A GATA Transcription Factor from Soybean (Glycine max) Regulates Chlorophyll Biosynthesis and Suppresses Growth in the Transgenic Arabidopsis thaliana
Abstract
1. Introduction
2. Results
2.1. Sequence Analysis of GmGATA58
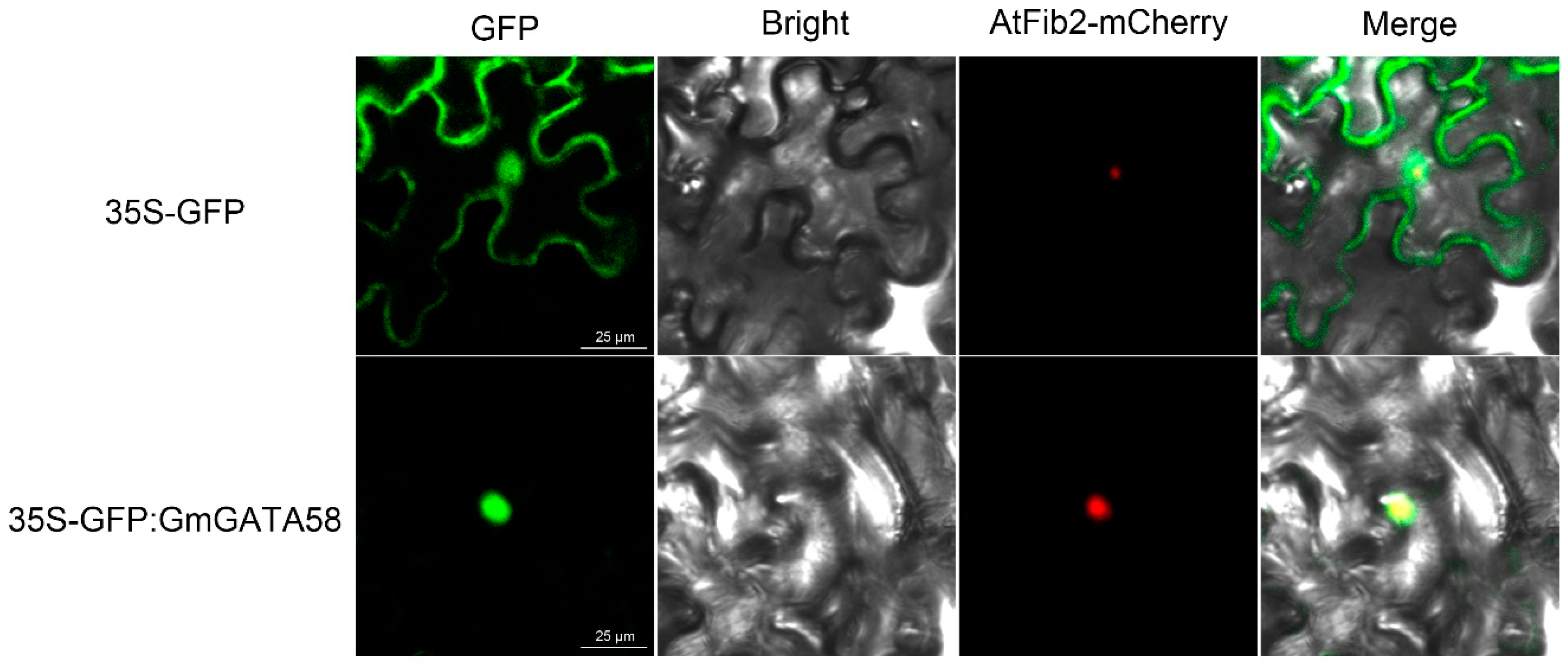
2.2. GmGATA58 Localizes in Nucleus
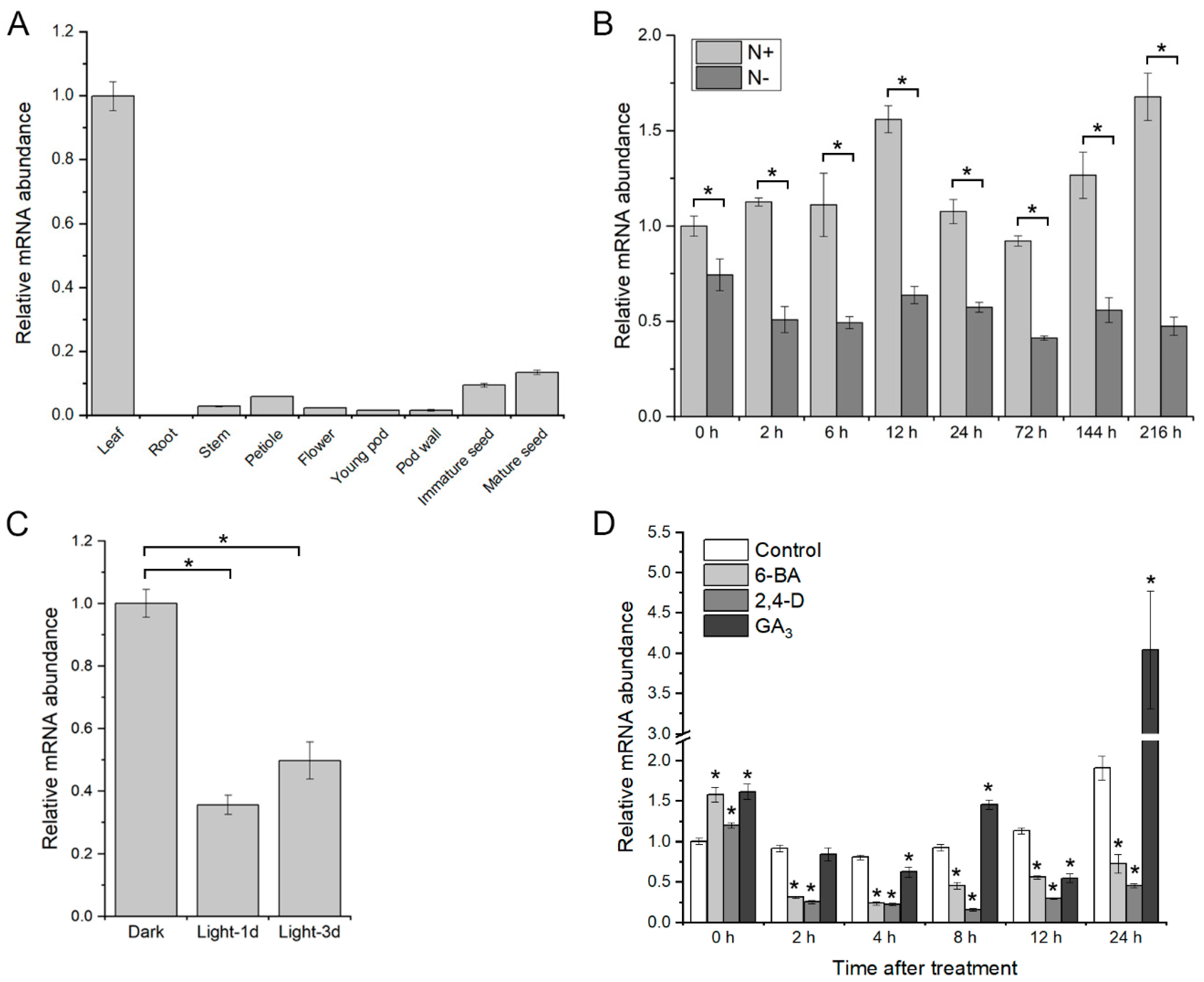
2.3. Expression Pattern of GmGATA58
2.4. Histochemical GUS Assay of Transgenic Arabidopsis Expressing GmGATA58 Promoter
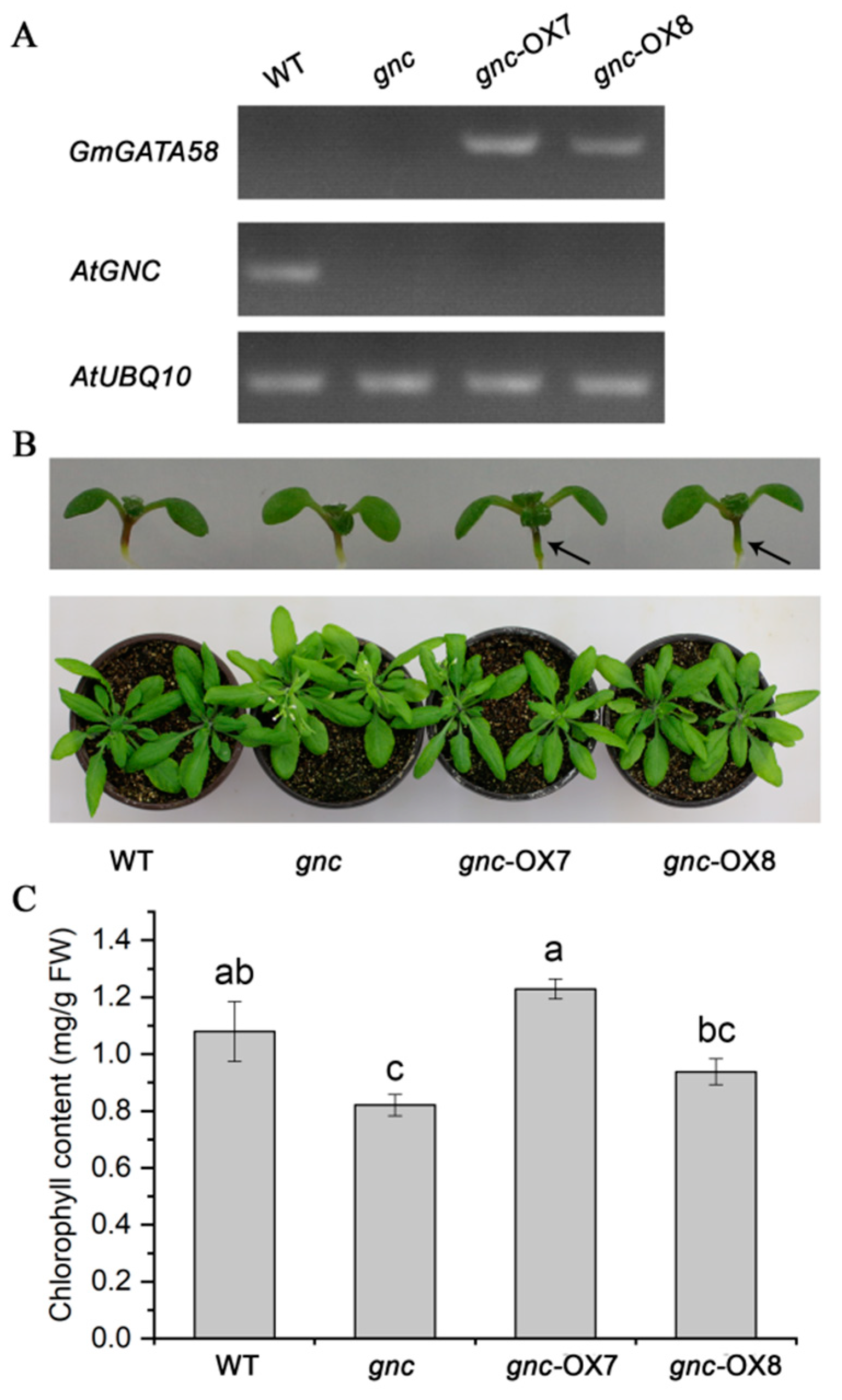
2.5. GmGATA58 Genetically Complements the gnc Mutant of Arabidopsis
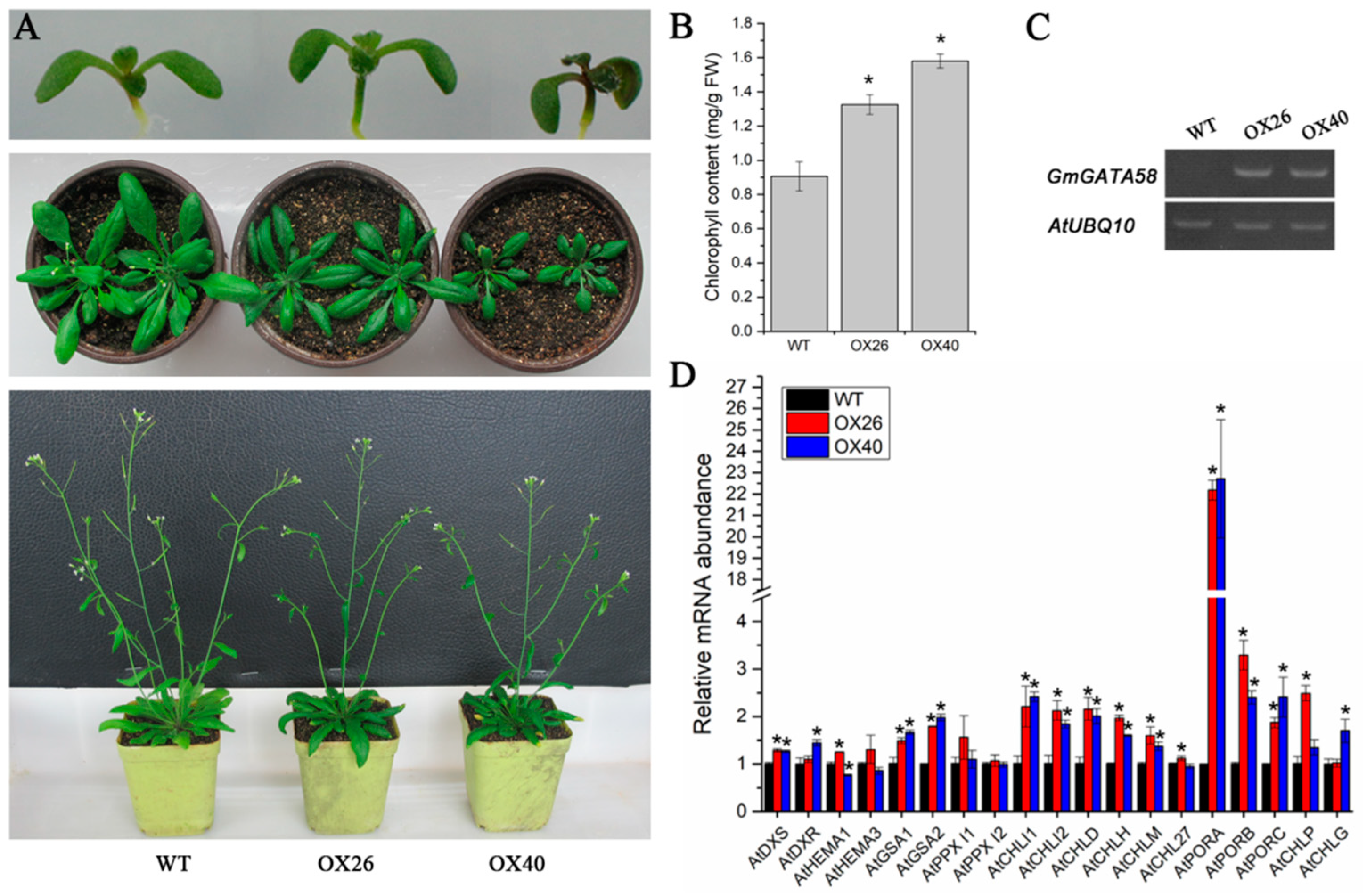
2.6. GmGATA58 Overexpression in Arabidopsis Increases Chlorophyll Content and Up-Regulates the Expression Levels of Chlorophyll Biosynthetic Genes
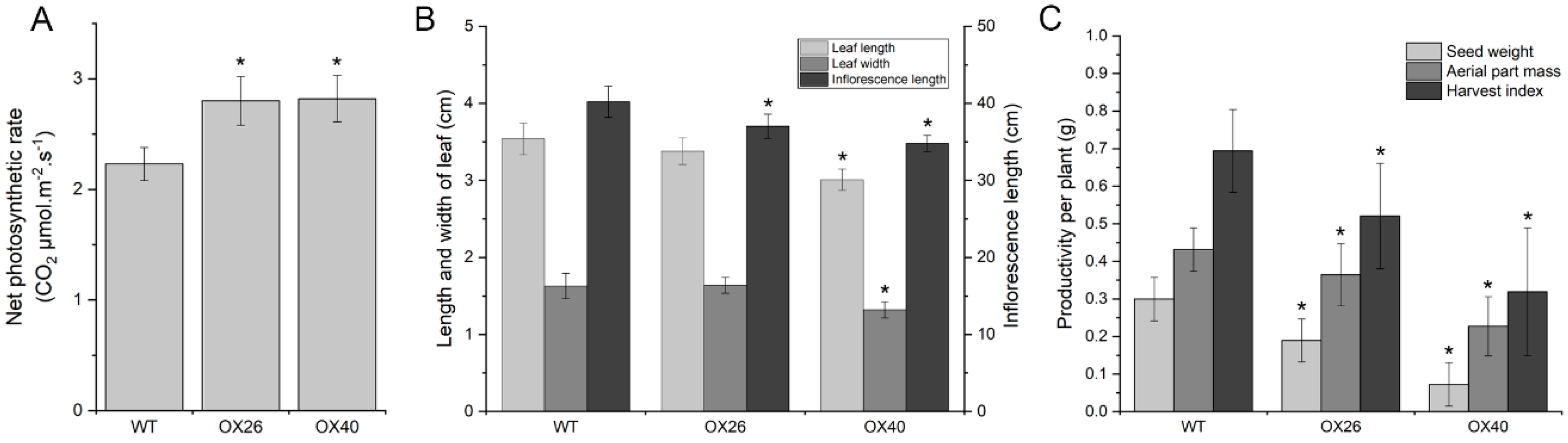
2.7. Plant Growth Vigor and Productivity of Arabidopsis Are Suppressed by Overexpression of GmGATA58
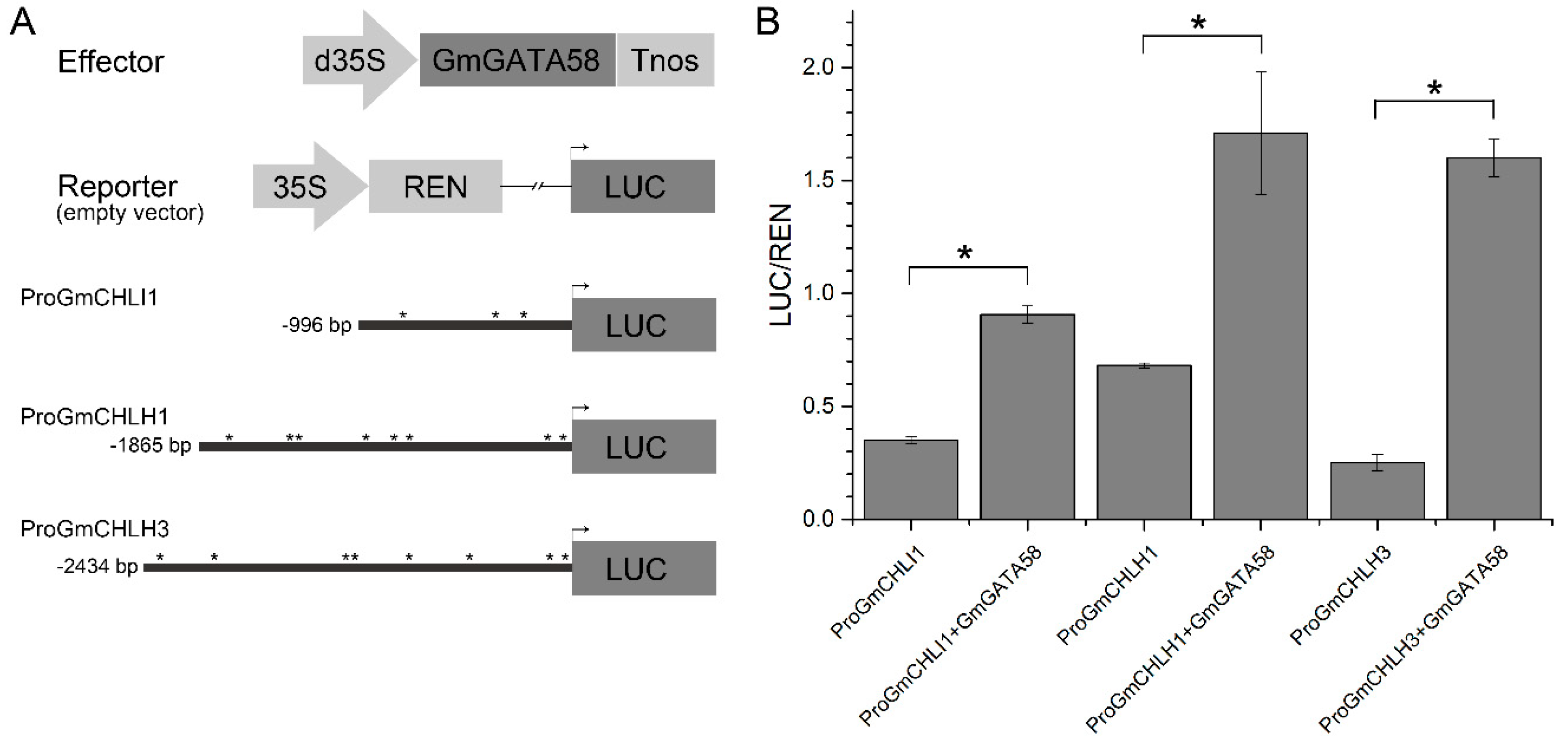
2.8. Transcription Activation of GmGATA58 against the Promoters of Chlorophyll Biosynthetic Genes from Soybean in Transiently Transformed Arabidopsis Protoplast
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. DNA and RNA Extraction
4.3. Isolation of GmGATA58 gene
4.4. Subcellular Localization of GmGATA58
4.5. Isolation and Analysis of GmGATA58 Promoter
4.6. Overexpression Vector Construction and Arabidopsis Transformation
