Abstract
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is one of the most extensively antibiotic-resistant pathogens encountered in the clinical setting today. A few studies to-date suggest that CRKP and carbapenem-susceptible K. pneumoniae (CSKP) differ from one another not only with respect to their underlying genetics, but also their transcriptomic and metabolomic fingerprints. Within this context, we characterize the fatty acid methyl ester (FAME) profiles of these pathogens in vitro. Specifically, we evaluated the FAME profiles of six Klebsiella pneumoniae carbapenemase (KPC)-producing isolates belonging to the CC258 lineage (KPC+/258+), six KPC-producing isolates belonging to non-CC258 lineages (KPC+/258−), and six non-KPC-producing isolates belonging to non-CC258 lineages (KPC−/258−). We utilized a single-step sample preparation method to simultaneously lyse bacterial cells and transesterify the lipid fraction, and identified 14 unique FAMEs using gas chromatography-mass spectrometry. The machine learning algorithm Random Forest identified four FAMEs that were highly discriminatory between CC258 and non-CC258 isolates (9(Z)-octadecenoate, 2-phenylacetate, pentadecanoate, and hexadecanoate), of which three were also significantly different in relative abundance between these two groups. These findings suggest that distinct differences exist between CC258 and non-CC258 K. pneumoniae isolates with respect to the metabolism of both fatty acids and amino acids, a hypothesis that is supported by previously-acquired transcriptomic data.
1. Introduction
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is one of the most extensively antibiotic-resistant pathogens encountered in the clinical setting today, with some isolates exhibiting resistance to all available antibiotic compounds, for example [1,2,3]. Infections involving CRKP isolates that produce carbapenemases (i.e., enzymes that hydrolyze carbapenem antibiotics) result in significantly increased morbidity and mortality relative to infections caused by isolates that have developed resistance via alternative mechanisms [4]. Klebsiella pneumoniae carbapenemase (KPC) is the most commonly-encountered carbapenemase throughout much of the Americas, Mediterranean Europe, and parts of the Middle East [5], and most KPC-producing isolates derive from relatively few clonal lineages, of which clonal complex 258 (CC258) is dominant [6]. The mechanisms underlying CC258′s dominance as a carbapenemase-producing lineage remain elusive, although it has been hypothesized that both chromosomal and plasmid-encoded factors may contribute [7,8,9].
A small number of studies have suggested that CRKP and carbapenem-susceptible K. pneumoniae (CSKP) differ from one another not only with respect to their underlying genetics, but also their transcriptomic and metabolomic fingerprints. For example, Bruchmann and colleagues demonstrated that K. pneumoniae isolates belonging to CC258 produced a distinct transcriptomic fingerprint relative to CSKP isolates belonging to a range of other sequence types (STs) [10]. Rees and colleagues demonstrated that CRKP isolates belonging to CC258 produced a distinct volatile metabolic signature relative to both CSKP isolates belonging to other STs, as well as CRKP isolates belonging to non-CC258 lineages [11]. Low and colleagues demonstrated that CRKP and CSKP produced distinct soluble metabolic profiles using a population of resistant isolates that were likely neither CC258 nor KPC-producers based on the geographic region from which they were obtained [12]. Taken together, these findings suggest that carbapenemase-producing K. pneumoniae isolates arising from dominant lineages are transcriptionally and metabolically distinct from non-carbapenemase-producing isolates arising from a diverse range of other, non-dominant genetic lineages.
In the present study, we sought to characterize the fatty acid methyl ester (FAME) profiles of both carbapenemase-producing and non-carbapenemase-producing K. pneumoniae, in vitro. Specifically, we evaluated the FAME profiles of six KPC-producing isolates belonging to the CC258 lineage (KPC+/258+), six KPC-producing isolates belonging to non-CC258 lineages (KPC+/258−), and six non-KPC-producing isolates belonging to non-CC258 lineages (KPC−/258−). We utilized a single-step sample preparation method to simultaneously lyse bacterial cells and transesterify the lipid fraction, according to a procedure previously reported by Müller and colleagues [13]. The FAME profiles were then analyzed using gas chromatography-mass spectrometry (GC-MS) and discriminatory FAMEs were identified using the machine learning algorithm Random Forest (RF) [14]. To the best of our knowledge, the present study is the first to evaluate FAME profiles of K. pneumoniae isolates in the context of antibiotic resistance.
