Kmt2c/Mll3 Haploinsufficiency Causes Autism-like Behavioral Deficits in Mice
Abstract
1. Introduction
2. Materials and Methods
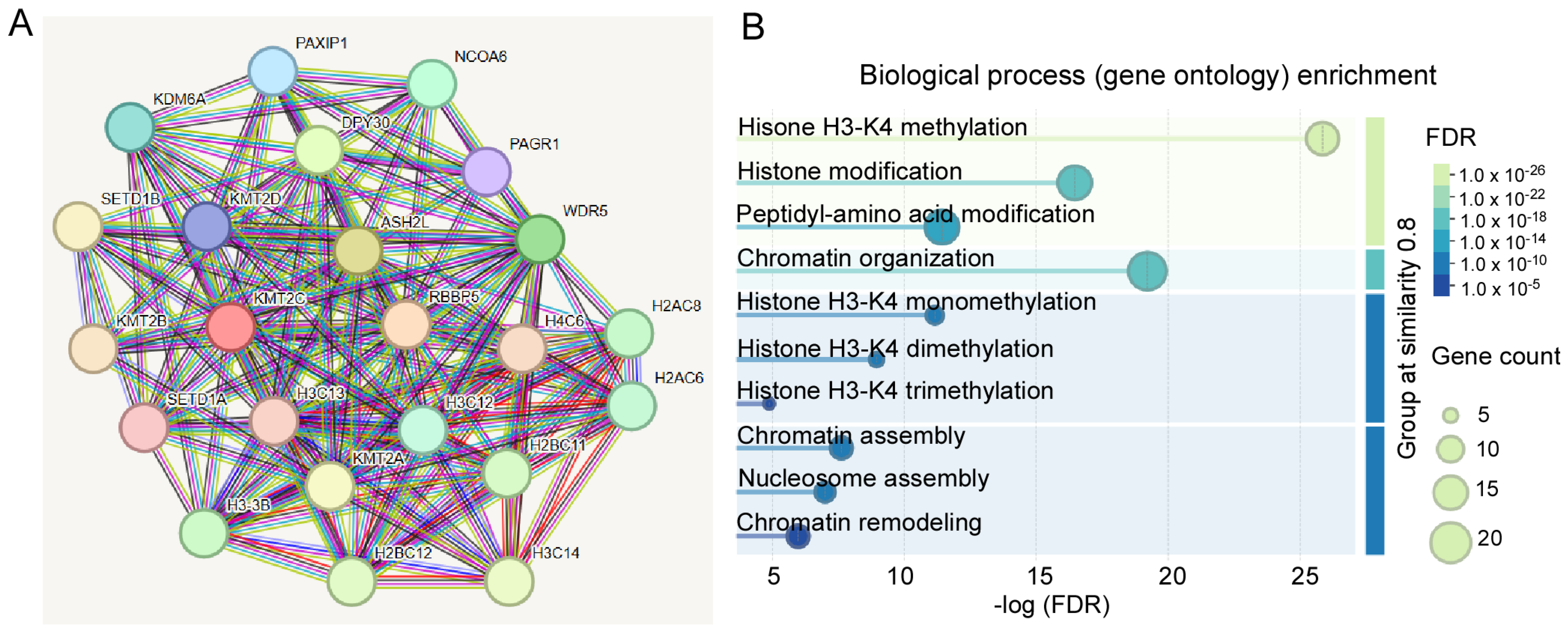
2.1. Protein–Protein Interaction (PPI) Network and Functional Annotation Analysis
2.2. Animal Care and Husbandry
2.3. Behavioral Tests
2.4. Statistical Analysis
3. Results
3.1. KMT2C/Kmt2c Is Spatiotemporally Expressed in the Human/Mouse Brain
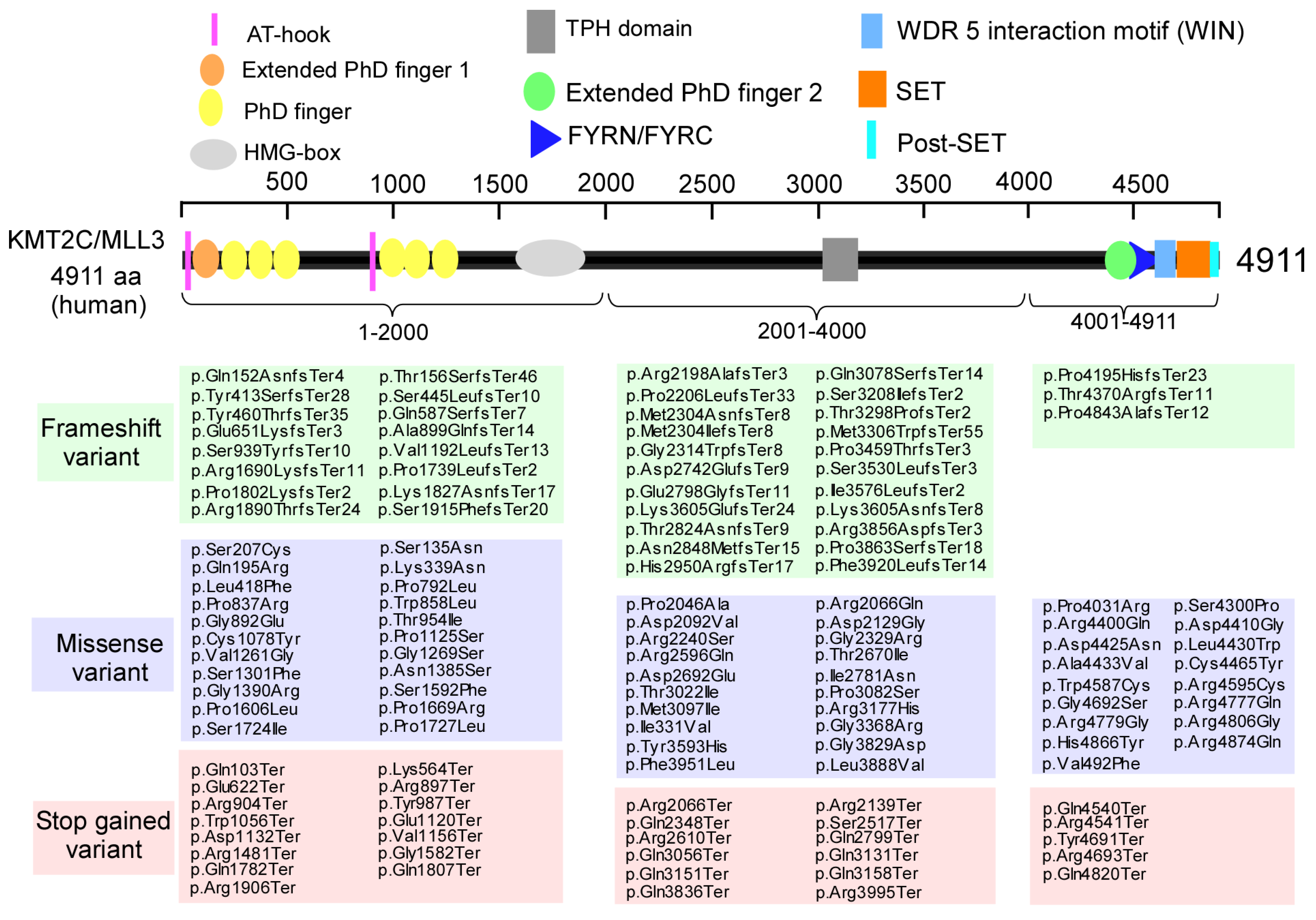
3.2. The Biological Roles of KMT2C in the Brain
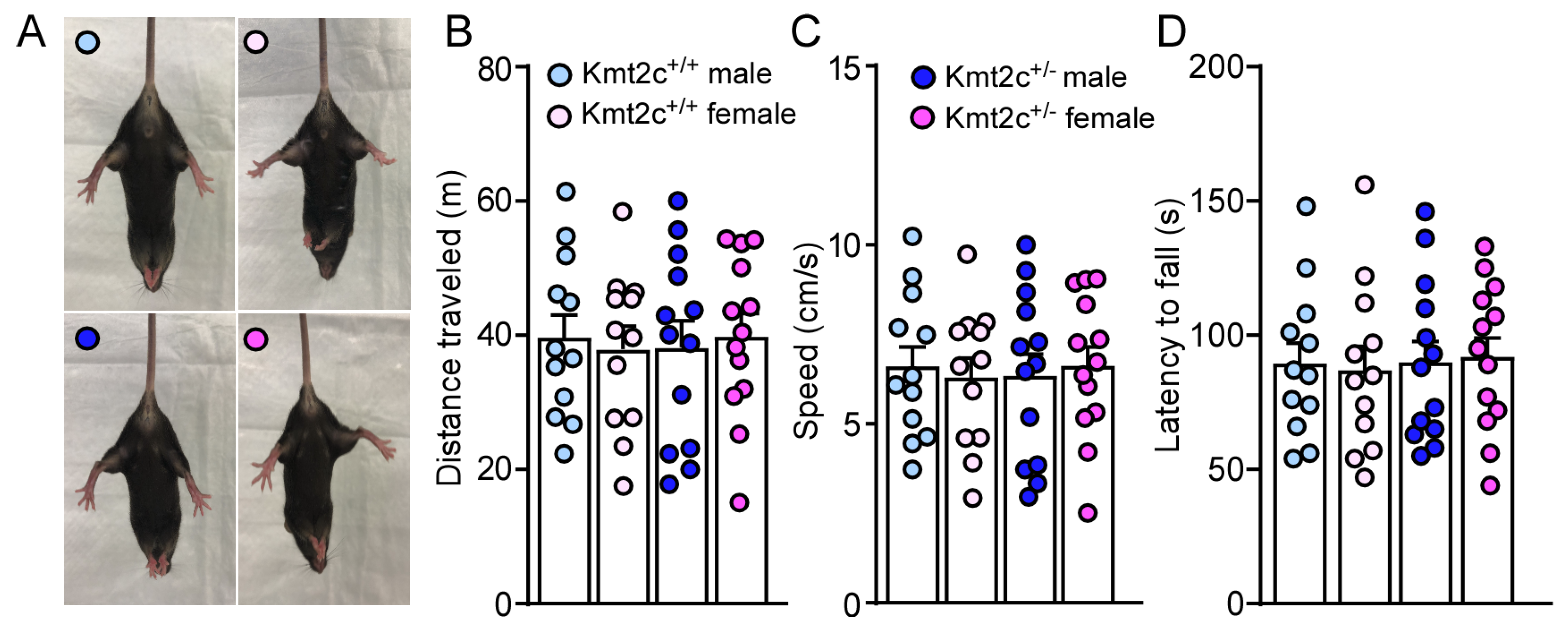
3.3. Kmt2c Haploinsufficiency Mice Have Normal Motor Function Without Anxiety-like Behaviors
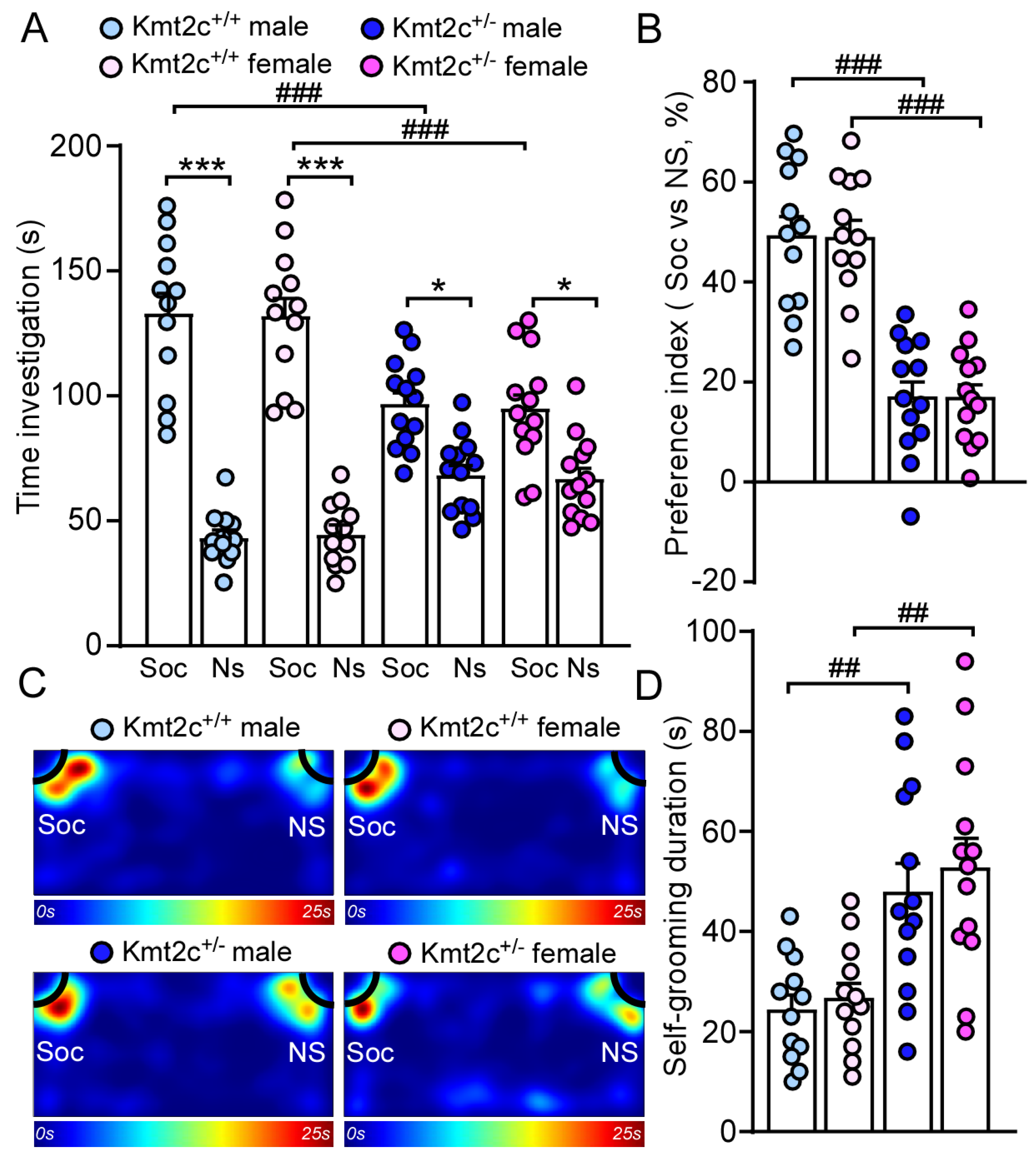
3.4. Kmt2c Haploinsufficiency Mice Exhibit Autism-like Behavioral Deficits
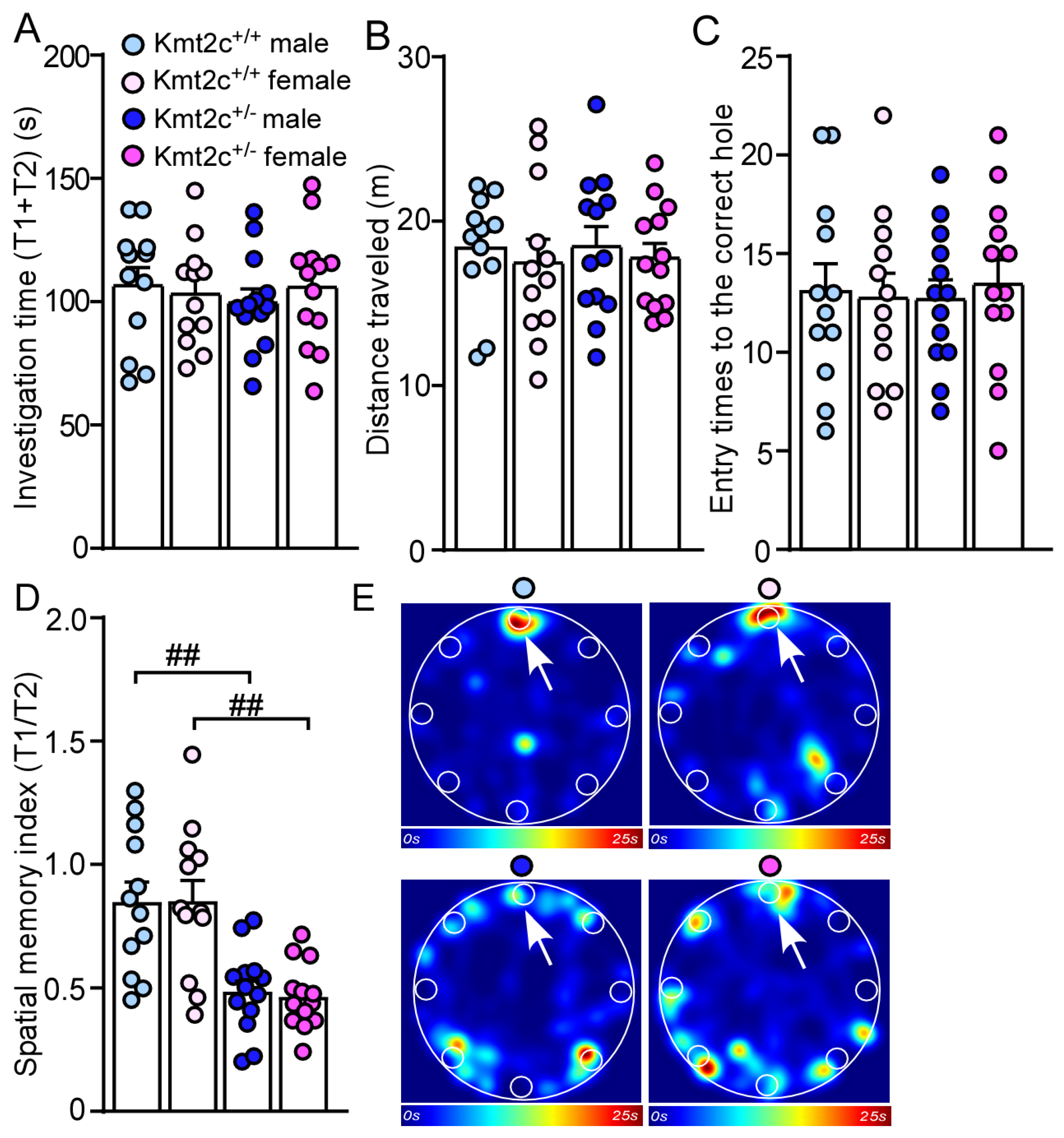
3.5. Kmt2c Haploinsufficiency Mice Display Cognitive Deficits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASD | Autism spectrum disorder |
| NDD | Neurodevelopmental delay |
| ID | Intellectual disability |
| CDC | Centers for Diseases Control and Prevention ear dichroism |
| PFC | Prefrontal cortex |
| DIV | Days in vitro |
| PPI | Protein–protein interaction |
| SFARI | Simons Foundation Autism Research Initiative |
References
- Matelski, L.; Van de Water, J. Risk factors in autism: Thinking outside the brain. J. Autoimmun. 2016, 67, 1–7. [Google Scholar] [CrossRef]
- Love, C.; Sominsky, L.; O’Hely, M.; Berk, M.; Vuillermin, P.; Dawson, S.L. Prenatal environmental risk factors for autism spectrum disorder and their potential mechanisms. BMC Med. 2024, 22, 393. [Google Scholar] [CrossRef]
- Bailey, A.; Le Couteur, A.; Gottesman, I.; Bolton, P.; Simonoff, E.; Yuzda, E.; Rutter, M. Autism as a strongly genetic disorder: Evidence from a British twin study. Psychol. Med. 1995, 25, 63–77. [Google Scholar] [CrossRef]
- Tick, B.; Bolton, P.; Happé, F.; Rutter, M.; Rijsdijk, F. Heritability of autism spectrum disorders: A meta-analysis of twin studies. J. Child Psychol. Psychiatry 2016, 57, 585–595. [Google Scholar] [CrossRef]
