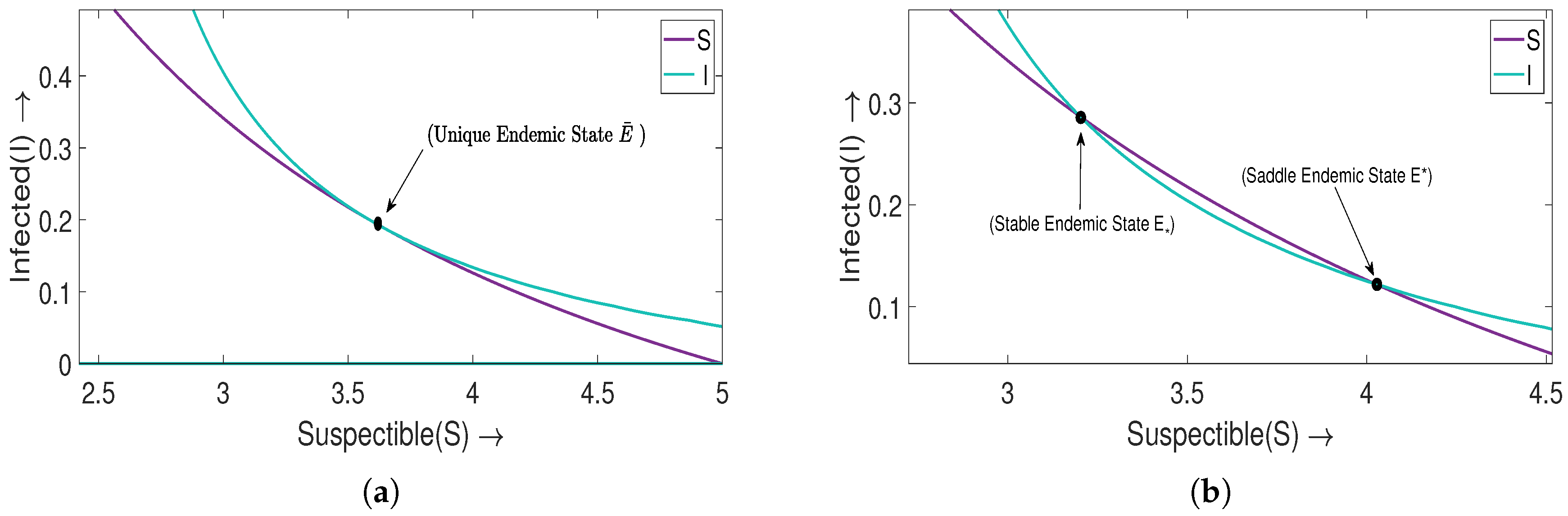1. Introduction
The theory of competing risks, which analyzes causes of death, was initially explored by Graunt in 1662. This theory has since gained firm footing among contemporary epidemiologists [
1]. In 1760, Daniel Bernoulli examined the propagation of smallpox within a community using a mathematical model. In the early 20th century, William Hamer [
2] and Ronald Ross [
3] applied the law of mass action to elucidate the dissemination of epidemics among individuals. However, a groundbreaking advancement in the understanding of disease transmission emerged shortly after the introduction of the epidemic model proposed by Kermack and McKendrick in 1927 [
4]. This model delineates the correlation between susceptible and infected individuals within a population.
The majority of researchers all around the world focus on the temporal development of epidemics with bilinear incidence rates, saturated types of incidence, and many other disease-generated interactions [
5,
6,
7]. Additionally, researchers explore some control techniques, such as treatment and vaccines [
8,
9,
10]. The model we consider is a SIR-type model in which the population is divided into three compartments: S (susceptible), individuals who can contract the disease; I (infected), individuals who have contracted the disease and can transmit it; and R (recovered/removed), individuals who have either recovered with immunity or died and hence do not participate in further disease transmission. Recently, Zhang et al. [
7] performed a temporal analysis, including several bifurcations (backward and forward), of an SIR epidemic model, which is further explored by Gupta and Kumar [
8] under the following three assumptions: (i) The susceptible and infected populations are homogeneously distributed in space; that is, these individuals are only time-dependent. (ii) The total recruitment in
S-compartment is taken as constant and denoted as
, and (iii) both the incidence and the treatment rates are of saturated types. A flow chart for an SIR model with saturated treatment is given as follows
Figure 1.
Thus, the proposed SIR model is given as follows:
with the initial condition
The epidemiologically meaning of the parameters used in the system (
1) are given in the
Table 1.
The saturated treatment rate comes into account due to the limitations of medical resources or the availability of the treatment for a particular disease. Here, it is notable that the parameter is the saturation factor, which quantifies the infected individual being delayed for treatment. When the number of infected individuals is low, i.e., , the term approximates linear treatment, , and the healthcare system operates effectively. As the number of infectives increases, the denominator grows, reducing the effective treatment rate per infected individual, which realistically reflects the situation when medical resources become inadequate or overloaded. The parameter thus regulates the decline in per-capita treatment efficacy as infection levels rise. A higher value of means faster saturation, indicating that even a moderate number of infections can overwhelm treatment capacity. This term is especially important when studying outbreaks in regions with limited healthcare infrastructure.
Due to biological constraints, all of these parameters are assumed to be non-negative. Gupta and Kumar [
8] reduced the proposed system (
1) by assuming that the third variable,
R, is not present in the first two equations. They also introduced a new parameter,
, that measures the disease-induced death rate after the recovery, which remains non-negative. Thus, the reduced model is given as follows:
In the context of epidemiological modeling, spatial heterogeneity can play a significant role in understanding the dynamics of virus outbreaks. Spatial heterogeneity refers to variations in disease spread, population density, and other factors across different geographic regions [
11,
12]. If virus outbreaks were always spatially homogeneous, it would mean that the disease spreads uniformly and identically everywhere, which is rarely the case. A deterministic epidemic model in a constrained one-dimensional context, with self-diffusive spatial motion, was proposed by Webb in 1981 [
13]. The author’s analysis revealed that, if the initial facts are positive, the solutions will always be well posed and positive. In order to forecast the spread of an infectious disease, the spatio-temporal dynamics of an epidemic model may be taken into account. As a result, time- and space-based mathematical models are useful to understand the spread of epidemics [
12,
14,
15,
16,
17]. Thus, a reaction–diffusion process in disease dynamics deals with a disease spreading over space and time through the movement of individuals.