2. Materials and Methods
2.1. Chemicals and Reagents
Methanolic trimethylsulfonium hydroxide solution (0.25 M), a 37 component fatty acid methyl esters (FAMEs) mixture (Supelco® 37 Component FAME Mix), a bacterial fatty acid methyl ester mixture (Bacterial Acid Methyl Ester (BAME) Mix, Supelco), and an alkane mixture (C7–C30) were kindly provided by Supelco (Bellefonte, PA, USA). Methanol and tert-butyl-methyl ether were of GC-grade and were purchased from Sigma-Aldrich (Bellefonte, PA, USA).
2.2. Bacterial Strains, Culture Conditions, and Sample Preparation
Eighteen clinical isolates of K. pneumoniae were preserved at −80 °C in a 20% glycerol solution. A small aliquot of bacterial cells was extracted from this frozen stock and spread over the surface of a solid growth media comprised of TrypticaseTM Soy Broth (Becton Dickinson, Franklin Lakes, NJ, USA) and agar powder (1.5% final agar concentration, Alfa Aesar, Haverhill, MA, USA). Due to the rapid doubling time of K. pneumoniae, cultures were incubated at room temperature overnight to prevent overgrowth. Of the 18 isolates evaluated, six were KPC-positive and belonged to CC258 (KPC+/258+), six were KPC-positive and belonged to lineages other than CC258 (KPC+/258−), and six were KPC-negative and belonged to lineages other than CC258 (KPC−/258−). KPC-negative isolates belonging to CC258 (KPC−/258+) were not evaluated, due to their scarcity in the clinical setting. STs were identified via multilocus sequence typing (MLST), as described previously by Diancourt and colleagues [15]. KPC production was verified both genotypically via the amplification of the blaKPC gene [16], and phenotypically via the ETEST® assay using either meropenem or ertapenem (BioMérieux, Marcy-l’Étoile, France) [11]. Additional information about the strains evaluated in this study is presented in Table 1. These isolates have also been described previously [11].

Table 1.
Klebsiella pneumoniae clinical isolates evaluated in the present study.
Approximately 15 mg of bacterial cells were harvested from the surface of the agar plates and transferred to 1.5 mL Eppendorf tubes. Cells were suspended in 10 µL of distilled water, and then 30 µL of methanolic trimethylsulfonium hydroxide solution (0.25 M) was added to simultaneously lyse cells and transesterify the lipid fraction, according to the procedure reported by Müller and colleagues [13]. The reaction mixture was dried under a nitrogen stream, dissolved in 200 µL of a tert-butyl-methyl ether/methanol (MeOH) mixture (10:1 v/v), and directly injected into the GC-MS system. Three biological replicates were prepared per strain, for a total of 54 samples.
2.3. Analytical Instrumentation
All GC-MS applications were carried out on a Shimadzu GC2010 instrument and a TQ8050 triple quadrupole mass spectrometer (Shimadzu Scientific Instruments, Inc., Columbia, MD, USA) equipped with an AOC-6000 autosampler. Only the single quadrupole acquisition mode was exploited on the TQ8050 MS. Data were acquired by using the GCMS solution software ver. 4.45 (Shimadzu). The column employed was an SLB-5ms [(silphenylene polymer, practically equivalent in polarity to poly (5% diphenyl/95% methylsiloxane)], with the following dimensions: 30 m × 0.25 mm ID × 0.25 μm df (Supelco, Bellefonte, USA). The GC temperature program was, as follows: 50 °C hold for one minute, then +5 °C/min ramp from 50 °C to 300 °C, and +20 °C/min ramp from 300 °C to 350 °C. Helium head pressure (constant linear velocity mode 35 cm/s) was 48 kPa. Injection temperature, mode, and volume were 280 °C, split ratio 1:50, and 1 μL, respectively. The MS system was run in full scan conditions: Scan speed 2000 amu/s; mass range 45–400 m/z. Interface and ion source temperatures: 200 and 250 °C.
2.4. Data Elaboration and Statistical Analysis
Chromatographic data were processed using GCMSsolution (Shimadzu) and then manually aligned across the 54 samples, using a maximum retention time shift of 3 s. A signal-to-noise (S/N) cutoff of 10:1 was used. Unless specified (see Table 2), the resulting peaks were putatively identified based on the combination of a dual filter, namely: (1) The MS similarity with the NIST17 library (≥80%) and, 2) the experimental linear retention index (LRI) within a ±5 range compared to the NIST database and the literature (mainly from [17,18,19]). Contaminants and artifacts (e.g., siloxane, phthalates) were removed, and only compounds with mass spectral fragmentation typical for methylated carboxylic acids were retained. The data matrix obtained was first normalized according to the exact weight of each bacterial sample and then the relative abundance of compounds across chromatograms was further normalized using Probabilistic Quotient Normalization (PQN) [20]. The data were log10-transformed, and the arithmetic mean was calculated for each compound within each set of biological replicates (n = 3 per isolate), resulting in a 14 × 18 data matrix (number of compounds × number of strains).