4.7. Semi-Quantitative RT-PCR and Quantitative PCR Analysis
4.8. Determination of Chlorophyll Content, Photosynthetic Rate and Growth in Arabidopsis
4.9. Dual-Luciferase Assay in Transiently Transformed Protoplast of Arabidopsis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Eckhardt, U.; Grimm, B.; Hortensteiner, S. Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol. Biol. 2004, 56, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Buttery, B.R.; Buzzell, R.I.; Findlay, W.I. Relationships among photosynthetic rate, bean yield and other characters in field-grown cultivars of soybean. Can. J. Plant Sci. 1981, 61, 190–197. [Google Scholar] [CrossRef]
- Maekawa, T.; Kokubun, M. Correlation of leaf nitrogen, chlorophyll and rubisco contents with photosynthesis in a supernodulating soybean genotype Sakukei 4. Plant Prod. Sci. 2005, 8, 419–426. [Google Scholar] [CrossRef]
- Zhu, X.-G.; Long, S.P.; Ort, D.R. Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 2010, 61, 235–261. [Google Scholar] [CrossRef] [PubMed]
- Zelitch, I. The close relationship between net photosynthesis and crop yield. BioScience 1982, 32, 796–802. [Google Scholar] [CrossRef]
- Ainsworth, E.A.; Yendrek, C.R.; Skoneczka, J.A.; Long, S.P. Accelerating yield potential in soybean: Potential targets for biotechnological improvement. Plant Cell Environ. 2012, 35, 38–52. [Google Scholar] [CrossRef]