- Satterstrom, F.K.; Kosmicki, J.A.; Wang, J.; Breen, M.S.; De Rubeis, S.; An, J.Y.; Peng, M.; Collins, R.; Grove, J.; Klei, L.; et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 2020, 180, 568–584.e523. [Google Scholar] [CrossRef]
- De Rubeis, S.; He, X.; Goldberg, A.P.; Poultney, C.S.; Samocha, K.; Cicek, A.E.; Kou, Y.; Liu, L.; Fromer, M.; Walker, S.; et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014, 515, 209–215. [Google Scholar] [CrossRef]
- Wang, T.; Guo, H.; Xiong, B.; Stessman, H.A.; Wu, H.; Coe, B.P.; Turner, T.N.; Liu, Y.; Zhao, W.; Hoekzema, K.; et al. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat. Commun. 2016, 7, 13316. [Google Scholar] [CrossRef]
- Koemans, T.S.; Kleefstra, T.; Chubak, M.C.; Stone, M.H.; Reijnders, M.R.F.; de Munnik, S.; Willemsen, M.H.; Fenckova, M.; Stumpel, C.; Bok, L.A.; et al. Functional convergence of histone methyltransferases EHMT1 and KMT2C involved in intellectual disability and autism spectrum disorder. PLoS Genet. 2017, 13, e1006864. [Google Scholar] [CrossRef]
- Stessman, H.A.; Xiong, B.; Coe, B.P.; Wang, T.; Hoekzema, K.; Fenckova, M.; Kvarnung, M.; Gerdts, J.; Trinh, S.; Cosemans, N.; et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat. Genet. 2017, 49, 515–526. [Google Scholar] [CrossRef]
- Chen, C.H.; Huang, A.; Huang, Y.S.; Fang, T.H. Identification of a Rare Novel KMT2C Mutation That Presents with Schizophrenia in a Multiplex Family. J. Pers. Med. 2021, 11, 1254. [Google Scholar] [CrossRef]
- Whitford, W.; Taylor, J.; Hayes, I.; Smith, W.; Snell, R.G.; Lehnert, K.; Jacobsen, J.C. A novel 11 base pair deletion in KMT2C resulting in Kleefstra syndrome 2. Mol. Genet. Genom. Med. 2023, e2350. [Google Scholar] [CrossRef]
- Rots, D.; Choufani, S.; Faundes, V.; Dingemans, A.J.M.; Joss, S.; Foulds, N.; Jones, E.A.; Stewart, S.; Vasudevan, P.; Dabir, T.; et al. Pathogenic variants in KMT2C result in a neurodevelopmental disorder distinct from Kleefstra and Kabuki syndromes. Am. J. Hum. Genet. 2024, 111, 1626–1642. [Google Scholar] [CrossRef]
- Lavery, W.J.; Barski, A.; Wiley, S.; Schorry, E.K.; Lindsley, A.W. KMT2C/D COMPASS complex-associated diseases [K(CD)COM-ADs]: An emerging class of congenital regulopathies. Clin. Epigenetics 2020, 12, 10. [Google Scholar] [CrossRef]
