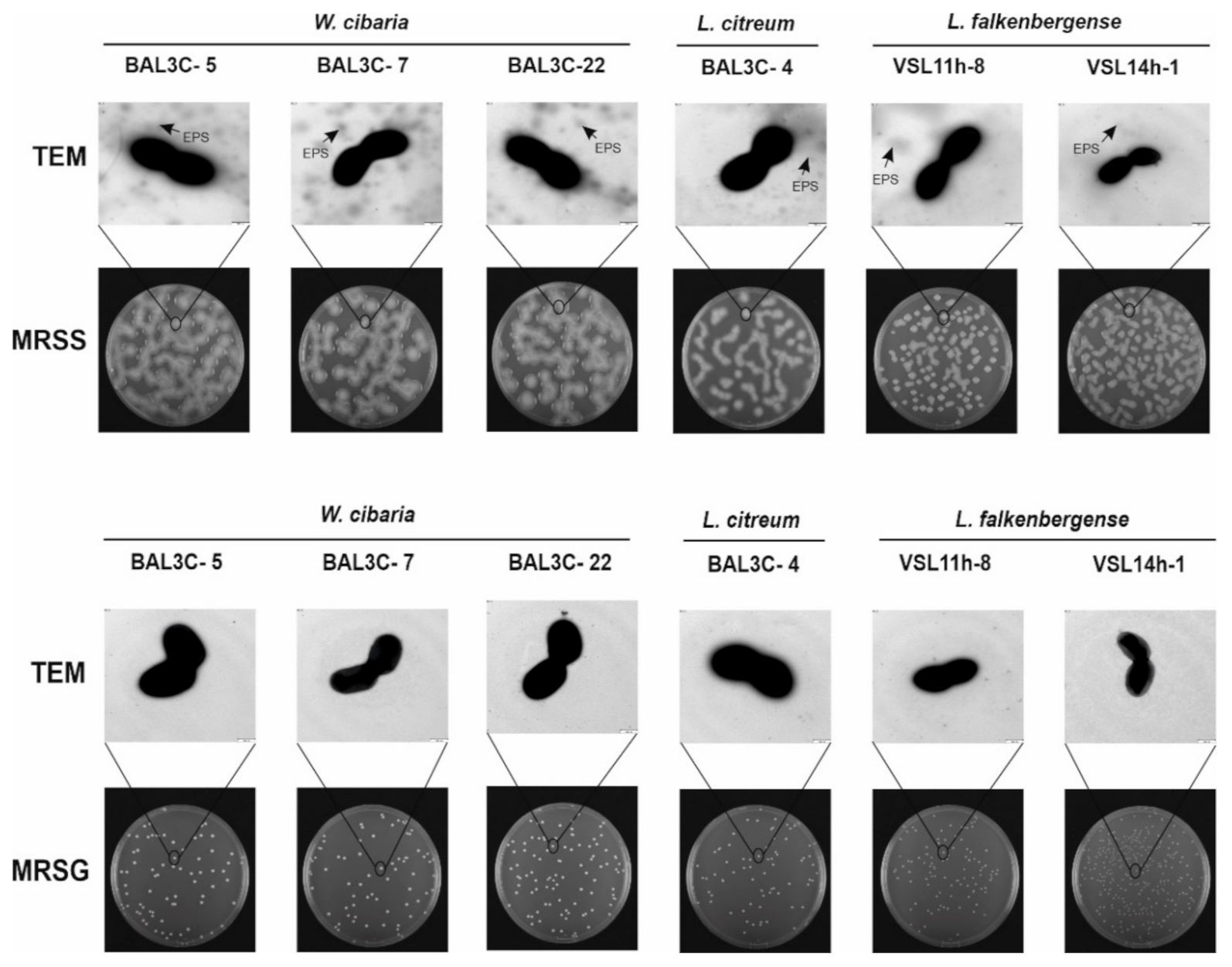
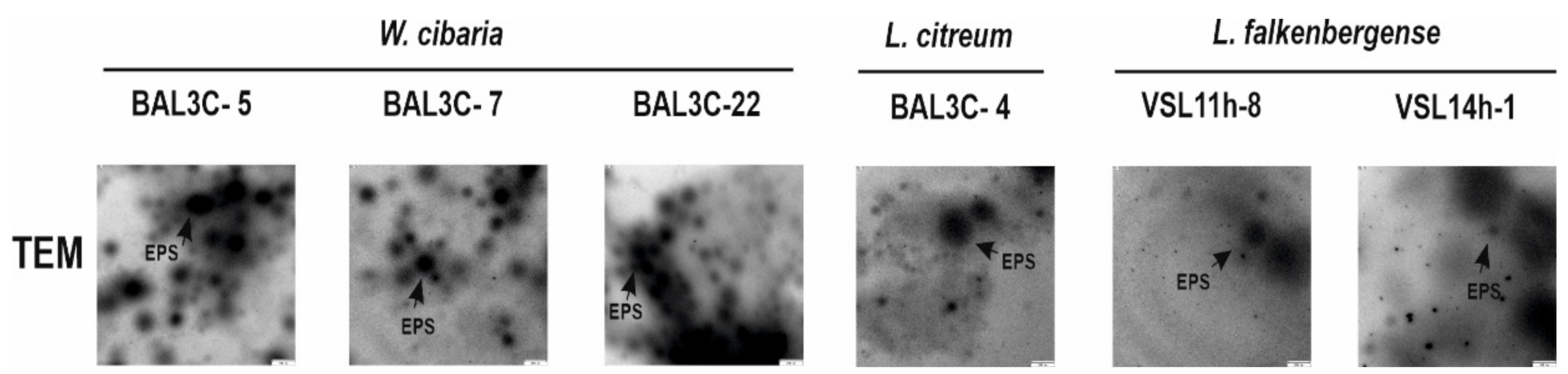


| Strains Codes | RAPD Type | Closest LAB Type Strains | rrs Gene Similarity | pheS Gene Similarity |
|---|---|---|---|---|
| VSL11h-8 | A | L. falkenbergense LMG 10779T | 100% | 99.2% |
| VSL14h-1 | B | L. falkenbergense LMG 10779T | 100% | 99.2% |
| mBAL21_1 | C | L. mesenteroides subsp. jonggajibkimchii JCM 6124T (=LMG 6893T) | 99.9% | 100% |
| BAL3C-4 | D | L. citreum JCM 9698T (=LMG 9849T) | 99.9% | 100% |
| BAL3C-3 | E | W. cibaria JCM 12495T (=LMG 17699T) | 99.9% | 98.9% |
| BAL3C-5 | F | W. cibaria JCM 12495T (=LMG 17699T) | 99.9% | 98.9% |
| BAL3C-6 | G | W. cibaria JCM 12495T (=LMG 17699T) | 99.9% | 98.9% |
| BAL3C-7 | H | W. cibaria JCM 12495T (=LMG 17699T) | 99.9% | 98.9% |
| BAL3C-9 | I | W. cibaria JCM 12495T (=LMG 17699T) | 99.9% | 98.9% |
| BAL3C-11 | J | W. cibaria JCM 12495T (=LMG 17699T) | 99.9% | 98.9% |
| BAL3C-12 | K | W. cibaria JCM 12495T (=LMG 17699T) | 99.9% | 98.9% |
| BAL3C-13 | L | W. cibaria JCM 12495T (=LMG 17699T) | 99.9% | 98.9% |
| BAL3C-22 | M | W. cibaria JCM 12495T (=LMG 17699T) | 99.9% | 98.9% |
| Strain | 1 EPS (g/L) | 2 Riboflavin (μg/L) |
|---|---|---|
| W. cibaria BAL3C-5 | 6.5 ± 0.9 a | 650.3 ± 8.8 b |