- Buttery, B.R.; Buzzell, R.I. The relationship between chlorophyll content and rate of photosynthesis in soybeans. Can. J. Plant Sci. 1977, 57, 1–5. [Google Scholar] [CrossRef]
- Dordas, C.; Sioulas, C. Safflower yield, chlorophyll content, photosynthesis, and water use efficiency response to nitrogen fertilization under rainfed conditions. Ind. Crops Prod. 2008, 27, 75–85. [Google Scholar] [CrossRef]
- Kim, S.; Schlicke, H.; Van Ree, K.; Karvonen, K.; Subramaniam, A.; Richter, A.; Grimm, B.; Braam, J. Arabidopsis chlorophyll biosynthesis: An essential balance between the methylerythritol phosphate and tetrapyrrole pathways. Plant Cell 2013, 25, 4984–4993. [Google Scholar] [CrossRef]
- Brzezowski, P.; Richter, A.S.; Grimm, B. Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim. Biophys. Acta 2015, 1847, 968–985. [Google Scholar] [CrossRef]
- Kobayashi, K.; Masuda, T. Transcriptional regulation of tetrapyrrole biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 1811. [Google Scholar] [CrossRef] [PubMed]
- Hudson, D.; Guevara, D.; Yaish, M.W.; Hannam, C.; Long, N.; Clarke, J.D.; Bi, Y.M.; Rothstein, S.J. GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS ONE 2011, 6, e26765. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.H.; Zubo, Y.O.; Tapken, W.; Kim, H.J.; Lavanway, A.M.; Howard, L.; Pilon, M.; Kieber, J.J.; Schaller, G.E. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 2012, 160, 332–348. [Google Scholar] [CrossRef] [PubMed]