- Chen, R.; Okeyo-Owuor, T.; Patel, R.M.; Casey, E.B.; Cluster, A.S.; Yang, W.; Magee, J.A. Kmt2c mutations enhance HSC self-renewal capacity and convey a selective advantage after chemotherapy. Cell Rep. 2021, 34, 108751. [Google Scholar] [CrossRef]
- Sze, C.C.; Shilatifard, A. Mll3/Mll4/Compass Family on Epigenetic Regulation of Enhancer Function and Cancer. Cold Spring Harb. Perspect. Med. 2016, 6, a026427. [Google Scholar] [CrossRef]
- Lee, S.; Roeder, R.G.; Lee, J.W. Roles of Histone H3-Lysine 4 Methyltransferase Complexes in NR-Mediated Gene Transcription. Prog. Mol. Biol. Transl. Sci. 2009, 87, 343–382. [Google Scholar]
- Gala, K.; Li, Q.; Sinha, A.; Razavi, P.; Dorso, M.; Sanchez-Vega, F.; Chung, Y.R.; Hendrickson, R.; Hsieh, J.J.; Berger, M.; et al. KMT2C Mediates the Estrogen Dependence of Breast Cancer Through Regulation of ERα Enhancer Function. Oncogene 2018, 37, 4692–4710. [Google Scholar] [CrossRef]
- Rampias, T.; Karagiannis, D.; Avgeris, M.; Polyzos, A.; Kokkalis, A.; Kanaki, Z.; Kousidou, E.; Tzetis, M.; Kanavakis, E.; Stravodimos, K.; et al. The Lysine-Specific Methyltransferase KMT2C/MLL3 Regulates DNA Repair Components in Cancer. EMBO Rep. 2019, 20, e46821. [Google Scholar] [CrossRef]
- Jiao, Y.; Lv, Y.; Liu, M.; Liu, Y.; Han, M.; Xiong, X.; Zhou, H.; Zhong, J.; Kang, X.; Su, W. The Modification Role and Tumor Association with a Methyltransferase: KMT2C. Front. Immunol. 2024, 15, 1444923. [Google Scholar] [CrossRef]
- Brauer, B.; Merino-Veliz, N.; Ahumada-Marchant, C.; Arriagada, G.; Bustos, F.J. KMT2C Knockout Generates ASD-Like Behaviors in Mice. Front. Cell Dev. Biol. 2023, 11, 1227723. [Google Scholar] [CrossRef]
- Nakamura, T.; Yoshihara, T.; Tanegashima, C.; Kadota, M.; Kobayashi, Y.; Honda, K.; Ishiwata, M.; Ueda, J.; Hara, T.; Nakanishi, M.; et al. Transcriptomic Dysregulation and Autistic-Like Behaviors in Kmt2c Haploinsufficient Mice Rescued by an LSD1 Inhibitor. Mol. Psychiatry 2024, 29, 2888–2904. [Google Scholar] [CrossRef]