Table 2.
Methyl carboxylic acids detected in K. pneumoniae.
All statistical analyses were performed using R v3.3.2 (R Foundation for Statistical Computing, Vienna, Austria). The Random Forest (RF) algorithm was used to identify the most highly discriminatory features [14]. Five-fold cross-validation was performed, and this was repeated 10 times for each comparison. Mean decrease in accuracy (MDA) was used as the measure of variable importance. Hierarchical clustering analysis (HCA) was used to visualize the relationships between strains, using the Euclidean distance as the distance metric. A Kruskal-Wallis (KW) test [21], with post-hoc Dunn’s test [22] and Benjamini-Hochberg (BH) correction [23] was used to identify metabolites that were significantly different between groups, with p = 0.05 defined as the threshold for statistical significance.
3. Results and Discussion
3.1. The FAME Profile of Klebsiella pneumoniae Clinical Isolates
We reported on a total of 14 unique FAMEs across the 18 isolates evaluated (Table 2), of which 10 were confirmed by pure chemical standards, three putatively identified based on the MS similarity search and LRI value, and one attributed based only on the MS similarity match. The latter was reported as methyl pentadecenoate, since it was recognized as a monounsaturated fatty acid, but it was not possible to locate the position of the double bond. Of these, six were derived from saturated carboxylic acids (dodecanoate (C12), tetradecanoate (C14), pentadecanoate (C15), hexadecanoate (C16), heptadecanoate (C17), and octadecanoate (C18)), four from monounsaturated carboxylic acids (pentadecenoate, 9(Z)-hexadecenoate, 10-heptadecenoate, and 9(Z)-octadecenoate), and four from more complex carboxylic acids (3-methylsulfanylpropanoate, 2-phenylacetate, 3-hydroxytetradecanoate, and cis-9,10-methyleneoctadecanoate). The origins of the even-chain saturated carboxylic acids (dodecanoate, tetradecanoate, hexadecanoate, and octadecanoate) are well-characterized in bacteria, such as K. pneumoniae, and result principally from the biosynthesis and/or degradation of fatty acids in two-carbon units [24]. Two of the unsaturated species that we identified (9(Z)-hexadecenoate and 9(Z)-octadecenoate) are also generated via these same processes. The odd-chain saturated carboxylic acids (pentadecanoate and heptadecanoate) are postulated to originate from either lipid biosynthesis using propanoate (C3) as a starting material (instead of acetate, C2) or via decarboxylation of hexadecanoic acid and octadecanoic acid, respectively [24]. It is, therefore, reasonable to hypothesize that the two odd-chain monounsaturated carboxylic acids that we identified (pentadecenoate and 10-heptadecenoate) may originate from this same process, in a manner analogous to the production of 9(Z)-hexadecenoate and 9(Z)-octadecenoate during fatty acid biosynthesis and/or degradation in two-carbon units.
The metabolism of sulfur-containing amino acids (i.e., cysteine and methionine) may be responsible for the production of 3-methylsulfanylpropanoic acid, while phenylalanine metabolism is plausibly related to the production of 2-phenylacetic acid [24]. Cis-9,10-methyleneoctadecanoate likely originates from the methylation of the respective cis-monosaturated fatty acid (9(Z)-octadecanoate) with a methyl group derived from S-adenosyl methionine [24]. The formation of such cyclopropane-containing fatty acids is thought to occur in the context of phosphatidylethanolamine metabolism [25]. Finally, 3-hydroxytetradecanoate has been identified as a common component of the endotoxic lipopolysaccharide (Lipid A) located on the outer membrane of Gram-negative bacteria, generated predominantly under anaerobic conditions [25]. In summary, the FAMEs identified in the present study are likely to originate predominantly from fatty acid biosynthesis and/or degradation, while a smaller subset also arises from the metabolism of amino acids and other cell membrane components.
