Effect of Marination Time on the Antioxidant Properties of Peptides Extracted from Organic Dry-Fermented Beef
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Extraction and Identification of Peptides
2.3. Electrophoretic Analysis
2.4. Chromatographic Analysis
2.5. Determination of Antioxidant Activity
2.6. Determination of Color Stability
2.7. Statistical Analysis
3. Results
3.1. Peptide Profile
3.2. Peptide Composition
3.3. Determination of Antioxidant Activity
4. Discussion
4.1. Peptide Profile and Composition
4.2. Determination of Antioxidant Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lytou, A.E.; Panagou, E.Z.; Nychas, G.J.E. Effect of different marinating conditions on the evolution of spoilage microbiota and metabolomic profile of chicken breast fillets. Food Microbiol. 2017, 66, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Yusop, S.M.; O’Sullivan, M.G.; Kerry, J.F.; Kerry, J.P. Effect of marinating time and low pH on marinade performance and sensory acceptability of poultry meat. Meat Sci. 2010, 85, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Berge, P.; Ertbjerg, P.; Larsen, L.M.; Astruc, T.; Vignon, X.; Møller, A.J. Tenderization of beef by lactic acid injected at different times post mortem. Meat Sci. 2001, 57, 347–357. [Google Scholar] [CrossRef]
- Wronkowska, M.; Jadacka, M.; Soral-Śmietana, M.; Zander, L.; Dajnowiec, F.; Banaszczyk, P.; Jeliński, T.; Szmatowicz, B. Acid whey concentrated by ultrafiltration a tool for modeling bread properties. LWT-Food Sci. Technol. 2015, 61, 172–176. [Google Scholar] [CrossRef]
- Rzepkowska, A.; Zielińska, D.; Ołdak, A.; Kołożyn-Krajewska, D. Organic whey as a source of Lactobacillus strains with selected technological and antimicrobial properties. Int. J. Food Sci. Technol. 2017, 52, 1983–1994. [Google Scholar] [CrossRef]
- Wójciak, K.M.; Karwowska, M.; Dolatowski, Z.J. Use of acid whey and mustard seed to replace nitrites during cooked sausage production. Meat Sci. 2014, 96, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Wójciak, K.M.; Dolatowski, Z.J. Effect of acid whey on nitrosylmyoglobin concentration in uncured fermented sausage. LWT-Food Sci. Technol. 2015, 64, 713–719. [Google Scholar] [CrossRef]