- Bastakis, E.; Hedtke, B.; Klermund, C.; Grimm, B.; Schwechheimer, C. LLM-domain B-GATA transcription factors play multifaceted roles in controlling greening in Arabidopsis. Plant Cell 2018, 30, 582–599. [Google Scholar] [CrossRef]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004, 134, 1718–1732. [Google Scholar] [CrossRef]
- Behringer, C.; Schwechheimer, C. B-GATA transcription factors - insights into their structure, regulation, and role in plant development. Front. Plant Sci. 2015, 6, 90. [Google Scholar] [CrossRef]
- An, Y.; Zhou, Y.; Han, X.; Shen, C.; Wang, S.; Liu, C.; Yin, W.; Xia, X. The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar. J. Exp. Bot. 2019, 71, 1969–1984. [Google Scholar] [CrossRef]
- Bi, Y.M.; Zhang, Y.; Signorelli, T.; Zhao, R.; Zhu, T.; Rothstein, S. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 2005, 44, 680–692. [Google Scholar] [CrossRef]
- Zhang, Z.; Ren, C.; Zou, L.; Wang, Y.; Li, S.; Liang, Z. Characterization of the GATA gene family in Vitis vinifera: Genome-wide analysis, expression profiles, and involvement in light and phytohormone response. Genome 2018, 61, 713–723. [Google Scholar] [CrossRef]
- Zhang, C.; Hou, Y.; Hao, Q.; Chen, H.; Chen, L.; Yuan, S.; Shan, Z.; Zhang, X.; Yang, Z.; Qiu, D.; et al. Genome-wide survey of the soybean GATA transcription factor gene family and expression analysis under low nitrogen stress. PLoS ONE 2015, 10, e0125174. [Google Scholar] [CrossRef]