In particular, diffusion is known to seek an equilibrium solution in which populations are evenly dispersed [
10,
17,
18,
19]. The reaction–diffusion equations, on the other hand, may cause Turing instability and the Turing pattern [
20], which is a non-equilibrium phenomenon. The authors of the articles [
15,
16,
17,
21] used epidemic models with saturated types of incidence rates and constant removal rates to study the cumulative behavior of the spatial distribution across space and time with self-diffusion. In particular, Sun et al. [
15] explored an SI model with a nonlinear incidence rate and performed a Turing analysis to ensure the appearance of strip- and spot-type patterns via self-diffusion. Liu et al. [
17] examined the dynamics of an epidemic model with a constant removal rate and studied how diffusion affects the emergence of spatial patterns. Further, Wang et al. [
16] studied the SI model with logistic growth in susceptible individuals, using a bilinear incidence rate and diffusion to observe various types of patterns.
On the one hand, the susceptible populations always have a propensity to avoid the infected individuals in a natural setting, provided that they can recognize the infected ones due to symptoms. On the other hand, the gradient in population density of the infected compartment may cause a flux that affects the diffusion of a susceptible population. This indicates that the diffusion in one compartment affects the diffusion of the other. Cross-diffusion in mathematical models describes the effect on the motility of one species due to the presence of the other. Sun et al. [
22] studied the influence of cross-diffusion on the spread of an epidemic with the help of a spatio-temporal model. According to Wang et al. [
23], no Turing pattern emerges with self-diffusion for an epidemic SI model, whereas cross-diffusion develops such patterns. Even with the nonlinearity taken into account in cross-diffusive terms, its impact on various populations is still similar. In order to replicate this scenario in our study, we have created novel, nonlinear, complex cross-diffusion with both quadratic and rational factors for both the susceptible and infected classes.
In general, a vulnerable individual takes precautions to avoid getting into contact with a person infected with a communicable disease like COVID-19, AIDS, etc. Additionally, the mobility of people is impacted by media coverage and health ministry awareness, which cannot be captured through simple cross-diffusion. These elements cannot be disregarded if one wants to learn more about disease transmission. Though many researchers consider nonlinear cross-diffusion using the standard heat equation, very few researchers focus on disease dissemination in an environment with nonlinear cross-diffusion. Fortunately, some research studies have been suggested that have examined nonlinear cross-diffusion for a set of reaction–diffusion equations for various interacting population models [
24,
25,
26,
27,
28,
29]. In particular, cross-diffusion and self-diffusion models were originally proposed by Shigesada et al. [
27] to model the spatial segregation of two competing species. The key characteristic of this model was to demonstrate that, if random diffusion alone is unable to result in the spatial segregation of two competing species, then perhaps a nonlinear dispersal approach can. Further, Ni et al. [
30] proposed a non-linear cross-diffusion and analyzed the spike-layer equilibrium state. In the article [
26], the authors also studied the effect of cross-diffusion in a second species and showed that the flux of the second species is directed towards the decreasing population density of the first species. Recently, Zhou et al. [
29] analyzed a cooperative two-species Lotka–Volterra model to demonstrate that, if birth rates are high and cross-diffusions are suitably weak, the model has at least one coexisting state. This literature suggests that the proposed nonlinear cross-diffusion terms may be constituents that are quadratic [
31], rational [
24], or a combination of both [
29].
A variety of diffusion rates across many decades of mathematical modeling have been utilized. Motivated by the above-cited works, we have proposed nonlinear cross-diffusion, on the basis of which the dissemination of susceptible and infected populations depends on the movement of each class, as follows:
Here,
(for
),
,
,
,
,
, and
are the positive diffusive constants; due to these parameters, the influences on self- and cross-diffusion is affected. Also, we notice the following:
For
, cross-diffusion is similar to that described in the article [
25];
For
, the diffusion is the same as in the article [
31];
For
, the diffusion is of the self-type. Many researchers [
15,
16,
17] studied the effect of self-diffusion in the distribution of various species in many branches of mathematical modeling. Here, it is notable that the roles of
S and
I will be changed according to the species of these models.
In the diffusion term
the first term
represents self-diffusion, as it influences the term
of the gradient. In an interacting environment, the self-diffusion for the movement of a susceptible population depends on the density of infected individuals and vice versa. The cross-diffusion term
influences the interaction between susceptible and infected populations. Crowding’s hindrance of movement can be incorporated by introducing the current population density of infected individuals in the denominator of the fractional part of the cross-diffusion term [
32]. Also, if the population density of either
S or
I increases, it will make movement difficult. The flux of
is
, which is replicated the same as in the earlier case.
Quadratic terms (, ): These account for density-dependent movement pressure. For example, the term in the first equation suggests that the movement of susceptible individuals increases nonlinearly with their own density, possibly due to crowding, competition for resources, or social interaction.
Saturated terms (e.g., , , ): These describe limited or saturating behavioral responses. For instance, the rate at which susceptible individuals respond to infected individuals (via the gradient of I) is modulated through , implying that their reaction saturates as the infected population grows. This reflects realistic constraints such as a limited avoidance ability, immune fatigue, or bounded information processing.
Mixed-gradient terms (e.g., in and in ): These terms indicate the behavioral responses of one class to the spatial gradient of the other—for example, susceptible individuals moving in response to spatial variation in infected density. The saturated structure again ensures realistic movement bounds.
We have demonstrated, through flux calculations, that our cross-diffusion is an indication of the propensity of the vulnerable population to avoid the infected ones. As far as our knowledge goes, this form of cross-diffusion is not available in the literature.
As a result, the system (
2) with described nonlinear cross-diffusive functions over
(
) can be written as
with the following initial and boundary conditions:
Here, the boundary condition is taken as the Neumann type. The parameter
n denotes the outward normal unit vector for the domain
. In order to explore the system (
3) for the effect of cross-diffusion between susceptible and infected populations due to their movement, the remaining part of this manuscript is arranged in the following order. Next, in
Section 2, we analyze the boundedness of solutions to the system (
3). The existence and non-existence of non-constant equilibrium states and constant equilibrium states are determined in
Section 3.1 and
Section 3.2, respectively. Through the existence of codimension-2 Turing–Hopf bifurcation, followed by a spatial analysis and Turing instability, we determine the Turing space in the spatial domain in
Section 4. An extensive numerical simulation is performed in
Section 5 to obtain the various patterns, such as Turing, non-Turing, Turing–Hopf, and Turing–Bogdanov–Takens patterns. In the last part of the manuscript, we discuss the main results in
Section 6.