- Wójciak, K.M.; Kęska, P.; Okoń, A.; Solska, E.; Libera, J.; Dolatowski, Z.J. The influence of acid whey on the antioxidant peptides generated to reduce oxidation and improve colour stability in uncured roast beef. J. Sci. Food Agric. 2018, 98, 3728–3734. [Google Scholar] [CrossRef]
- Kęska, P.; Wójciak, K.M.; Stadnik, J. Bioactive peptides from beef products fermented by acid whey–in vitro and in silico study. Sci. Agric. 2019, 76, 311–320. [Google Scholar] [CrossRef]
- Mora, L.; Gallego, M.; Escudero, E.; Reig, M.; Aristoy, M.C.; Toldrá, F. Small peptides hydrolysis in dry-cured meats. Int. J. Food Microbiol. 2015, 212, 9–15. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Determination of the degree of hydrolysis of food protein hydrolysates by trinitrobenzenesulfonicacid. J. Agric. Food Chem. 1979, 27, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, Y.; Zou, X.; He, J.; Xu, X.; Zhou, G.; Li, C. In vitro protein digestibility of pork products is affected by the method of processing. Food Res. Int. 2017, 92, 88–94. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhou, G.; Bai, Y.; Wang, C.; Zhu, S.; Xu, X.; Li, C. The effect of meat processing methods on changes in disulfide bonding and alteration of protein structures: Impact on protein digestion products. RSC Adv. 2018, 8, 17595–17605. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Rad Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Oyaizu, M. Studies on products of browning reaction. Jpn. J. Nutr. Dietetics 1986, 44, 307–315. [Google Scholar] [CrossRef]
- Pikul, J.; Leszczynski, D.E.; Kummerow, F.A. Evaluation of three modified TBA methods for measuring lipid oxidation in chicken meat. J. Agr. Food Chem. 1989, 37, 1309–1313. [Google Scholar] [CrossRef]
- Nam, K.C.; Ahn, D.U. Effects of ascorbic acid and antioxidants on the color of irradiated ground beef. J. Food Sci. 2003, 68, 1686–1690. [Google Scholar] [CrossRef]
- Stadnik, J.; Stasiak, D.M. Effect of acid whey on physicochemical characteristics of dry-cured organic pork loins without nitrite. Int. J. Food Sci. Technol. 2016, 51, 970–977. [Google Scholar] [CrossRef]