- Shansky, R.M.; Woolley, C.S. Considering Sex as a Biological Variable Will Be Valuable for Neuroscience Research. J. Neurosci. Off. J. Soc. Neurosci. 2016, 36, 11817–11822. [Google Scholar] [CrossRef]
- Zhu, J.W.; Li, Y.F.; Wang, Z.T.; Jia, W.Q.; Xu, R.X. Toll-Like Receptor 4 Deficiency Impairs Motor Coordination. Front. Neurosci. 2016, 10, 33. [Google Scholar] [CrossRef]
- Ma, K.; Taylor, C.; Williamson, M.; Newton, S.S.; Qin, L. Diminished Activity-Dependent Bdnf Signaling Differentially Causes Autism-Like Behavioral Deficits in Male and Female Mice. Front. Psychiatry 2023, 14, 1182472. [Google Scholar] [CrossRef]
- Ma, K.; McDaniel, K.; Zhang, D.; Webb, M.; Qin, L. Chemogenetic Inhibition of Prefrontal Cortex Ameliorates Autism-Like Social Deficits and Absence-Like Seizures in a Gene-Trap Ash1l Haploinsufficiency Mouse Model. Genes 2024, 15, 1619. [Google Scholar] [CrossRef]
- Ma, K.; Qin, L.; Matas, E. Histone Deacetylase Inhibitor MS-275 Restores Social and Synaptic Function in a Shank3-Deficient Mouse Model of Autism. Neuropsychopharmacology 2018, 43, 1779–1788. [Google Scholar] [CrossRef]
- Kang, H.J.; Kawasawa, Y.I.; Cheng, F.; Zhu, Y.; Xu, X.; Li, M.; Sousa, A.M.; Pletikos, M.; Meyer, K.A.; Sedmak, G.; et al. Spatio-temporal Transcriptome of the Human Brain. Nature 2011, 478, 483–489. [Google Scholar] [CrossRef]
- Johnson, M.B.; Kawasawa, Y.I.; Mason, C.E.; Krsnik, Z.; Coppola, G.; Bogdanović, D.; Geschwind, D.H.; Mane, S.M.; State, M.W.; Sestan, N. Functional and Evolutionary Insights into Human Brain Development Through Global Transcriptome Analysis. Neuron 2009, 62, 494–509. [Google Scholar] [CrossRef]
- Herz, H.M.; Garruss, A.; Shilatifard, A. Set for Life: Biochemical Activities and Biological Functions of Set Domain-Containing Proteins. Trends Biochem. Sci. 2013, 38, 621–639. [Google Scholar] [CrossRef]
- Cottam, N.C.; Ofori, K.; Stoll, K.T.; Bryant, M.; Rogge, J.R.; Hekmatyar, K.; Sun, J.; Charvet, C.J. From Circuits to Lifespan: Translating Mouse and Human Timelines with Neuroimaging-Based Tractography. J. Neurosci. Off. J. Soc. Neurosci. 2025, 45, e1429242025. [Google Scholar] [CrossRef]