5. Pattern Formation
For a boundary value problem, the Dirichlet boundary condition prescribes the value of a variable at the domain boundary, while the Neumann boundary condition necessitates the value of the variable’s derivative at the domain boundary. In practice, populations often interact with their surroundings in more complex ways, and boundary conditions are used to simplify the mathematical representation of these interactions. Therefore, Neumann conditions allow for more flexibility in terms of boundary behavior and may be used in situations where specifying values on the boundary is not feasible or meaningful. Therefore, we employ the numerical simulations for the system (
3) to comprehend the phenomenon of pattern development through cross-diffusion in a two-dimensional space.
The diffusion part of the system (
3) is numerically solved using the semi-implicit finite difference technique [
46], and for the temporal part, we used finite difference technique. All numerical simulations incorporate non-zero initial conditions and zero-flux boundary conditions within a closed habitat measuring
units. The habitat discretization is performed around the stationary state
, utilizing spatial coordinates
and
, where
, with a time step size of
. In recent years, numerous researchers [
18,
32,
35,
47] have explored random perturbations inspired by diverse natural phenomena (such as disease seasonality) to uncover novel patterns and underscore the significance of perturbation effects. Building upon these insights, a perturbation of magnitude
is introduced as follows:
Here, the terms and describe sinusoidal and cosinusoidal variations along the axis and , with a wavelength of 100 units in the and direction (since the period of the sine function is ). Physically, the perturbation represents localized fluctuations in the population densities due to external or internal factors, such as population clustering or environmental influences.
We aim to illustrate the significant impact of the cross-diffusion coefficients
and
on pattern formation through extensive numerical simulations involving perturbation
. The distribution of
S and
I individuals in the SI-plane gives rise to a variety of intriguing patterns, including “holes,” “strips,” and “labyrinthine,” as evidenced in the findings of several researchers [
18,
23,
32,
38,
47]. We select appropriate diffusion coefficients to satisfy the conditions for Turing instability around unstable and stable endemic states, resulting in the emergence of non-Turing and Turing patterns, respectively [
16,
32]. In the subsequent subsection, we expand this discussion.
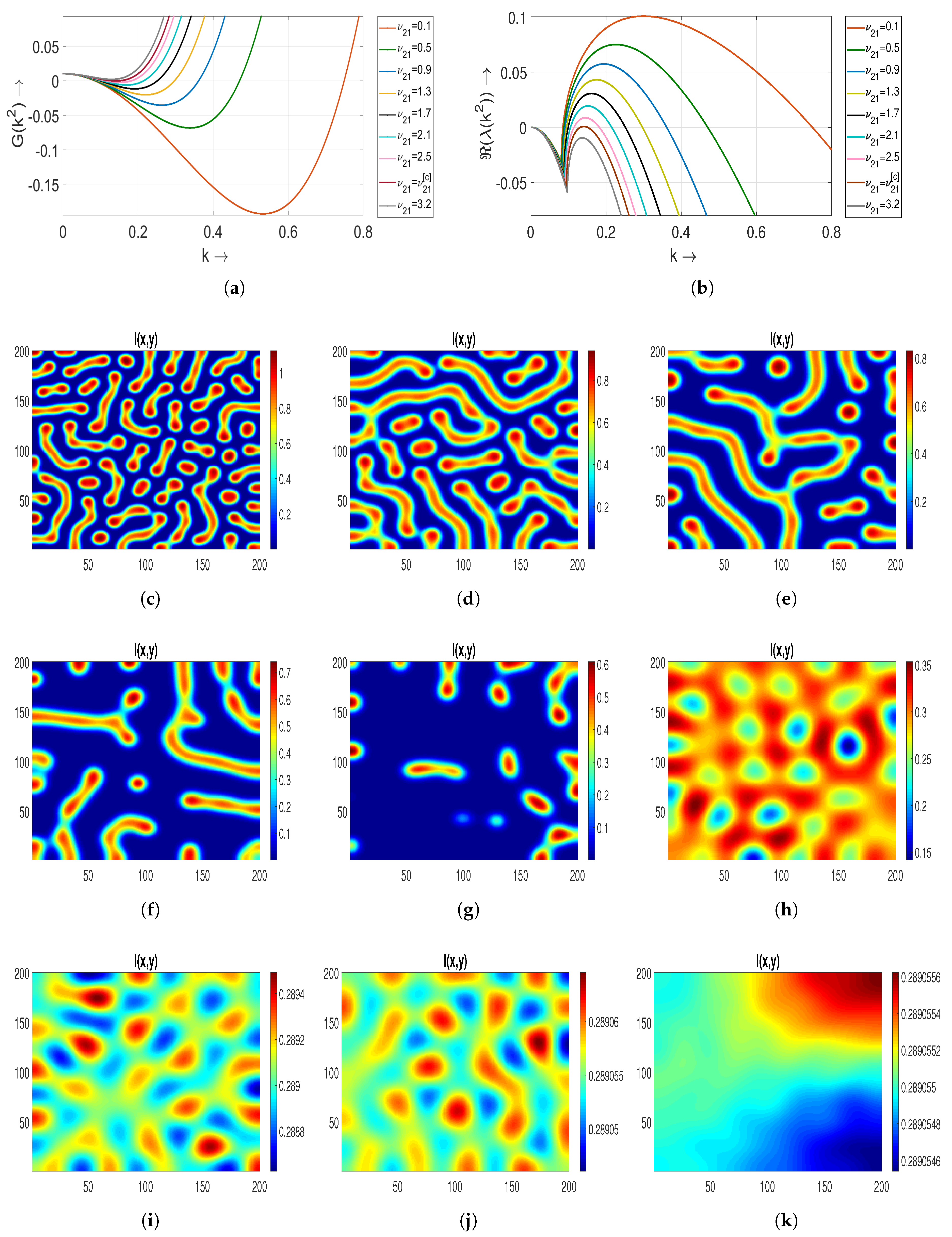
5.1. Non-Turing Patterns
The term non-Turing patterns refers to patterns derived within the vicinity of an unstable equilibrium state concerning the associated ordinary system of a diffusive model. To determine such a type of pattern for the system (
3), we choose the parameters
. If we take
, the equilibrium state
is unstable. For the diffusion coefficients mentioned in
Table 2, it is clear from
Figure 4a that the determinant of
at the endemic state
is negative. Also, the interval of wave number
k, that is,
decreases as we increase
in
. In
Figure 4b, we notice that the interval of a positive real part of the eigenvalues of
also decreases with an increasing value of
. In
Figure 4, we illustrated non-Turing patterns within the
region at time
. In
Figure 4c, a non-Turing pattern is observed for
, where the two endemic states,
and
, merge to form
.