- Castellano, P.; Aristoy, M.C.; Sentandreu, M.Á.; Vignolo, G.; Toldrá, F. Peptides with angiotensin I converting enzyme (ACE) inhibitory activity generated from porcine skeletal muscle proteins by the action of meat-borne Lactobacillus. J. Proteom. 2013, 89, 183–190. [Google Scholar] [CrossRef]
- López, C.M.; Sentandreu, M.A.; Vignolo, G.M.; Fadda, S.G. Low molecular weight peptides derived from sarcoplasmic proteins produced by an autochthonous starter culture in a beaker sausage model. EuPA Open Proteom. 2015, 7, 54–63. [Google Scholar] [CrossRef]
- Montowska, M.; Rao, W.; Alexander, M.R.; Tucker, G.A.; Barrett, D.A. Tryptic digestion coupled with ambient desorption electrospray ionization and liquid extraction surface analysis mass spectrometry enabling identification of skeletal muscle proteins in mixtures and distinguishing between beef, pork, horse, chicken, and turkey meat. Anal. Chem. 2014, 86, 4479–4487. [Google Scholar] [CrossRef]
- Montowska, M.; Fornal, E. Detection of peptide markers of soy, milk and egg white allergenic proteins in poultry products by LC-Q-TOF-MS/MS. LWT-Food Sci. Technol. 2018, 87, 310–317. [Google Scholar] [CrossRef]
- Di Luccia, A.; Picariello, G.; Cacace, G.; Scaloni, A.; Faccia, M.; Liuzzi, V.; Alviti, G.; Musso, S.S. Proteomic analysis of water soluble and myofibrillar protein changes occurring in dry-cured hams. Meat Sci. 2005, 69, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Hughes, M.C.; Kerry, J.P.; Arendt, E.K.; Kenneally, P.M.; McSweeney, P.L.H.; O’neill, E.E. Characterization of proteolysis during the ripening of semi-dry fermented sausages. Meat Sci. 2002, 62, 205–216. [Google Scholar] [CrossRef]
- Picariello, G.; De Martino, A.; Mamone, G.; Ferranti, P.; Addeo, F.; Faccia, M.; SpagnaMusso, M.; Di Luccia, A. Proteomic study of muscle sarcoplasmic proteins using AUT-PAGE/SDS-PAGE as two-dimensional gel electrophoresis. J. Chromatogr B 2006, 833, 101–108. [Google Scholar] [CrossRef]
- Candogan, K.; Wardlaw, F.B.; Acton, J.C. Effect of starter culture on proteolytic changes during processing of fermented beef sausages. Food Chem. 2009, 116, 731–737. [Google Scholar] [CrossRef]