- Komada, M.; Takao, K.; Miyakawa, T. Elevated Plus Maze for Mice. J. Vis. Exp. JoVE 2008, 22, 1088. [Google Scholar]
- Qin, L.; Williams, J.B.; Tan, T.; Liu, T.; Cao, Q.; Ma, K.; Yan, Z. Deficiency of autism risk factor ASH1L in prefrontal cortex induces epigenetic aberrations and seizures. Nat. Commun. 2021, 12, 6589. [Google Scholar] [CrossRef]
- Kim, H.; Lim, C.-S.; Kaang, B.-K. Neuronal mechanisms and circuits underlying repetitive behaviors in mouse models of autism spectrum disorder. Behav. Brain Funct. 2016, 12, 3. [Google Scholar] [CrossRef]
- Pitts, M.W. Barnes Maze Procedure for Spatial Learning and Memory in Mice. Bio. Protoc. 2018, 8, e2744. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Schwartz, C.E. Intellectual Disability and Autism Spectrum Disorders: Causal Genes and Molecular Mechanisms. Neurosci. Biobehav. Rev. 2014, 46, 161–174. [Google Scholar] [CrossRef]
- Charvet, C.J. Closing the Gap from Transcription to the Structural Connectome Enhances the Study of Connections in the Human Brain. Dev. Dyn. 2020, 249, 1047–1061. [Google Scholar] [CrossRef]
- Ashokkumar, D.; Zhang, Q.; Much, C.; Bledau, A.S.; Naumann, R.; Alexopoulou, D.; Dahl, A.; Goveas, N.; Fu, J.; Anastassiadis, K.; et al. MLL4 is Required After Implantation, Whereas Mll3 Becomes Essential During Late Gestation. Development 2020, 147, dev186999. [Google Scholar] [CrossRef]
- Xie, G.; Lee, J.E.; Senft, A.D.; Park, Y.K.; Jang, Y.; Chakraborty, S.; Thompson, J.J.; McKernan, K.; Liu, C.; Macfarlan, T.S.; et al. Mll3/Mll4 Methyltransferase Activities Control Early Embryonic Development and embryonic Stem Cell Differentiation in a Lineage-Selective Manner. Nat. Genet. 2023, 55, 693–705. [Google Scholar] [CrossRef]
- Vallianatos, C.N.; Iwase, S. Disrupted Intricacy of Histone H3K4 Methylation in neurodevelopmental Disorders. Epigenomics 2015, 7, 503–519. [Google Scholar] [CrossRef]
- Wang, H.; Helin, K. Roles of H3k4 Methylation in Biology and Disease. Trends Cell Biol. 2025, 35, 115–128. [Google Scholar] [CrossRef]
- Cheung, I.; Shulha, H.P.; Jiang, Y.; Matevossian, A.; Wang, J.; Weng, Z.; Akbarian, S. Developmental Regulation and Individual Differences of Neuronal H3K4me3 Epigenomes in the Prefrontal Cortex. Proc. Natl. Acad. Sci. USA 2010, 107, 8824–8829. [Google Scholar]