| W. cibaria BAL3C-7 | 6.7 ± 0.8 a | 684.5 ± 9.3 a |
| W. cibaria BAL3C-22 | 7.4 ± 0.9 a | 584.3 ± 8.1 c |
| L. citreum BAL3C-4 | 4.9 ± 0.7 b | 325.2 ± 7.3 e |
| L. falkenbergense VSL11h-8 | 0.58 ± 0.05 c | 548.2 ± 9.6 d |
| L. falkenbergense VSL14h-1 | 0.25 ± 0.04 c | 203.5 ± 7.6 f |
| Strain | Supernatant 1 | After Precipitation and Dialysis 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EPS (mg/mL) | DNA 3 (μg/mL) | RNA 3 (ng/mL) | Protein (μg/mL) | Total EPS mg | EPS (mg/mL) | DNA 3 (ng/mL) | RNA 3 (ng/mL) | Protein 3 (μg/mL) | Total EPS mg | |
| W. cibaria BAL3C-5 | 7.8 ± 0.01 | 1.38 | <20 | 110 | 182 | 1.0 ± 0.10 | 60 | <20 | <1.0 | 159 |
| W. cibaria BAL3C-7 | 8.2 ± 0.01 | 0.80 | <20 | <1.0 | 190 | 0.8 ± 0.05 | <0.5 | <20 | <1.0 | 166 |
| W. cibaria BAL3C-22 | 8.6 ± 0.1 | 2.51 | <20 | 144 | 200 | 0.94 ± 0.12 | <0.5 | <20 | <1.0 | 175 |
| L. citreum BAL3C-4 | 5.0 ± 0.6 | 1.07 | <20 | <1.0 | 122 | 0.98 ± 0.05 | 256 | <20 | <1.0 | 107 |
| L. falkenbergense VSL11h-8 | 0.7 ± 0.1 | 0.63 | <20 | <1.0 | 17 | 0.94 ± 0.13 | 798 | <20 | <1.0 | 13 |
| L. falkenbergense VSL14h-1 | 0.3 ± 0.1 | 0.30 | <20 | <1.0 | 7 | 0.84 ± 0.16 | 726 | <20 | <1.0 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llamas-Arriba, M.G.; Hernández-Alcántara, A.M.; Mohedano, M.L.; Chiva, R.; Celador-Lera, L.; Velázquez, E.; Prieto, A.; Dueñas, M.T.; Tamame, M.; López, P. Lactic