- Stoeva, S.; Byrne, C.E.; Mullen, A.M.; Troy, D.J.; Voelter, W. Isolation and identification of proteolytic fragments from TCA soluble extracts of bovine M. longissimus dorsi. Food Chem. 2000, 69, 365–370. [Google Scholar] [CrossRef]
- Mora, L.; Sentandreu, M.A.; Fraser, P.D.; Toldrá, F.; Bramley, P.M. Oligopeptides arising from the degradation of creatine kinase in Spanish dry-cured ham. J. Agric. Food Chem. 2009, 57, 8982–8988. [Google Scholar] [CrossRef]
- Mora, L.; Sentandreu, M.A.; Toldrá, F. Identification of small troponin T peptides generated in dry-cured ham. Food Chem. 2010, 123, 691–697. [Google Scholar] [CrossRef]
- Ferranti, P.; Nitride, C.; Nicolai, M.A.; Mamone, G.; Picariello, G.; Bordoni, A.; Valli, V.; Di Nunzio, M.; Babini, E.; Marcolini, E.; et al. In vitro digestion of Bresaola proteins and release of potential bioactive peptides. Food Res. Int. 2014, 63, 157–169. [Google Scholar] [CrossRef]
- Morzel, M.; Chambon, C.; Hamelin, M.; Santé-Lhoutellier, V.; Sayd, T.; Monin, G. Proteome changes during pork meat ageing following use of two different pre-slaughter handling procedures. Meat Sci. 2004, 67, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J.; Kim, Y.-S.; Hwang, J.-W.; Kim, E.-K.; Moon, S.-H.; Jeon, B.-T.; Park, P.-J. Purification and characterization of a novel antioxidative peptide from duck skin by-products that protects liver against oxidative damage. Food Res. Int. 2012, 49, 285–295. [Google Scholar] [CrossRef]
- Estévez, M. Protein carbonyls in meat systems: A review. Meat Sci. 2011, 89, 259–279. [Google Scholar] [CrossRef] [PubMed]
- Gallego, M.; Mora, L.; Aristoy, M.C.; Toldrá, F. The use of label-free mass spectrometry for relative quantification of sarcoplasmic proteins during the processing of dry-cured ham. Food Chem. 2016, 196, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Mora, L.; Escudero, E.; Aristoy, M.C.; Toldrá, F. A peptidomic approach to study the contribution of added casein proteins to the peptide profile in Spanish dry-fermented sausages. Int. J. Food Microbiol. 2015, 212, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, V.; Estévez, M.; Ventanas, J.; Ventanas, S. Impact of lipid content and composition on lipid oxidation and protein carbonylation in experimental fermented sausages. Food Chem. 2014, 147, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Brewer, S. Irradiation effects on meat color–a review. Meat Sci. 2004, 68, 1–17. [Google Scholar] [CrossRef]