3.2. Discrimination between CC258 and Non-CC258 via FAME Profiles
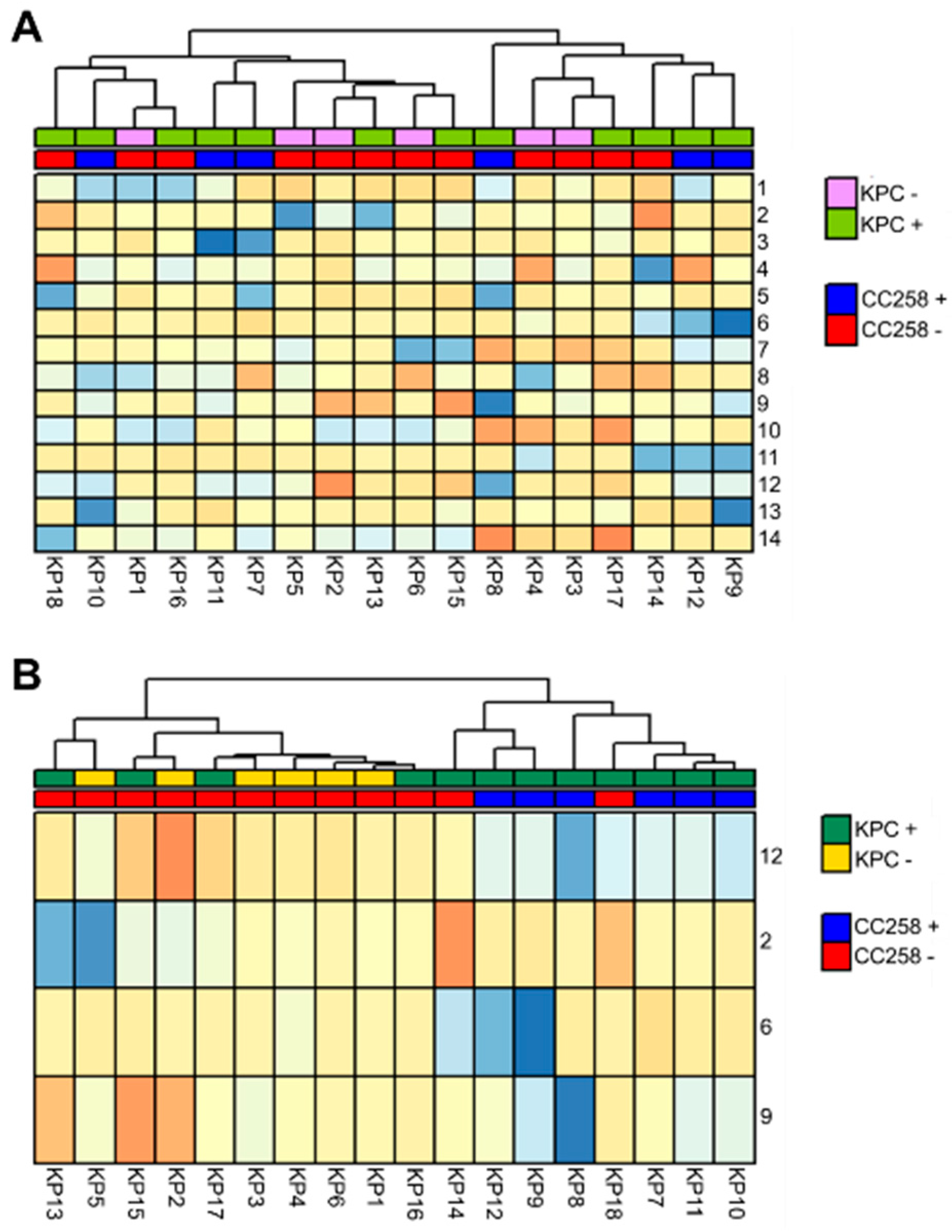
We sought to determine whether the FAME profiles of these K. pneumoniae isolates could distinguish between isolates belonging to CC258 (n = 6) and those belonging to other lineages (non-CC258, n = 12), as well as between KPC-positive (n = 12) and KPC-negative (n = 6) K. pneumoniae isolates, irrespective of sequence type. We first analyzed the data using the unsupervised approach of hierarchical clustering analysis (HCA), which did not reveal obvious differences between any of the three experimental groups (KPC+/258+, KPC+/258−, and KPC−/258−) (Figure 1A).

Figure 1.
Heatmaps generated using hierarchical clustering analysis (HCA) for the differentiation between CC258 and non-CC258 K. pneumoniae. (A, top) HCA performed using all 14 compounds did not reveal any obvious clustering pattern. (B, bottom) HCA performed using the four compounds identified as highly discriminatory between CC258 and non-CC258 reveals two distinct clusters, one of which includes 10 of 12 non-ST258 K. pneumoniae (left), and the other includes all six ST258 K. pneumoniae and two of 12 non-ST258 K. pneumoniae (right). Euclidean distance was used as the distance metric between samples. Isolate numbers (columns) are listed in Table 1. The numbering system for compounds (rows) are listed in Table 2. Cell color is on an arbitrary scale ranging from low abundance (blue) to high abundance (orange).