For
, high-density regions in the distribution of susceptible population manifest as blue holes against a reddish brown background of low density. Similarly, in the distribution of an infected population, red holes indicative of low density appear within a blue background, signifying a situation of heightened transmission [
Figure 4d]. Moreover, as
increases from
to
, these individual holes coalesce, gradually giving rise to stripes. Consequently, a novel category of non-Turing patterns, characterized by a combination of strips and holes, emerges, as depicted in
Figure 4e. Lastly, for
,
Figure 4f portrays predominant strip-like non-Turing patterns. It is observed that, for the system (
2), Hopf bifurcation occurs at
, and the equilibrium state
starts to stabilize with further increments in the value of
. In the following subsection, we will proceed to investigate the Turing patterns.
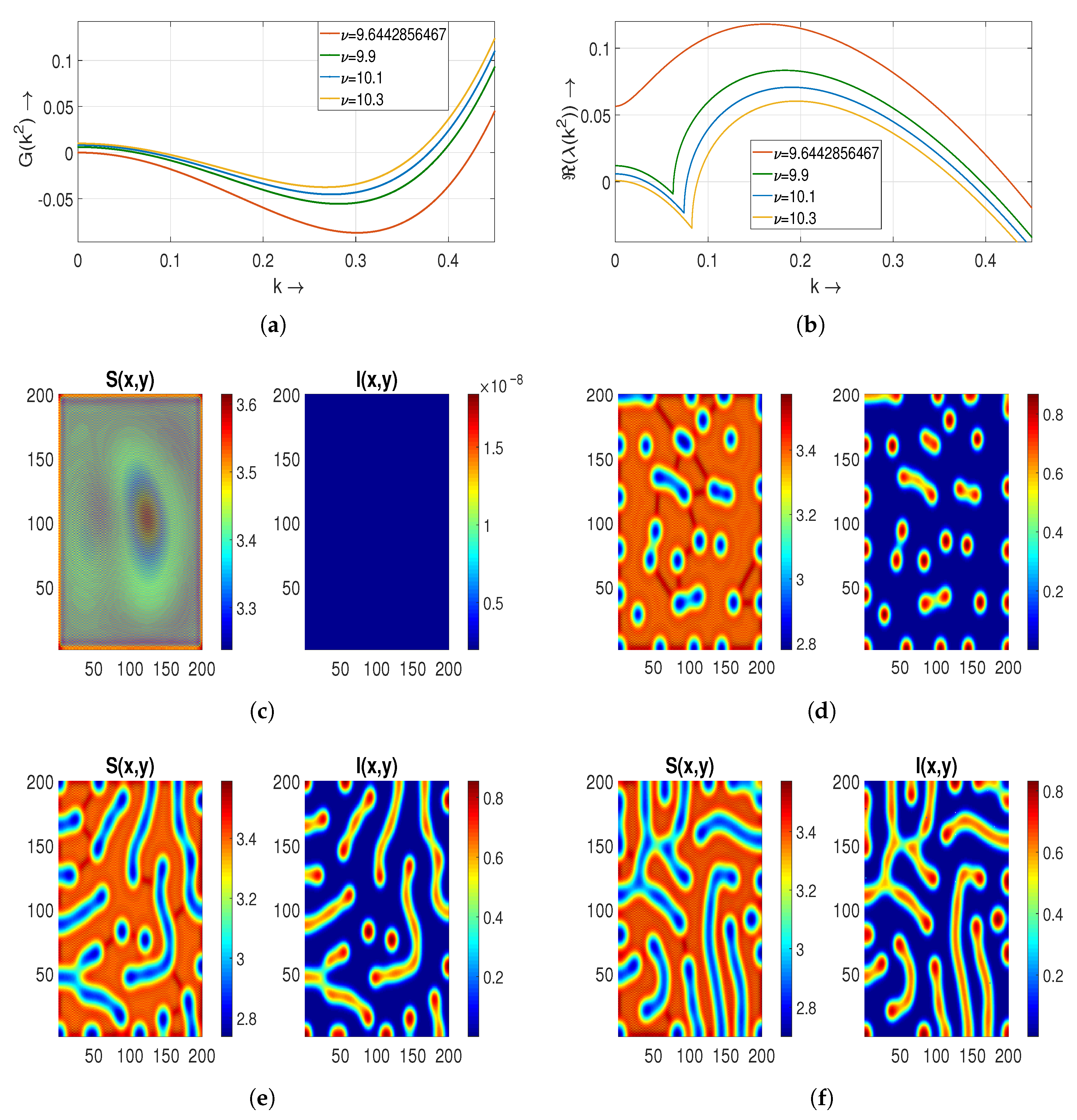
5.2. Turing Patterns
Continuing from the preceding
Section 5.1, we ascertain that the equilibrium state
remains stable for
. Furthermore, the condition for Turing instability persists within the same range of diffusion coefficients. Consequently, the resulting patterns in this scenario are classified as Turing patterns. We meticulously investigate these patterns across diverse values of the treatment parameter
and the diffusion coefficient
.
5.2.1. Effect of Treatment Parameter on Turing Patterns
In
Figure 5a,b, we plot
and
with respect to the wave number
k to determine the Turing patterns at time
. It shows that the interval of
decreases as
increases. As the equilibrium state
is now stable and the conditions of Turing instability also hold, we obtain Turing patterns. Strips emerge with a blue color into the red background in
Figure 5c,d for
(corresponding to the Hopf bifurcation threshold) and
in the distribution of the
S individual. Meanwhile, in the distribution of
I, we notice that red strips come out in the blue background. Hence, we conclude that the spread of a disease is high in this case. In order to determine the effect of saturation parameter
, we further choose two more values of
as
and
. For both the values of
, the roles of strips are now interchangeable in both cases compared to the above case. This results in the appearance of blue strips with a high-density region in a red background [
Figure 5e,f].