- Dincer, A.; Gavin, D.P.; Xu, K.; Zhang, B.; Dudley, J.T.; Schadt, E.E.; Akbarian, S. Deciphering H3K4me3 Broad Domains Associated with Gene-Regulatory Networks and Conserved Epigenomic Landscapes in the Human Brain. Transl. Psychiatry 2015, 5, e679. [Google Scholar] [CrossRef]
- Werling, D.M.; Geschwind, D.H. Sex Differences in Autism Spectrum Disorders. Curr. Opin. Neurol. 2013, 26, 146–153. [Google Scholar] [CrossRef]
- Loomes, R.; Hull, L.; Mandy, W.P.L. What Is the Male-to-Female Ratio in Autism Spectrum Disorder? A Systematic Review and Meta-Analysis. J. Am. Acad. Child Adolesc. Psychiatry 2017, 56, 466–474. [Google Scholar] [CrossRef]
- Antunes, M.; Biala, G. The Novel Object Recognition Memory: Neurobiology, Test Procedure, and Its Modifications. Cogn. Process 2012, 13, 93–110. [Google Scholar] [CrossRef]
- Arnsten, A.F.; Rubia, K. Neurobiological Circuits Regulating Attention, Cognitive Control, Motivation, and Emotion: Disruptions in Neurodevelopmental Psychiatric Disorders. J. Am. Acad. Child Adolesc. Psychiatry 2012, 51, 356–367. [Google Scholar] [CrossRef]
- Fortier, A.V.; Meisner, O.C.; Nair, A.R.; Chang, S.W.C. Prefrontal Circuits Guiding Social Preference: Implications in Autism Spectrum Disorder. Neurosci. Biobehav. Rev. 2022, 141, 104803. [Google Scholar] [CrossRef]
- Wang, X.; Bey, A.L.; Katz, B.M.; Badea, A.; Kim, N.; David, L.K.; Duffney, L.J.; Kumar, S.; Mague, S.D.; Hulbert, S.W.; et al. Altered mGluR5-Homer Scaffolds and Corticostriatal Connectivity in a Shank3 Complete Knockout Model of Autism. Nat. Commun. 2016, 7, 11459. [Google Scholar] [CrossRef]
- Duffney, L.J.; Zhong, P.; Wei, J.; Matas, E.; Cheng, J.; Qin, L.; Ma, K.; Dietz, D.M.; Kajiwara, Y.; Buxbaum, J.D.; et al. Autism-like Deficits in Shank3-Deficient Mice Are Rescued by Targeting Actin Regulators. Cell Rep. 2015, 11, 1400–1413. [Google Scholar] [CrossRef]
- Lee, E.; Lee, J.; Kim, E. Excitation/Inhibition Imbalance in Animal Models of Autism Spectrum Disorders. Biol. Psychiatry 2016, 81, 838–847. [Google Scholar] [CrossRef]