We next employed the supervised machine learning algorithm Random Forest (RF) in an attempt to identify patterns of FAMEs that were associated with specific experimental groups. RF was unable to identify a pattern of FAMEs that could reliably differentiate between KPC-positive and KPC-negative K. pneumoniae isolates, with an average five-fold cross-validation accuracy of only 65%. In contrast, RF able to differentiate between CC258 and non-CC258 (irrespective of carbapenem susceptibility) with an average five-fold cross-validation accuracy of 86%. Four compounds were identified as highly discriminatory between CC258 and non-CC258 isolates: 9(Z)-octadecenoate, 2-phenylacetate, pentadecanoate, and hexadecanoate. The mean decreases in accuracy (MDA, a measure of variable importance generated from RF) for these compounds ranged between 0.76 and 1.28, which was notably higher than the MDAs of the remaining 10 compounds, which ranged from 0.19 to 0.51.
HCA performed using only these four FAMEs revealed two distinct clusters, one of which included 10 of the 12 non-CC258 isolates, and the other included all six CC258 isolates, as well as the remaining two non-CC258 isolates (Figure 1B). The two non-CC258 isolates that clustered with the six isolates belonging to CC258 included one from ST42 and one from ST307. Notably, our collection of isolates included two others from ST42 and one other from ST307, and these clustered correctly. It is therefore unclear as to why these two specific isolates clustered incorrectly, as it does not appear to be a lineage-specific phenomenon. Instead, it may represent strain-specific metabolic differences.
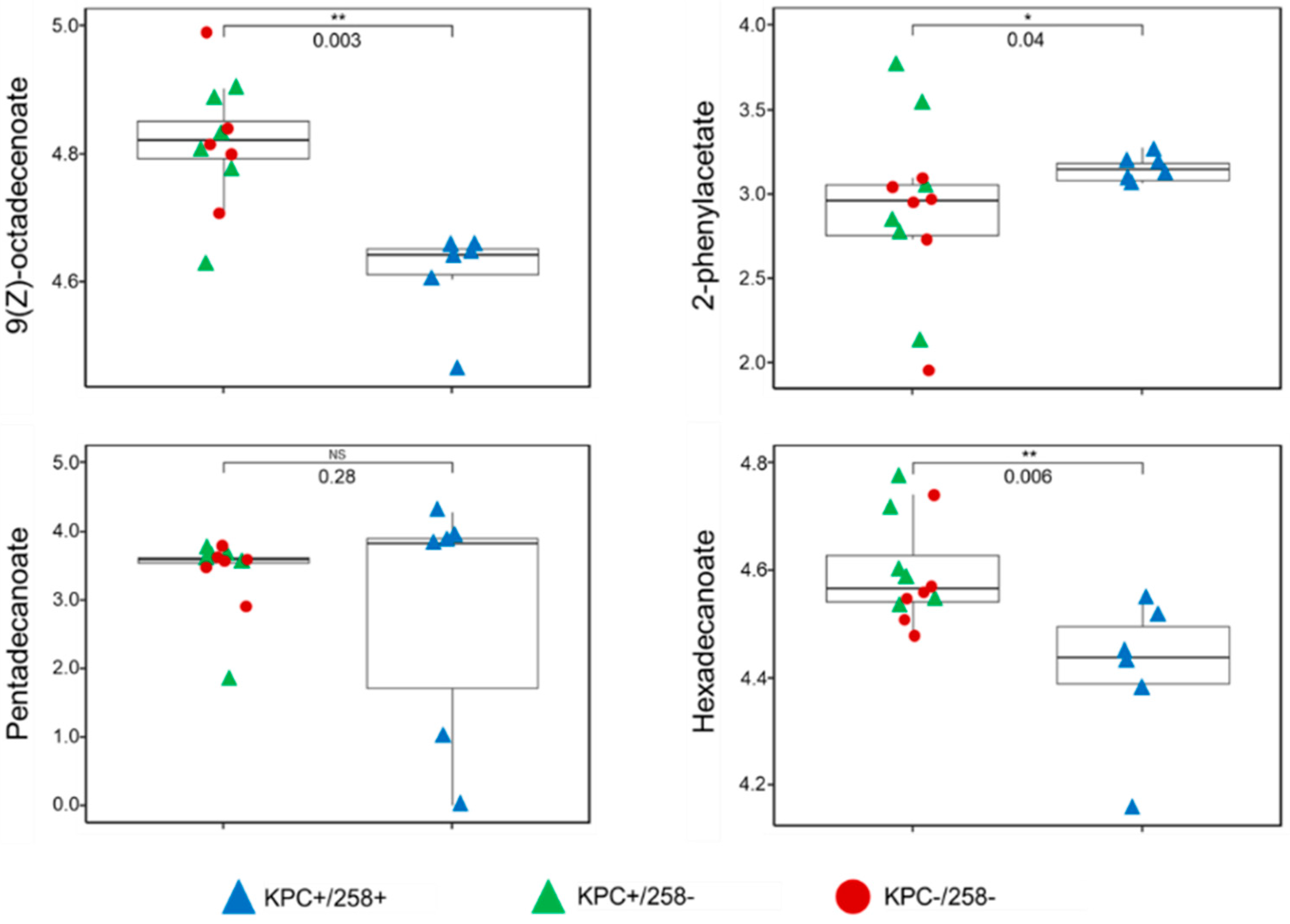
Of the four discriminatory compounds, three were significantly different (p < 0.05) in relative abundance between CC258 and non-CC258 K. pneumoniae. 