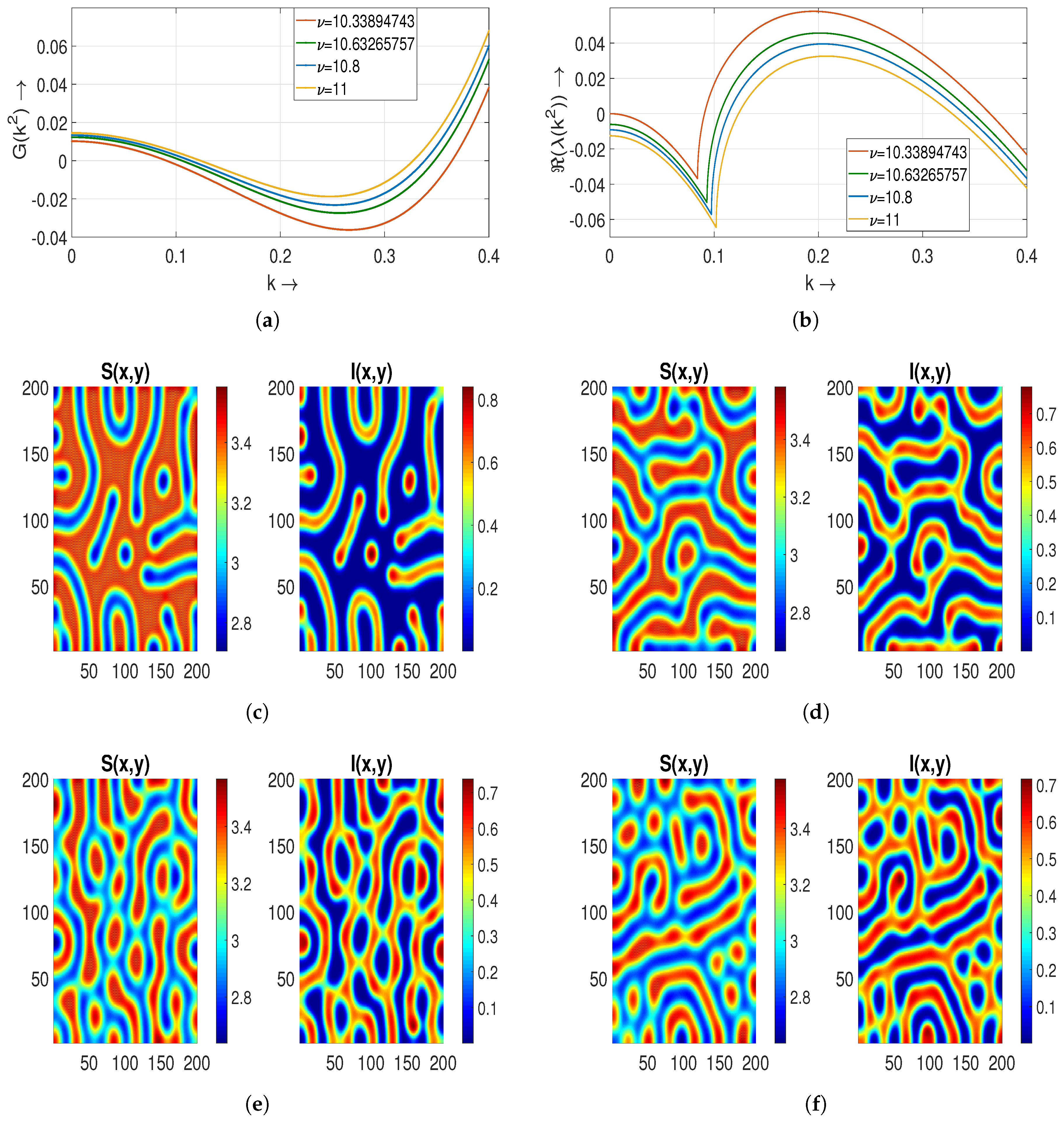
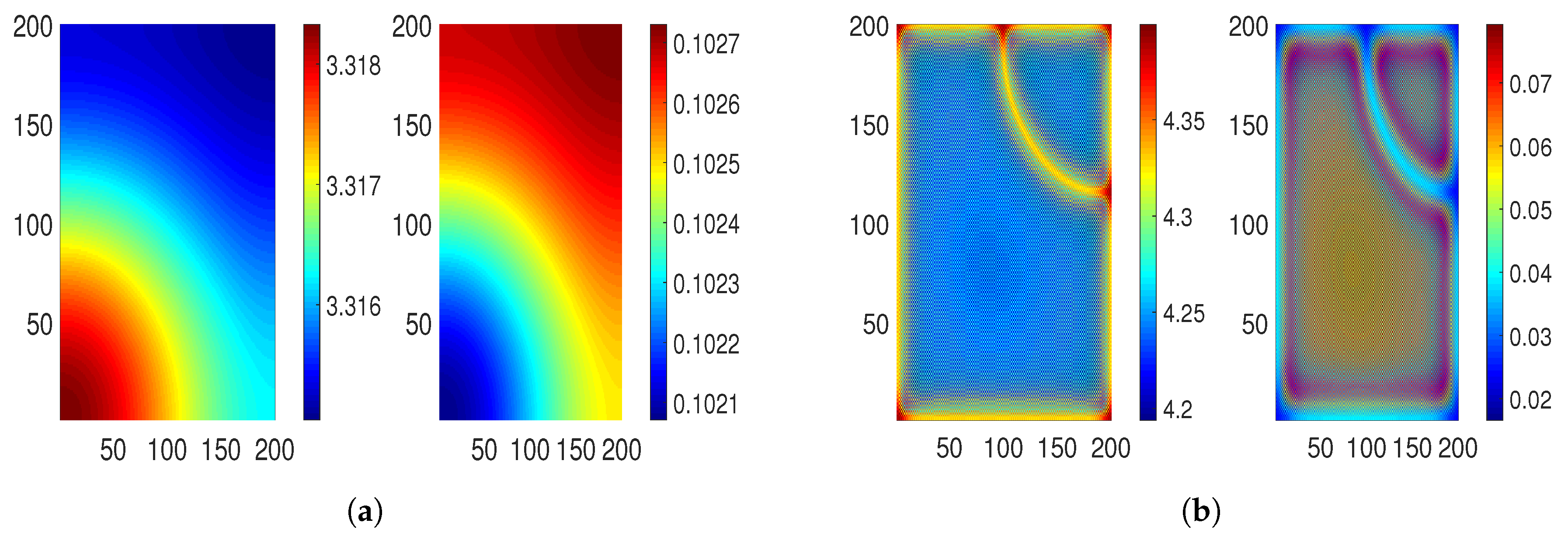
5.2.2. Effect of Diffusion Coefficient on Turing Patterns
For the numerical values of parameters given in
Table 2, we determine the effect of the diffusion coefficient
on Turing patterns.