- Cooper, J.V.; Suman, S.P.; Wiegand, B.R.; Schumacher, L.; Lorenzen, C.L. Impact of Light Source on Color and Lipid Oxidative Stabilities from a Moderately Color-Stable Beef Muscle during Retail Display. Meat Muscle Biol. 2018, 2, 102–110. [Google Scholar] [CrossRef]
- Wu, W.; Gao, X.G.; Dai, Y.; Fu, Y.; Li, X.M.; Dai, R.T. Post-mortem changes in sarcoplasmic proteome and its relationship to meat color traits in M. semitendinosus of Chinese Luxi yellow cattle. Food Res. Int. 2015, 72, 98–105. [Google Scholar] [CrossRef]
- Cava, R.; Ladero, L.; González, S.; Carrasco, A.; Ramírez, M.R. Effect of pressure and holding time on colour, protein and lipid oxidation of sliced dry-cured Iberian ham and loin during refrigerated storage. Innov. Food Sci. Emerg. Technol. 2009, 10, 76–81. [Google Scholar] [CrossRef]
- Yu, Q.; Wu, W.; Tian, X.; Hou, M.; Dai, R.; Li, X. Unraveling proteome changes of Holstein beef M. semitendinosus and its relationship to meat discoloration during post-mortem storage analyzed by label-free mass spectrometry. J. Proteom. 2017, 154, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Sayd, T.; Morzel, M.; Chambon, C.; Franck, M.; Figwer, P.; Larzul, C.; Le Roy, P.; Monin, G.; Chérel, P.; Laville, E. Proteome analysis of the sarcoplasmic fraction of pig semimembranosus muscle: Implications on meat color development. J. Agric. Food Chem. 2006, 54, 2732–2737. [Google Scholar] [CrossRef] [PubMed]
- Canto, A.C.; Suman, S.P.; Nair, M.N.; Li, S.; Rentfrow, G.; Beach, C.M.; Silva, T.J.; Wheeler, T.L.; Shackelford, S.D.; Grayson, A.; et al. Differential abundance of sarcoplasmic proteome explains animal effect on beef Longissimus lumborum color stability. Meat Sci. 2015, 102, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Nair, M.N.; Suman, S.P.; Chatli, M.K.; Li, S.; Joseph, P.; Beach, C.M.; Rentfrow, G. Proteome basis for intramuscular variation in color stability of beef semimembranosus. Meat Sci. 2016, 113, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Joseph, P.; Suman, S.P.; Rentfrow, G.; Li, S.; Beach, C.M. Proteomics of muscle-specific beef color stability. J. Agric. Food Chem. 2012, 60, 3196–3203. [Google Scholar] [CrossRef]