- Hao, Q.N.; Zhou, X.A.; Sha, A.H.; Wang, C.; Zhou, R.; Chen, S.L. Identification of genes associated with nitrogen-use efficiency by genome-wide transcriptional analysis of two soybean genotypes. BMC Genom. 2011, 12, 525. [Google Scholar] [CrossRef] [PubMed]
- Buttery, B.R.; Buzzell, R.I. Soybean leaf nitrogen in relation to photosynthetic rate and yield. Can. J. Plant Sci. 1988, 68, 793–795. [Google Scholar] [CrossRef]
- Lawlor, D.W.; Lemaire, G.; Gastal, F. Nitrogen, Plant Growth and Crop Yield. In Plant Nitrogen; Lea, P.J., Morot-Gaudry, J.-F., Eds.; Springe: Berlin/Heidelberg, Germany, 2001; pp. 343–367. [Google Scholar] [CrossRef]
- Fritschi, F.B.; Ray, J.D. Soybean leaf nitrogen, chlorophyll content, and chlorophyll a/b ratio. Photosynthetica 2007, 45, 92–98. [Google Scholar] [CrossRef]
- Hawkins, J.A.; Sawyer, J.E.; Barker, D.W.; Lundvall, J.P. Using relative chlorophyll meter values to determine nitrogen application rates for corn. Agron. J. 2007, 99, 1034–1040. [Google Scholar] [CrossRef]
- Behringer, C.; Bastakis, E.; Ranftl, Q.L.; Mayer, K.F.; Schwechheimer, C. Functional diversification within the family of B-GATA transcription factors through the leucine-leucine-methionine domain. Plant Physiol. 2014, 166, 293–305. [Google Scholar] [CrossRef]