9(Z)-octadecenoate and hexadecanoate were significantly more abundant in non-CC258 relative to CC258, while 2-phenylacetate was significantly more abundant in CC258 relative to non-CC258 (Figure 2). Neither pentadecanoate (the fourth discriminatory compound) nor any of the other 10 compounds were significantly different in relative abundance between CC258 and non-CC258. In addition, no compounds were significantly different in relative abundance between KPC-positive and KPC-negative K. pneumoniae. Taken together, these findings suggest that CC258 and non-CC258 K. pneumoniae isolates differ from one another with respect to fundamental aspects of metabolism, including the metabolism of amino acids (2-phenylacetate) and fatty acids (9(Z)-octadecenoate, hexadecanoate, and pentadecanoate). Such fundamental metabolic differences are in agreement with the conclusions that have been drawn from prior studies, for example [10,11,12].
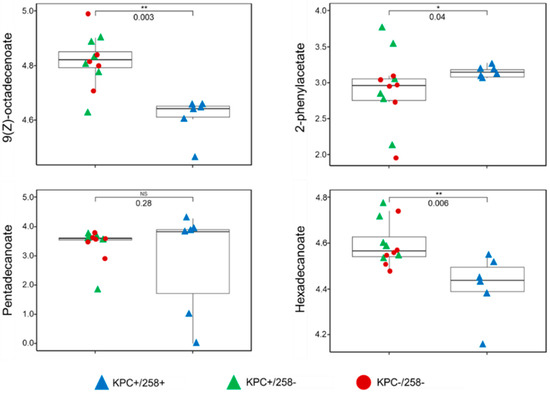
Figure 2.
Boxplots depicting the relative abundances of the four compounds that were discriminatory between ST258 and non-ST258. 9(Z)-octadecenoate, 2-phenylacetate, and hexadecanoate were significantly different between the two experimental groups, while pentadecanoate was not. Blue triangles: KPC+/258+; green triangles: KPC+/258−; red circles: KPC−/258−; * p-value < 0.05; ** p-value < 0.01.
To the best of our knowledge, the present study is the first to evaluate differences between carbapenem-resistant and carbapenem-susceptible K. pneumoniae isolates with respect to their FAME profiles. However, prior studies have considered differences in bacterial FAME composition in the context of other antibiotic resistance phenotypes. The majority of these studies have focused on resistance phenotypes that are seemingly directly mediated by alterations to membrane lipid composition, such as resistance to polymyxins or tetracyclines, for example [25,26]. Importantly, our findings do not support a relationship between KPC-production and FAME composition, but instead suggest that certain high-risk endemic K. pneumoniae clones (specifically CC258) may produce a distinct FAME profile relative to other K. pneumoniae lineages. These findings are consistent with the transcriptomic data reported by Bruchmann and colleagues, who demonstrated differences between CC258 and non-CC258 in the expression of genes associated with the metabolism of alpha-amino acids and carboxylic acids [10].