- Liu, F.; Xu, Q.; Dai, R.; Ni, Y. Effects of natural antioxidants on colour stability, lipid oxidation and metmyoglobin reducing activity in raw beef patties. Acta. Sci. Pol. Technol. Aliment. 2015, 14, 37–44. [Google Scholar] [CrossRef]
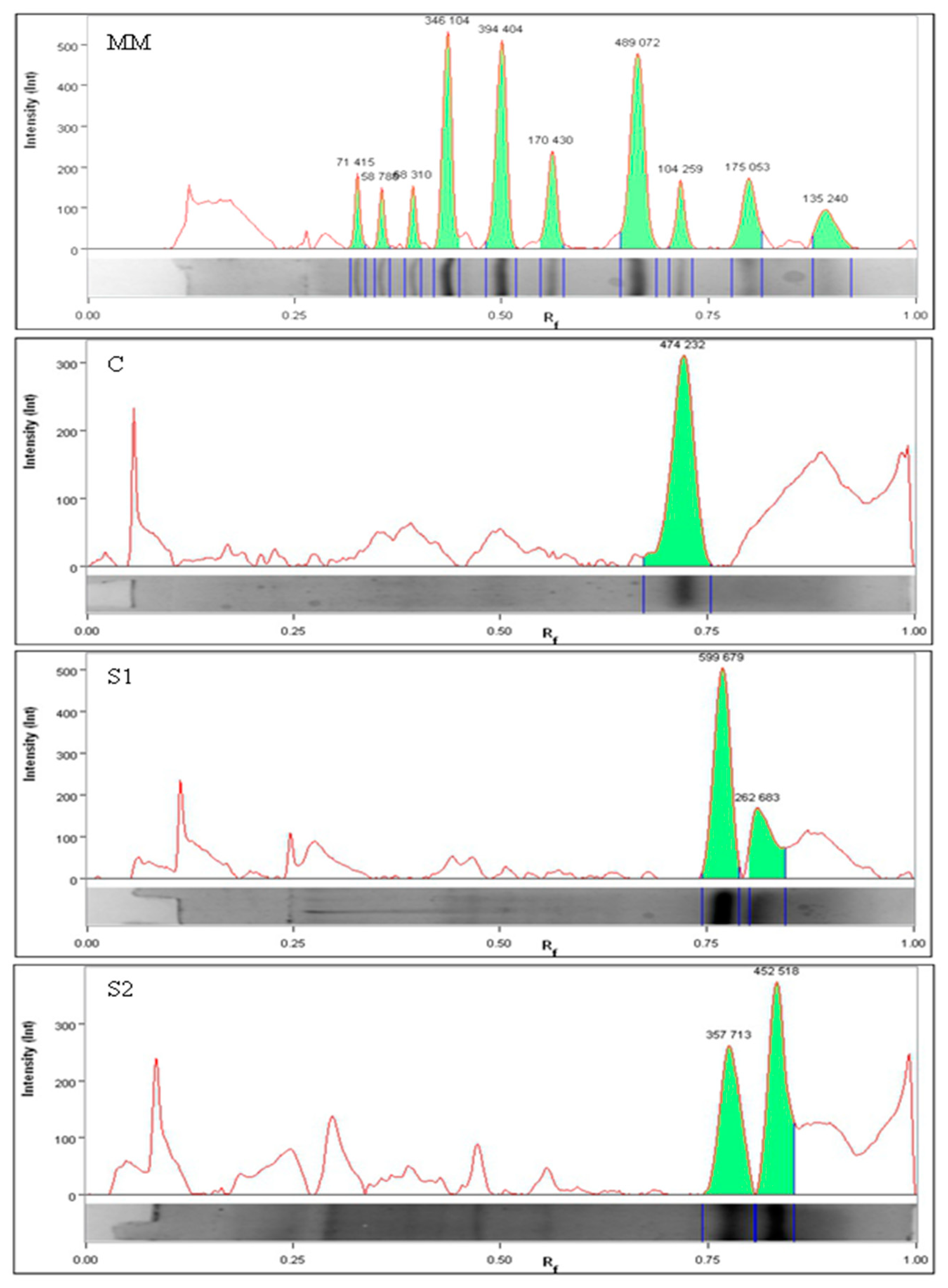
| Batch | Time [days] | Peptide Content [mg/mL] | Antioxidant Activity | Color Parameters | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ABTS IC50 [µg/mL] | RP [A700] | TBARS [mg MDA/kg] | ORP [mV] | L* | a* | b* | ΔR | |||
| C | 31 | 1.18 ± 0.05 Aa | 7.46 ± 0.71 Aa | 0.97 ± 0.07 Aa | 2.35 ± 0.62 Aa | 252.03 ± 15.95 Aa | 36.03 ± 2.40 Aa | 7.30 ±1.05 Aa | 5.27 ±1.58 Aa | 2.14 ± 0.26 Aa |
| 62 | 4.73 ± 0.51 Ba | 23.05 ± 1.91 Ba | 1.78 ± 0.28 Ba | 2.05 ± 0.53 Aa | 347.77 ± 19.51 Ba | 33.34 ± 2.25 Ba | 7.33 ± 2.04 Aa | 5.76 ± 1.65 Aab | 2.11 ± 0.33 Aa | |
| S1 | 31 | 1.21 ± 0.17 Aab | 6.62 ± 0.21 Aa | 1.19 ± 0.09 Ab | 2.03 ± 0.70 Aa | 250.79 ± 8.24 Aa | 44.00 ±3.46 Ab | 9.11 ±1.42 Ab | 6.81 ±1.26 Aa | 2.21± 0.16 Aa |
| 62 | 3.95 ± 0.22 Bb | 19.40 ± 1.72 Bb | 1.25 ± 0.11 Ab | 1.64 ± 0.68 Aa | 302.93 ± 20.57 Bb | 39.57 ±3.49 Bb | 9.40 ± 1.41 Ab | 7.66 ± 2.42 Ab | 2.23 ± 0.25 Aa | |
| S2 | 31 | 1.29 ± 0.05 Ab | 13.88 ± 1.41 Ab | 1.04 ± 0.13 Aa | 2.17 ± 0.94 Aa | 258.52 ± 15.72 Aa | 43.78 ±2.81 Ab | 9.01 ±2.00 Ab | 6.75 ±1.98 Aa | 2.15 ± 0.31 Aa |
| 62 | 4.10 ± 0.48 Bb | 19.77 ± 1.34 Bb | 1.41 ± 0.12 Bc | 1.87 ± 0.91 Aa | 301.82 ± 12.47 Bb | 36.08 ± 4.67 Ba | 7.22 ± 1.87 Aa | 4.22 ± 1.75 Ba | 2.00 ± 0.37 Aa | |
| Batch | Band No | Molecular Weight [kDa] | Band Intensities | Bands [%] |
|---|---|---|---|---|
| C | 1 | 14.73 ± 0.08 a | 2.59 ± 0.82 a | 100 ± 0.00 |
| 2 | nd * | nd | nd | |
| S1 | 1 | 14.68 ± 0.37 a | 3.03 ± 0.96 a | 33.25 ± 6.96 |
| 2 | 13.78 ± 0.16 b | 1.79 ± 0.79 a | 66.75 ± 6.03 | |
| S2 | 1 | 11.76 ± 0.49 c | 1.44 ± 0.42 a | 37.30 ± 6.35 |
| 2 | 10.07 ± 0.09 d | 2.91 ± 0.42 a | 62.70 ± 6.35 |
| (A) | C | Protein sequence coverage: 7%. | ||||