- Missbach, S.; Weis, B.L.; Martin, R.; Simm, S.; Bohnsack, M.T.; Schleiff, E. 40S ribosome biogenesis co-factors are essential for gametophyte and embryo development. PLoS ONE 2013, 8, e54084. [Google Scholar] [CrossRef]
- Richter, R.; Behringer, C.; Muller, I.K.; Schwechheimer, C. The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 2010, 24, 2093–2104. [Google Scholar] [CrossRef]
- Mara, C.D.; Irish, V.F. Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol. 2008, 147, 707–718. [Google Scholar] [CrossRef]
- Ranftl, Q.L.; Bastakis, E.; Klermund, C.; Schwechheimer, C. LLM-domain containing B-GATA factors control different aspects of cytokinin-regulated development in Arabidopsis thaliana. Plant Physiol. 2016, 170, 2295–2311. [Google Scholar] [CrossRef]
- Hudson, D.; Guevara, D.R.; Hand, A.J.; Xu, Z.; Hao, L.; Chen, X.; Zhu, T.; Bi, Y.M.; Rothstein, S.J. Rice cytokinin GATA transcription Factor1 regulates chloroplast development and plant architecture. Plant Physiol. 2013, 162, 132–144. [Google Scholar] [CrossRef]
- Richter, R.; Bastakis, E.; Schwechheimer, C. Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering time in Arabidopsis. Plant Physiol. 2013, 162, 1992–2004. [Google Scholar] [CrossRef] [PubMed]