3.3. Study Strengths and Limitations
The present work represents the first to investigate FAME composition in the context of antibiotic-resistant K. pneumoniae, with a particular focus on compositional differences between KPC-positive isolates belonging to CC258, and a collection of KPC-positive/KPC-negative isolates belonging to a diverse collection of other sequence types. In addition, the present study evaluates both the phenotype-associated (i.e., carbapenem-resistant versus carbapenem-susceptible) and genotype-associated (i.e., CC258 versus non-CC258) contributions to the FAME profile. We acknowledge, however, that the number of isolates evaluated is relatively small and could be expanded upon in future studies. In addition, analysis of non-KPC-producing isolates belonging to CC258 (KPC−/258+) would represent an important comparison group to full elucidate both the phenotype-associated and genotype-associated contributions to the FAME profile. However, these isolates are rarely encountered in the clinical setting, and are therefore difficult to evaluate.
4. Conclusions
The present study demonstrates distinct differences between CC258 and non-CC258 K. pneumoniae with respect to their FAME profiles. These findings suggest that CC258 may produce a distinct fatty acid and amino acid profile relative to other K. pneumoniae lineages. This hypothesis is supported by previously-acquired transcriptomic data from Bruchmann and colleagues. The identification of a CC258-associated FAME profile could have therapeutic consequences via the identification of targetable pathways that contribute to the production of membrane lipids and amino acids. Furthermore, this approach could lead to the development of novel GC-based diagnostics for the rapid identification of K. pneumoniae isolates belonging to CC258. While the present study is largely exploratory, it represents an important step in improving our understanding of fatty acid or amino acid-associated metabolic differences between CC258 and non-CC258 K. pneumoniae.
Author Contributions
Conceptualization, C.A.R., G.P., M.B. and F.A.F.; methodology and validation, F.A.F., M.B. and G.P.; software, formal analysis and investigation, G.P. and C.A.R.; resources, G.P. and J.E.H.; data curation, C.A.R., F.A.F., M.B. and G.P.; writing—original draft preparation, C.A.R. and G.P.; writing—review and editing, C.A.R., G.P., F.A.F., M.B. and J.E.H.; visualization, C.A.R. and G.P.; supervision, G.P.; project administration and funding acquisition, G.P. and J.E.H.
Funding
This research received no external funding.
Acknowledgments
We thank Elizabeth B Hirsch, PharmD (University of Minnesota) for generously providing the strains used in this study. Christiaan A Rees was supported by the National Institutes of Health training grant T32 LM012204 (PI: Tor D Tosteson). The authors thank Shimadzu and Supelco for their support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, L.; Todd, R.; Kiehlbauch, J.; Walters, M.; Kallen, A. Pan-Resistant New Delhi Metallo-Beta-Lactamase-Producing Klebsiella pneumoniae-Washoe County, Nevada, 2016. MMWR Morb. Mortal. Wkly. Rep. 2017, 66, 33. [Google Scholar] [CrossRef]
- Elemam, A.; Rahimian, J.; Mandell, W. Infection with Panresistant Klebsiella pneumoniae: A Report of 2 Cases and a Brief Review of the Literature. Clin. Infect. Dis. 2009, 49, 271–274. [Google Scholar] [CrossRef]
- Sonnevend, Á.; Ghazawi, A.; Hashmey, R.; Haidermota, A.; Girgis, S.; Alfaresi, M.; Omar, M.; Paterson, D.L.; Zowawi, H.M.; Pál, T. Multihospital Occurrence of Pan-Resistant Klebsiella pneumoniae Sequence Type 147 with an ISEcp1-Directed blaOXA-181 Insertion in the mgrB Gene in the United Arab Emirates. Antimicrob. Agents Chemother. 2017, 61, e00418-17. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Goodman, K.E.; Harris, A.D.; Tekle, T.; Roberts, A.; Taiwo, A.; Simner, P.J.; Tamma, P.D.; Corresponding, M.H.S. Comparing the Outcomes of Patients with Carbapenemase-Producing and Non-Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae Bacteremia. Clin. Infect. Dis. 2017, 64, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Kitchel, B.; Rasheed, J.K.; Patel, J.B.; Srinivasan, A.; Navon-Venezia, S.; Carmeli, Y.; Brolund, A.; Giske, C.G. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: Clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 2009, 53, 3365–3370. [Google Scholar] [CrossRef] [PubMed]