| 10 | 20 | 30 | 40 | 50 | ||
| MAPSRKFFVG | GNWKMNGRKN | NLGELINTLN | AAKVPADTEV | VCAPPTAYID | ||
| 60 | 70 | 80 | 90 | 100 | ||
| FARQKLDPKI | AVAAQNCYKV | ANGAFTGEIS | PGMIKDLGAT | WVVLGHSERR | ||
| 110 | 120 | 130 | 140 | 150 | ||
| HVFGESDELI | GQKVAHALAE | GLGVIACIGE | KLDEREAGIT | EKVVFEQTKV | ||
| 160 | 170 | 180 | 190 | 200 | ||
| IADNVKDWSK | VVLAYEPVWA | IGTGKTATPQ | QAQEVHEKLR | GWLKSNVSDA | ||
| 210 | 220 | 230 | 240 | |||
| VAQSARIIYG | GSVTGATCKE | LASQPDVDGF LVGGASLKPE FVDIINAKQ | ||||
| S1 | Protein sequence coverage: 8% | |||||
| 10 | 20 | 30 | 40 | 50 | ||
| MAPSRKFFVG | GNWKMNGRKN | NLGELINTLN | AAKVPADTEV | VCAPPTAYID | ||
| 60 | 70 | 80 | 90 | 100 | ||
| FARQKLDPKI | AVAAQNCYKV | ANGAFTGEIS | PGMIKDLGAT | WVVLGHSERR | ||
| 110 | 120 | 130 | 140 | 150 | ||
| HVFGESDELI | GQKVAHALAE | GLGVIACIGE | KLDEREAGIT | EKVVFEQTKV | ||
| 160 | 170 | 180 | 190 | 200 | ||
| IADNVKDWSK | VVLAYEPVWA | IGTGKTATPQ | QAQEVHEKLR | GWLKSNVSDA | ||
| 210 | 220 | 230 | 240 | |||
| VAQSARIIYG | GSVTGATCKE | LASQPDVDGF LVGGASLKPE FVDIINAKQ | ||||
| S2 | Protein sequence coverage: 18% | |||||
| 10 | 20 | 30 | 40 | 50 | ||
| MAPSRKFFVG | GNWKMNGRKN | NLGELINTLN | AAKVPADTEV | VCAPPTAYID | ||
| 60 | 70 | 80 | 90 | 100 | ||
| FARQKLDPKI | AVAAQNCYKV ANGAFTGEIS PGMIKDLGAT WVVLGHSERR | |||||
| 110 | 120 | 130 | 140 | 150 | ||
| HVFGESDELI | GQKVAHALAE | GLGVIACIGE | KLDEREAGIT | EKVVFEQTKV | ||
| 160 | 170 | 180 | 190 | 200 | ||
| IADNVKDWSK | VVLAYEPVWA | IGTGKTATPQ | QAQEVHEKLR | GWLKSNVSDA | ||
| 210 | 220 | 230 | 240 | |||
| VAQSARIIYG | GSVTGATCKE LASQPDVDGF LVGGASLKPE FVDIINAKQ | |||||
| (B) | Peptides Sequence | Mass | C | S1 | S2 | |
| SQPDVDGFLVGGASLKPE | 1814.90506 | - | + | + | ||
| VGGASLKPEFVDIINAKQ | 1885.0309 | - | - | + | ||
| GAFTGEISPGMIKDLGATW | 1949.9557 | - | - | + | ||
| SQPDVDGFLVGGASLKPEF | 1961.97348 | + | + | + | ||
| ASQPDVDGFLVGGASLKPEF | 2033.01059 | - | - | + | ||
| ELASQPDVDGFLVGGASLKPEF | 2275.13724 | - | - | + | ||
| KVANGAFTGEISPGMIKDLGATW | 2362.19911 | - | - | + | ||
| ASQPDVDGFLVGGASLKPEFVD | 2247.10596 | - | + | - | ||
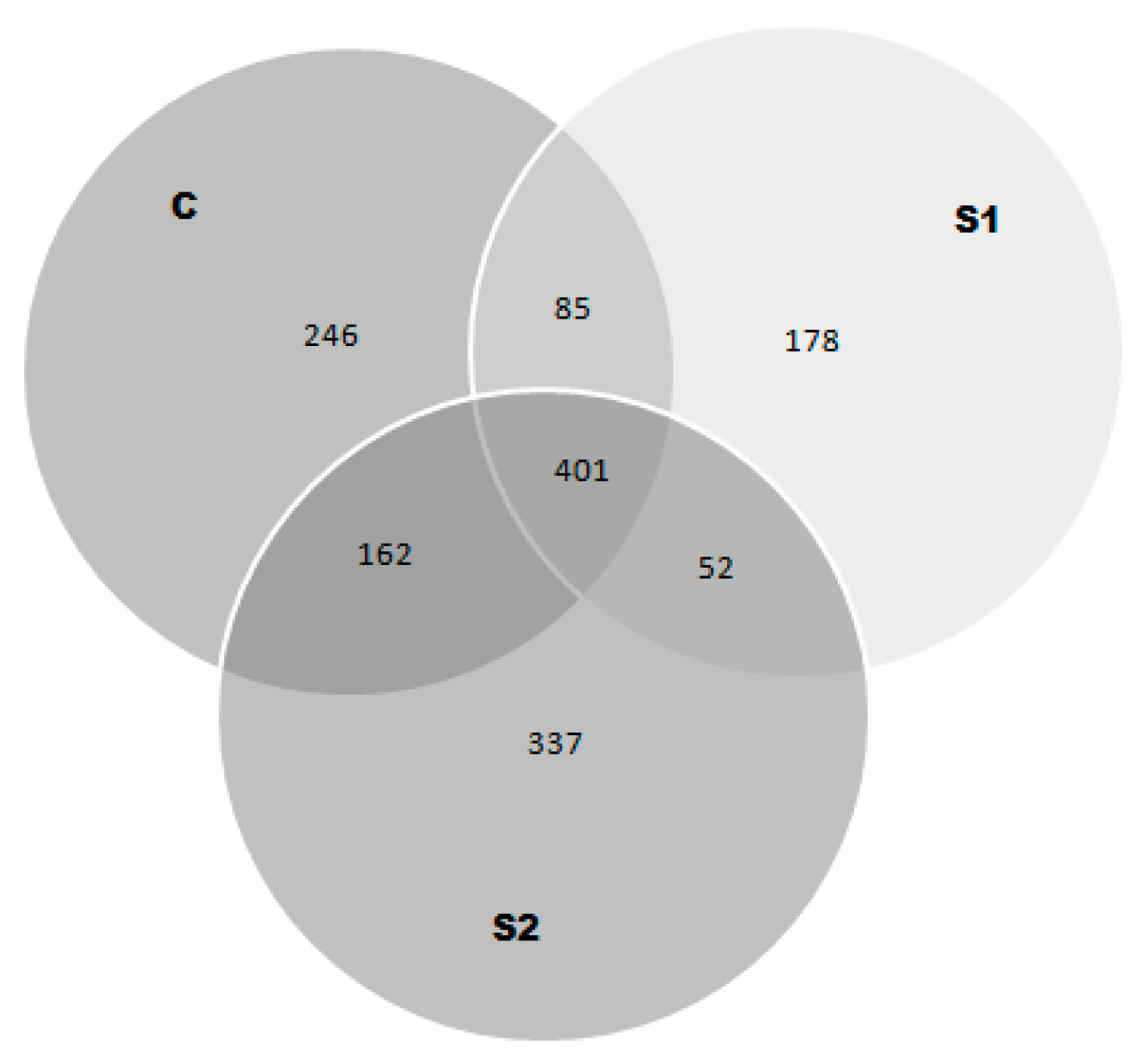
| Protein Name | ID a | Mass [Da] | C | S1 | S2 | |||
|---|---|---|---|---|---|---|---|---|