Figure 6a,b show that the instability conditions hold. We observe that some holes are merging to form small strips, indicating a hole–strip mixed type pattern for
[
Figure 6c].
For
and
, in
Figure 6d,e, it can be seen that these strips extend their size by mixing with holes, as well as strips. Thus, we obtain stripe-type patterns in which red strips with low-density regions appear in the blue background with high-density regions. Next, for
,
, and
, we notice that these strips start disappearing in the form of a blue background and then come with dull holes in a red background. These dull blue holes with a high-density region come into the red background [
Figure 6f–h]. This concludes that the region is safe for a certain time as the high-density region is small. Furthermore, for
and
(the numerical value of the Turing bifurcation threshold), these Turing patterns start to disappear as we get closer to the non-Turing patterns that we plot for
in
Figure 6i–k. Here,
Figure 6j represents a Turing–Hopf pattern.
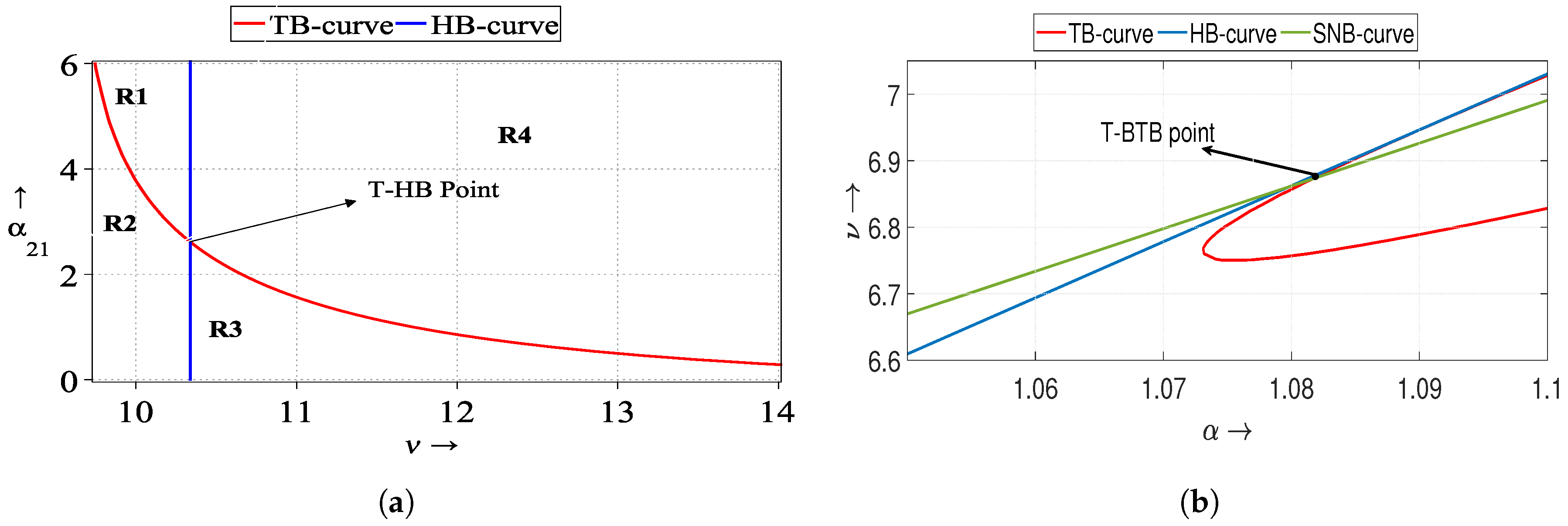
5.3. Turing–Bogdanov–Takens Patterns
In the article [
8], the authors explored the system (
2) with parameter values
,
,
,
, and
; the threshold values are
,
. Upon selecting specific diffusion coefficients in
Table 3, the threshold value of the parameter
, satisfied the conditions outlined in Remark 1 for the system (
3). At the Turing–BT bifurcation, we obtained the pattern given in
Figure 7a.
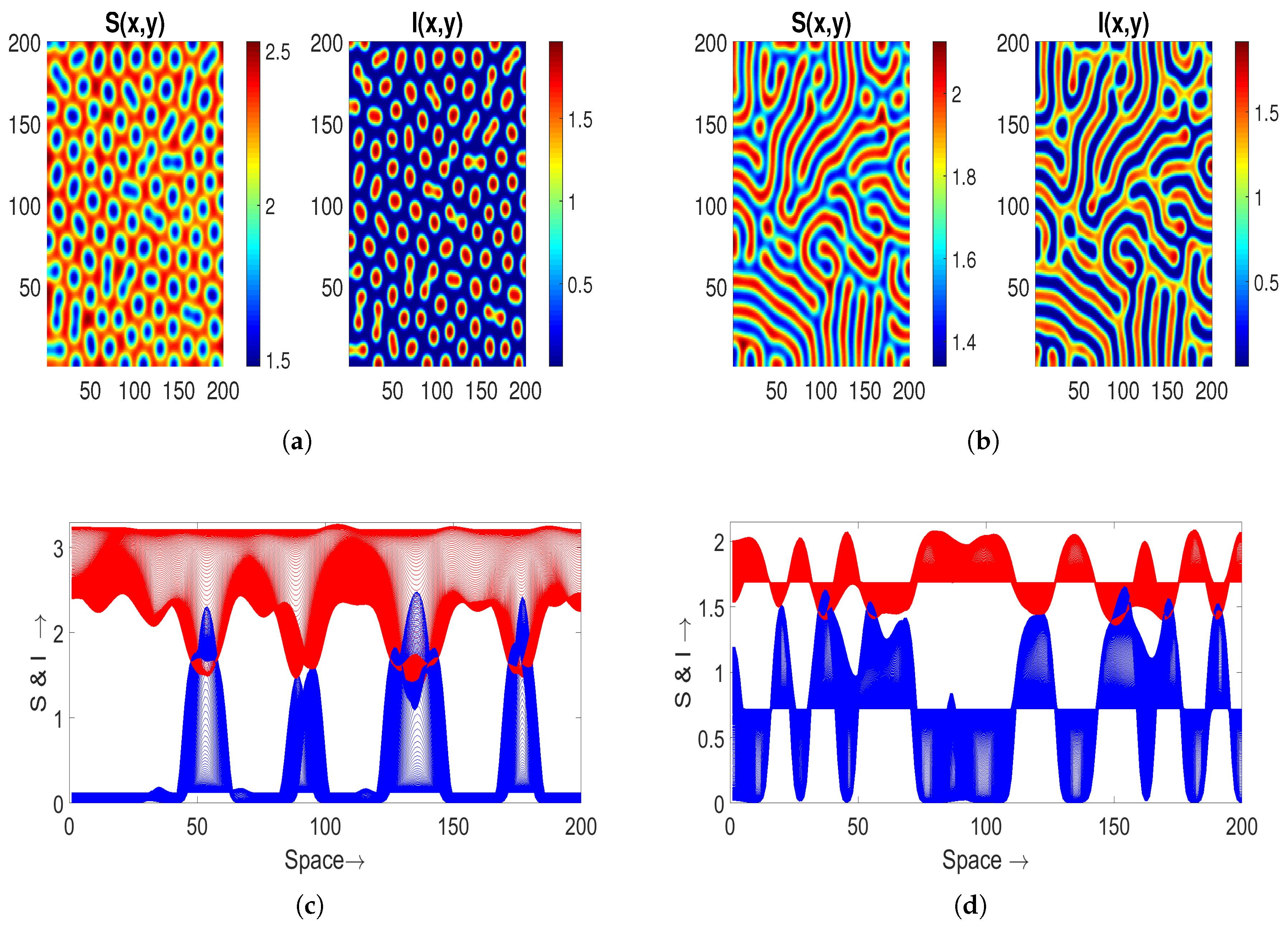
Further, with
we observe Turing–Bogdanov–Takens patterns in
Figure 8a. To examine the impact of treatment on the results, we increase the saturation factor
in both cases: from
to
. Notably, hole-type patterns manifest in
Figure 8a, which subsequently converge into strips within the Turing space for
(depicted in
Figure 8b). The corresponding spatial series for both populations
S (depicted in blue) and
I (depicted in red), as illustrated in
Figure 8c,d. These spatial series represent the associated 1D patterns.
To explore more about Turing–Bogdanov–Takens patterns, we take an alternative parameter set as