- Adler, A.; Paikin, S.; Sterlin, Y.; Glick, J.; Edgar, R.; Aronov, R.; Schwaber, M.J.; Carmelia, Y. A swordless knight: Epidemiology and molecular characteristics of the blaKPC-negative sequence type 258 Klebsiella pneumoniae clone. J. Clin. Microbiol. 2012, 50, 3180–3185. [Google Scholar] [CrossRef]
- Bowers, J.R.; Kitchel, B.; Driebe, E.M.; MacCannell, D.R.; Roe, C.; Lemmer, D.; De Man, T.; Rasheed, J.K.; Engelthaler, D.M.; Keim, P.; et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS ONE 2015, 10, e0133727. [Google Scholar] [CrossRef]
- Chmelnitsky, I.; Shklyar, M.; Hermesh, O.; Navon-Venezia, S.; Edgar, R.; Carmeli, Y. Unique genes identified in the epidemic extremely drug-resistant KPC-producing Klebsiella pneumoniae sequence type 258. J. Antimicrob. Chemother. 2013, 68, 74–83. [Google Scholar] [CrossRef]
- Bruchmann, S.; Muthukumarasamy, U.; Pohl, S.; Preusse, M.; Bielecka, A.; Nicolai, T.; Hamann, I.; Hillert, R.; Kola, A.; Gastmeier, P.; et al. Deep transcriptome profiling of clinical Klebsiella pneumoniae isolates reveals strain and sequence type-specific adaptation. Environ. Microbiol. 2015, 17, 4690–4710. [Google Scholar] [CrossRef]
- Rees, C.A.; Nasir, M.; Smolinska, A.; Lewis, A.E.; Kane, K.R.; Kossmann, S.E.; Sezer, O.; Zucchi, P.C.; Doi, Y.; Hirsch, E.B.; et al. Detection of high-risk carbapenem-resistant Klebsiella pneumoniae and Enterobacter cloacae isolates using volatile molecular profiles. Sci. Rep. 2018, 8, 13297. [Google Scholar] [CrossRef]
- Low, Y.M.; Yap, I.K.S.; Abdul Jabar, K.; Md Yusof, M.Y.; Chong, C.W.; Teh, C.S.J. Genotypic and metabolic approaches towards the segregation of Klebsiella pneumoniae strains producing different antibiotic resistant enzymes. Metabolomics 2017, 13, 65. [Google Scholar] [CrossRef]
- Müller, K.D.; Nalik, H.P.; Schmid, E.N.; Husmann, H.; Schomburg, G. Fast identification of mycobacterium species by GC analysis with trimethylsulfonium hydroxide (TMSH) for transesterification. J. High Resolut. Chromatogr. 1993, 16, 161–165. [Google Scholar] [CrossRef]
- Breiman, L.E.O. Random Forest. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Diancourt, L.; Passet, V.; Verhoef, J.; Grimont, P.S.B. Multilocus Sequence Typing of Klebsiella pneumoniae Nosocomial Isolates. J. Clin. Microbiol. 2005, 43, 4178–4182. [Google Scholar] [CrossRef] [PubMed]
- Tenover, F.C.; Kalsi, R.K.; Williams, P.P.; Carey, R.B.; Stocker, S.; Lonsway, D.; Rasheed, J.K.; Biddle, J.W.; McGowan, J.E.; Hanna, B. Carbapenem resistance in Klebsiella pneumoniae not detected by automated susceptibility testing. Emerg. Infect. Dis. 2006, 12, 1209–1213. [Google Scholar] [CrossRef]
- Tranchida, P.Q.; Franchina, F.A.; Dugo, P.; Mondello, L. A flow-modulated comprehensive gas chromatography-mass spectrometry method for the analysis of fatty acid profiles in marine and biological samples. J. Chromatogr. A 2012, 1255, 171–176. [Google Scholar] [CrossRef]
- Purcaro, G.; Tranchida, P.Q.; Dugo, P.; Camera, L.E.; Bisignano, G.; Conte, L.; Mondello, L. Characterization of bacterial lipid profiles by using rapid sample preparation and fast comprehensive two-dimensional gas chromatography in combination with mass spectrometry. J. Sep. Sci. 2010, 33, 2334–2340. [Google Scholar] [CrossRef]
- Beccaria, M.; Franchina, F.A.; Nasir, M.; Mellors, T.; Hill, J.E.; Purcaro, G. Investigation of mycobacteria fatty acid profile using different ionization energies in GC-MS. Anal. Bioanal. Chem. 2018, 410, 7987–7996. [Google Scholar] [CrossRef]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A.; Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Dunn, O.J. Multiple Comparisons Using Rank Sums. Technometrics 1964, 6, 241–252. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Fulco, A.J. Fatty acid metabolism in bacteria. Prog. Lipid Res. 1983, 22, 133–160. [Google Scholar] [CrossRef]
- Dunnick, J.K.; O’Leary, W.M. Correlation of bacteria lipid composition with antibiotic resistance. J. Bacteriol. 1970, 101, 892–900. [Google Scholar] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).