- Naito, T.; Kiba, T.; Koizumi, N.; Yamashino, T.; Mizuno, T. Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2007, 71, 1557–1560. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Han, X.; Tang, S.; Xia, X.; Yin, W. Poplar GATA transcription factor PdGNC is capable of regulating chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis under varying nitrogen levels. Plant Cell Tissue Organ Cult. 2014, 119, 313–327. [Google Scholar] [CrossRef]
- Richter, R.; Behringer, C.; Zourelidou, M.; Schwechheimer, C. Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2013, 110, 13192–13197. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.; Zhong, S. Interplay between light and plant hormones in the control of arabidopsis seedling chlorophyll biosynthesis. Front. Plant Sci. 2017, 8, 1433. [Google Scholar] [CrossRef]
- Springer, N.M. Isolation of plant DNA for PCR and genotyping using organic extraction and CTAB. Cold Spring Harb. Protoc. 2010, 2010, pdb-rot5515. [Google Scholar] [CrossRef]
- Chen, S.; Songkumarn, P.; Liu, J.; Wang, G.L. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 2009, 150, 1111–1121. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, J.; Zhang, Y.; Cai, X.; Gong, P.; Zhang, J.; Wang, T.; Li, H.; Ye, Z. Overexpression of SlGMEs leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Cell Rep. 2011, 30, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Yuxia, W.; Mingui, Z.; Jianxin, H. Response of photosynthesis function of salt cress and Arabidopsis to NaCl salt stress. Chin. Bull. Bot. 2007, 24, 154–160. [Google Scholar]