,
,
,
,
,
, and
. With the diffusion coefficients mentioned in
Table 3, we get the threshold value as
Again, these parametric values verify the fulfillment of the Turing–Bogdanov–Takens condition discussed in Remark 1 around the endemic state
, resulting in the jaggy pattern
Figure 7b. If we choose the value of
, then the disappearance of patterns is observed in
Figure 9a. The extinction indicates the decline of the infected population, aligning with the outcomes of the corresponding ODE model (
2), where solutions converge to the disease-free equilibrium state. In the scenario of increased saturation factor,
, hole-type Turing patterns reemerge in the distribution of both populations [
Figure 9b]. Corresponding surfaces are depicted in
Figure 9c,d, while spatial series are shown in
Figure 9e,f.
5.4. Effect of Perturbation on the Evolution of Patterns
The dynamics of an epidemiological reaction–diffusion model are profoundly shaped by the initial values. Consequently, the emergence of patterns is substantially governed via the influence of perturbations. Recent works [
41,
47] portrayed patterns depicting the spread of a specific disease originating from a central focal point, highlighting the crucial role of the point of origin. In alignment with this observation, we are motivated to investigate the behavior of the system (
3) under the same numerical parameter set as in the preceding scenarios. We aim to explore this behavior in the context of the introduced perturbation:
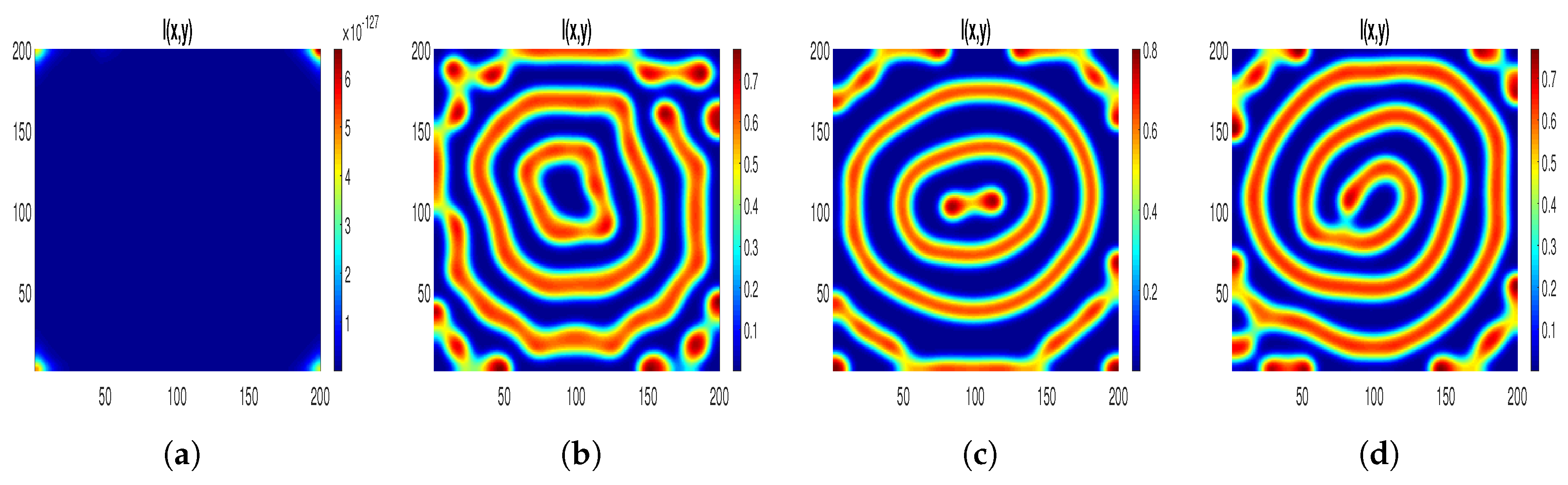
For the parameters mentioned in
Table 3, we present patterns with the
perturbation in
Figure 10. In order to comprehend the disease spread, our focus lies solely on the distribution of infected individuals. Notably, it is observed that low-density infected individuals are uniformly dispersed across the habitat, exhibiting blue coloration for
. This aligns with the extinction of the infected population, as indicated by the stability analysis of the disease-free state in the corresponding ODE system (
2).