| % Coverage b | Peptides Identified c | % Coverage b | Peptides identified | % Coverage b | Peptides Identified | |||
| Myoglobin | sp|P02192|MYG_BOVIN | 17.078 | 99.000 | 162 | 99.000 | 206 | 99.000 | 212 |
| Hemoglobin subunit alpha | sp|P01966|HBA_BOVIN | 15.184 | 65.000 | 20 | 46.000 | 10 | 51.000 | 13 |
| Creatine kinase type M | sp|Q9XSC6|KCRM_BOVIN | 42.989 | 59.000 | 113 | 64.000 | 130 | 52.000 | 108 |
| Beta-enolase | sp|Q3ZC09|ENOB_BOVIN | 47.096 | 45.000 | 49 | 45.000 | 65 | 56.000 | 65 |
| Actin, alpha skeletal muscle | sp|P68138|ACTS_BOVIN | 42.051 | 37.000 | 36 | 57.000 | 89 | 58.000 | 77 |
| Glyceraldehyde-3-phosphate dehydrogenase | sp|P10096|G3P_BOVIN | 35.868 | 23.000 | 34 | 36.000 | 39 | 36.000 | 43 |
| Myozenin-1 | sp|Q8SQ24|MYOZ1_BOVIN | 31.674 | 23.000 | 6 | 11.000 | 8 | 11.000 | 9 |
| Troponin T, fast skeletal muscle | sp|Q8MKI3|TNNT3_BOVIN | 32.126 | 22.000 | 18 | 30.000 | 34 | 30.000 | 26 |
| Phosphoglucomutase-1 | sp|Q08DP0|PGM1_BOVIN | 61.589 | 11.000 | 5 | 8.000 | 6 | 8.000 | 6 |
| Myosin-1 | sp|Q9BE40|MYH1_BOVIN | 222.290 | 10.000 | 53 | 11.000 | 66 | 12.000 | 54 |
| Triosephosphate isomerase | sp|Q5E956|TPIS_BOVIN | 26.690 | 7.000 | 1 | 8.000 | 3 | 18.000 | 7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kęska, P.; Wójciak, K.M.; Stadnik, J. Effect of Marination Time on the Antioxidant Properties of Peptides Extracted from Organic Dry-Fermented Beef. Biomolecules 2019, 9, 614. https://doi.org/10.3390/biom9100614
Kęska P, Wójciak KM, Stadnik J. Effect of Marination Time on the Antioxidant Properties of Peptides Extracted from Organic Dry-Fermented Beef. Biomolecules. 2019; 9(10):614. https://doi.org/10.3390/biom9100614
Chicago/Turabian StyleKęska, Paulina, Karolina M. Wójciak, and Joanna Stadnik. 2019. "Effect of Marination Time on the Antioxidant Properties of Peptides Extracted from Organic Dry-Fermented Beef" Biomolecules 9, no. 10: 614. https://doi.org/10.3390/biom9100614
APA StyleKęska, P., Wójciak, K. M., & Stadnik, J. (2019). Effect of Marination Time on the Antioxidant Properties of Peptides Extracted from Organic Dry-Fermented Beef. Biomolecules, 9(10), 614. https://doi.org/10.3390/biom9100614