- Gao, R.; Penzes, P. Common Mechanisms of Excitatory and Inhibitory Imbalance in Schizophrenia and Autism Spectrum Disorders. Curr. Mol. Med. 2015, 15, 146–167. [Google Scholar] [CrossRef]
- Yizhar, O.; Fenno, L.E.; Prigge, M.; Schneider, F.; Davidson, T.J.; O’Shea, D.J.; Sohal, V.S.; Goshen, I.; Finkelstein, J.; Paz, J.T.; et al. Neocortical Excitation/Inhibition Balance in Information Processing and Social Dysfunction. Nature 2011, 477, 171–178. [Google Scholar] [CrossRef]
- Spratt, P.W.E.; Ben-Shalom, R.; Keeshen, C.M.; Burke, K.J., Jr.; Clarkson, R.L.; Sanders, S.J.; Bender, K.J. The Autism-Associated Gene Scn2a Contributes to Dendritic Excitability and Synaptic Function in the Prefrontal Cortex. Neuron 2019, 103, 673–685.e675. [Google Scholar] [CrossRef] [PubMed]
- Frega, M.; Selten, M.; Mossink, B.; Keller, J.M.; Linda, K.; Moerschen, R.; Qu, J.; Koerner, P.; Jansen, S.; Oudakker, A.; et al. Distinct Pathogenic Genes Causing Intellectual Disability and Autism Exhibit a Common Neuronal Network Hyperactivity Phenotype. Cell Rep. 2020, 30, 173–186.e176. [Google Scholar] [CrossRef]
- Murugan, M.; Jang, H.J.; Park, M.; Miller, E.M.; Cox, J.; Taliaferro, J.P.; Parker, N.F.; Bhave, V.; Hur, H.; Liang, Y.; et al. Combined Social and Spatial Coding in a Descending Projection from the Prefrontal Cortex. Cell 2017, 171, 1663–1677.e1616. [Google Scholar] [CrossRef]
- Ferguson, B.R.; Gao, W.J. PV Interneurons: Critical Regulators of E/I Balance for Prefrontal Cortex-Dependent Behavior and Psychiatric Disorders. Front. Neural Circuits 2018, 12, 37. [Google Scholar] [CrossRef]
- Zhong, P.; Cao, Q.; Yan, Z. Selective Impairment of Circuits Between Prefrontal Cortex Glutamatergic Neurons and Basal Forebrain Cholinergic Neurons in a Tauopathy Mouse Model. Cereb. Cortex 2022, 32, 5569–5579. [Google Scholar] [CrossRef]
- Zhong, P.; Qin, L.; Yan, Z. Dopamine Differentially Regulates Response Dynamics of Prefrontal Cortical Principal Neurons and Interneurons to Optogenetic Stimulation of Inputs from Ventral Tegmental Area. Cereb. Cortex 2020, 30, 4402–4409. [Google Scholar] [CrossRef]