Furthermore, with increasing values of , a noteworthy transition unfolds: regions with a high-density infected population (depicted in blue) start to emerge within different circles, while areas with a low-density infected population (depicted in red) begin to manifest against a blue background.
In
Section 5.3, Turing–Bogdanov–Takens patterns were generated using three distinct sets of numerical parameters, along with the
perturbation. In this section, we assess the resulting patterns under the same diffusion coefficient values for these parameter sets, this time introducing the
perturbation. For the parameters
,
,
,
,
, and
, we examine two values of the treatment parameter
=
and
. The resulting patterns are illustrated in
Figure 11 at time
. These patterns indicate the formation of rings with varying radii in regions of low density. When the treatment parameter
is altered, a disease initiates from the center of the disk and spreads across the entire area. Our analysis reveals that the distribution exhibits distinct zones of low and high infection, forming a striped pattern within a circular configuration. In conclusion, we infer that disease transmission is expected to be limited within this geographical area.
Table 2.
Summary of patterns of
Figure 4,
Figure 5,
Figure 6 and
Figure 10. For biological parameters
and
and diffusion coefficients
.
Table 2.
Summary of patterns of
Figure 4,
Figure 5,
Figure 6 and
Figure 10. For biological parameters
and
and diffusion coefficients
.
| Sr. No. | Parameter Values | Nature of | Type of Patterns | Figure |
|---|
| , and | Unstable | Non-Turing | With -Figure 4c,d and with -Figure 10a,b |
| , and | Unstable | Non-Turing | With -Figure 4e,f |
| , | Stable | Turing | With -Figure 5c,d and with -Figure 10c,d |
| , and | Stable | Turing | With -Figure 5e,f |
| , | Stable | Turing | With -Figure 6c–i |
| , | Non-hyperbolic | Turing–Hopf | With -Figure 6j |
| , | Stable | Non-Turing | With -Figure 6k |
Table 3.
Summary of patterns of
Figure 8,
Figure 9 and
Figure 11. We fixed biological constants as
and the diffusion coefficients as
,
. Here, BT = Bogdanov–Takens, and
,
= perturbation.
Table 3.
Summary of patterns of
Figure 8,
Figure 9 and
Figure 11. We fixed biological constants as
and the diffusion coefficients as
,
. Here, BT = Bogdanov–Takens, and
,
= perturbation.
| Sr. No. | Parameter Values | Nature of | Type of Patterns | Figure |
|---|
| | double-zero equilibrium | Turing-B-T | With -Figure 8a,c |
| | Stable | Turing | With -Figure 8b,d |
| | double-zero equilibrium | Turing-B-T | With -Figure 9c,e and with -Figure 11a |
| | Stable | Turing | With -Figure 9d,f and with -Figure 11c |
6. Conclusions
In reality, an individual’s movement is influenced by a multitude of factors, including their disease-related precautions, access to medical care, and various other considerations. In light of these factors, we propose and employ a novel nonlinear cross-diffusion approach to investigate an SIR epidemic model featuring saturated-type treatment. Our proposed cross-diffusion mechanism, characterized by a combination of quadratic and rational components, offers enhanced realism compared to existing formulations in the literature. Importantly, the accuracy of this cross-diffusion representation is demonstrated concerning the populations’ mobility capacity through flux calculations. Utilizing coupled upper and lower solutions for elliptic partial differential equations, we establish the admissibility of at least one non-constant endemic state within the reduced SIR model augmented with the proposed cross-diffusion. This finding underscores the potential coexistence of susceptible and infected populations within their environment. To illustrate potential pattern formation scenarios driven by Turing instability, we numerically simulate the proposed model in a square environment of dimensions .
We modulate the treatment parameter to assess the impact of delaying treatment for infected individuals. We acknowledge the potential for a significant influence on pattern evolution based on variations in the initial disturbance. Therefore, in this study, we propose two novel perturbations, denoted as and , to explore diverse pattern types. We observe that, when the parameters associated with temporal models and diffusion coefficients are modified using the first perturbation , patterns such as holes, strips, and mixture-type patterns emerge. However, it is noteworthy that patterns obtained with the perturbation transition into circular strips under . This observation illustrates how a disease can propagate in the form of concentric stripe rings of varying radii when originating from a specific point, the disease’s center of origin. Additionally, through the identification of codimension-2 Turing–Hopf bifurcation, we determine the Turing space.
The authors [
8] investigate the existence of two pairs of Bogdanov–Takens bifurcation thresholds in the corresponding temporal model. For these parameter sets, a specific combination of diffusion coefficients is selected to observe Turing–Bogdanov–Takens patterns with both perturbations. It is observed that distinct patterns emerge for these parameter sets, and contrary to temporal predictions, the infection persists within the system. Consequently, a new set of relevant parameters is considered. It is concluded that, for lower values of
(which measures the delay in treatment for the infected population), the infected population becomes extinct due to the absence of a stable endemic state in the corresponding ODE model. This aligns with equilibrium stability results, where solutions converge towards the disease-free equilibrium state. Additionally, various non-Turing patterns around the unstable equilibrium state are plotted, meeting the criteria for Turing instability for different
values. When two endemic states are present, directed patterns suffer more significant disruption than in the case of a single endemic state. This implies that disease transmission worsens with increasing
values. The impact of the diffusion coefficient
is examined through numerical simulations to understand the influence of the suggested cross-diffusion. This work represents a modest advancement in comprehending the role of movement for both susceptible and infected populations in infectious disease modeling.
Model Limitations and Applicability: While this study highlights the influence of cross-diffusion and spatial heterogeneity under movement-restricted settings, its applicability to highly mobile populations—such as during the COVID-19 pandemic—is limited. The zero-flux boundary assumption does not reflect real-world mobility enabled by transportation networks like airports and highways. Moreover, the model assumes symptomatic transmission, which fails to capture diseases with significant asymptomatic or pre-symptomatic spread, such as COVID-19. In such cases, avoidance behavior is ineffective since infected individuals are not easily identifiable. To address these limitations, future work could incorporate nonlocal dispersal, mobility networks, and additional compartments for asymptomatic or undetected carriers to better reflect real-world transmission dynamics.