Acid Bacteria Isolated from Fermented Doughs in Spain Produce Dextrans and Riboflavin. Foods 2021, 10, 2004. https://doi.org/10.3390/foods10092004
Llamas-Arriba MG, Hernández-Alcántara AM, Mohedano ML, Chiva R, Celador-Lera L, Velázquez E, Prieto A, Dueñas MT, Tamame M, López P. Lactic Acid Bacteria Isolated from Fermented Doughs in Spain Produce Dextrans and Riboflavin. Foods. 2021; 10(9):2004. https://doi.org/10.3390/foods10092004
Chicago/Turabian StyleLlamas-Arriba, María Goretti, Annel M. Hernández-Alcántara, Mari Luz Mohedano, Rosana Chiva, Lorena Celador-Lera, Encarnación Velázquez, Alicia Prieto, María Teresa Dueñas, Mercedes Tamame, and Paloma López. 2021. "Lactic Acid Bacteria Isolated from Fermented Doughs in Spain Produce Dextrans and Riboflavin" Foods 10, no. 9: 2004. https://doi.org/10.3390/foods10092004
APA StyleLlamas-Arriba, M. G., Hernández-Alcántara, A. M., Mohedano, M. L., Chiva, R., Celador-Lera, L., Velázquez, E., Prieto, A., Dueñas, M. T., Tamame, M., & López, P. (2021). Lactic Acid Bacteria Isolated from Fermented Doughs in Spain Produce Dextrans and Riboflavin. Foods, 10(9), 2004. https://doi.org/10.3390/foods10092004