- Yoo, S.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Huang, Y.; Xiao, Z.; Yang, H.; Hao, Q.; Yuan, S.; Chen, H.; Chen, L.; Chen, S.; Zhou, X.; et al. A GATA Transcription Factor from Soybean (Glycine max) Regulates Chlorophyll Biosynthesis and Suppresses Growth in the Transgenic Arabidopsis thaliana. Plants 2020, 9, 1036. https://doi.org/10.3390/plants9081036
Zhang C, Huang Y, Xiao Z, Yang H, Hao Q, Yuan S, Chen H, Chen L, Chen S, Zhou X, et al. A GATA Transcription Factor from Soybean (Glycine max) Regulates Chlorophyll Biosynthesis and Suppresses Growth in the Transgenic Arabidopsis thaliana. Plants. 2020; 9(8):1036. https://doi.org/10.3390/plants9081036
Chicago/Turabian StyleZhang, Chanjuan, Yi Huang, Zhiyuan Xiao, Hongli Yang, Qingnan Hao, Songli Yuan, Haifeng Chen, Limiao Chen, Shuilian Chen, Xinan Zhou, and et al. 2020. "A GATA Transcription Factor from Soybean (Glycine max) Regulates Chlorophyll Biosynthesis and Suppresses Growth in the Transgenic Arabidopsis thaliana" Plants 9, no. 8: 1036. https://doi.org/10.3390/plants9081036
APA StyleZhang, C., Huang, Y., Xiao, Z., Yang, H., Hao, Q., Yuan, S., Chen, H., Chen, L., Chen, S., Zhou, X., & Huang, W. (2020). A GATA Transcription Factor from Soybean (Glycine max) Regulates Chlorophyll Biosynthesis and Suppresses Growth in the Transgenic Arabidopsis thaliana. Plants, 9(8), 1036. https://doi.org/10.3390/plants9081036