- Ferguson, B.R.; Gao, W.J. Thalamic Control of Cognition and Social Behavior Via Regulation of Gamma-Aminobutyric Acidergic Signaling and Excitation/Inhibition Balance in the Medial Prefrontal Cortex. Biol. Psychiatry 2018, 83, 657–669. [Google Scholar] [CrossRef]
- Gunaydin, L.A.; Grosenick, L.; Finkelstein, J.C.; Kauvar, I.V.; Fenno, L.E.; Adhikari, A.; Lammel, S.; Mirzabekov, J.J.; Airan, R.D.; Zalocusky, K.A.; et al. Natural Neural Projection Dynamics Underlying Social Behavior. Cell 2014, 157, 1535–1551. [Google Scholar] [CrossRef]
- Langen, M.; Bos, D.; Noordermeer, S.D.; Nederveen, H.; van Engeland, H.; Durston, S. Changes in the Development of Striatum are Involved in repetitive Behavior in Autism. Biol. Psychiatry 2014, 76, 405–411. [Google Scholar] [CrossRef]
- Peça, J.; Feliciano, C.; Ting, J.T.; Wang, W.; Wells, M.F.; Venkatraman, T.N.; Lascola, C.D.; Fu, Z.; Feng, G. Shank3 Mutant Mice Display Autistic-Like Behaviours and Striatal Dysfunction. Nature 2011, 472, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, C.; Chen, Q.; van der Goes, M.S.; Hawrot, J.; Yao, A.Y.; Gao, X.; Lu, C.; Zang, Y.; Zhang, Q.; et al. Striatopallidal Dysfunction Underlies Repetitive Behavior in Shank3-Deficient Model of Autism. J. Clin. Investig. 2017, 127, 1978–1990. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, K.; Webb, M.; Hayder, H.; Qin, L. Kmt2c/Mll3 Haploinsufficiency Causes Autism-like Behavioral Deficits in Mice. Biomolecules 2025, 15, 1547. https://doi.org/10.3390/biom15111547
Ma K, Webb M, Hayder H, Qin L. Kmt2c/Mll3 Haploinsufficiency Causes Autism-like Behavioral Deficits in Mice. Biomolecules. 2025; 15(11):1547. https://doi.org/10.3390/biom15111547
Chicago/Turabian StyleMa, Kaijie, Maria Webb, Haniya Hayder, and Luye Qin. 2025. "Kmt2c/Mll3 Haploinsufficiency Causes Autism-like Behavioral Deficits in Mice" Biomolecules 15, no. 11: 1547. https://doi.org/10.3390/biom15111547
APA StyleMa, K., Webb, M., Hayder, H., & Qin, L. (2025). Kmt2c/Mll3 Haploinsufficiency Causes Autism-like Behavioral Deficits in Mice. Biomolecules, 15(11), 1547. https://doi.org/10.3390/biom15111547






