The Emerging Role of Long Non-Coding RNAs in the Metastasis of Hepatocellular Carcinoma
Abstract
1. Introduction
2. LncRNAs as Novel Regulators in HCC Metastasis
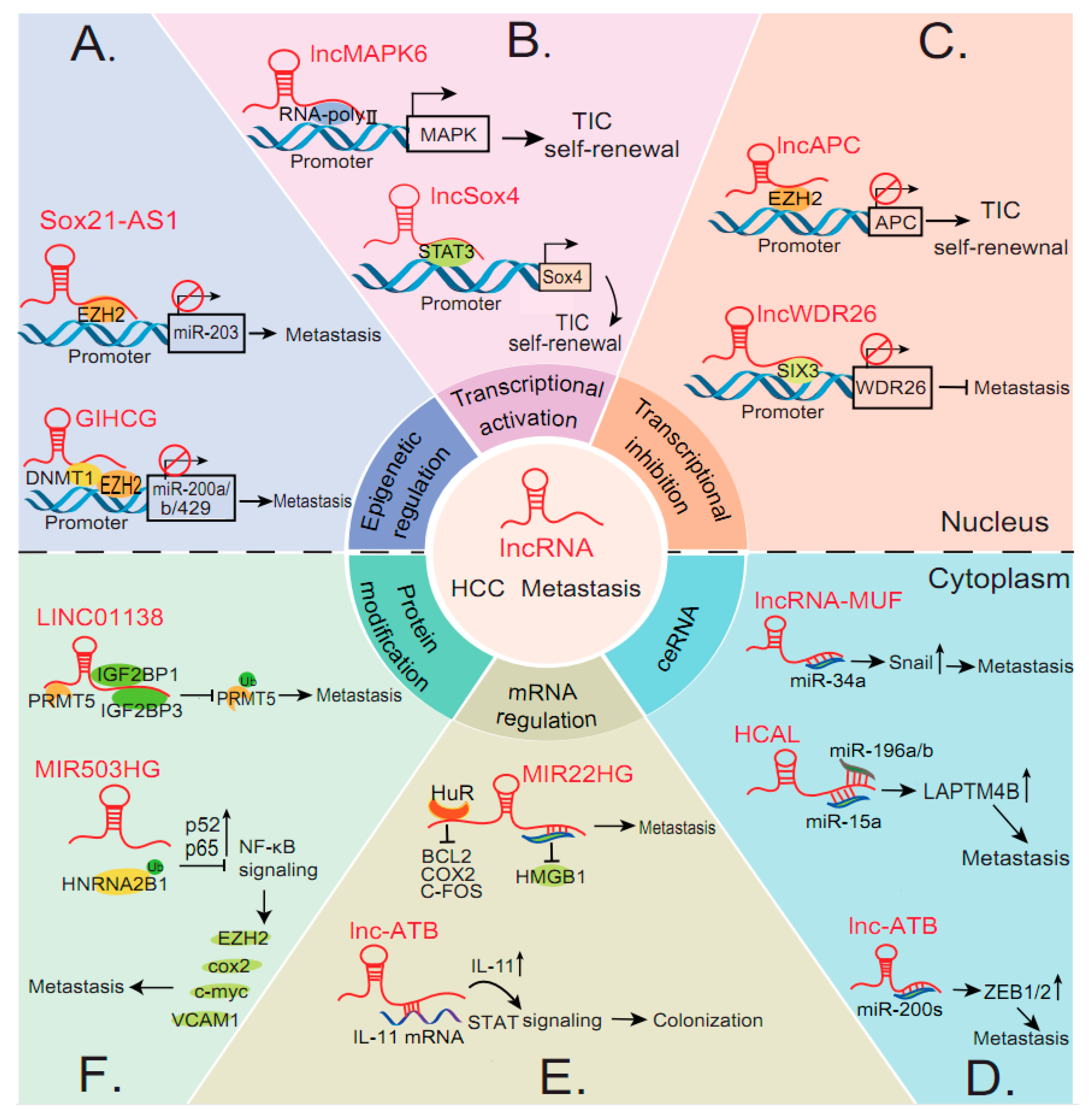
3. The Molecular Mechanisms of LncRNAs in HCC Metastasis
3.1. LncRNAs in HCC Metastasis at the Epigenetic and Transcriptional Level
3.1.1. Chromatin Modification and Regulation
3.1.2. Transcriptional Regulation
3.2. LncRNAs in HCC Metastasis at the Post-Transcriptional Level
3.2.1. Interactions with miRNAs
3.2.2. Interactions with mRNAs
3.2.3. Protein Modifications
4. Emerging Paradigms on HCC Metastasis
4.1. Pathways Controlled by LncRNAs in HCC Metastasis
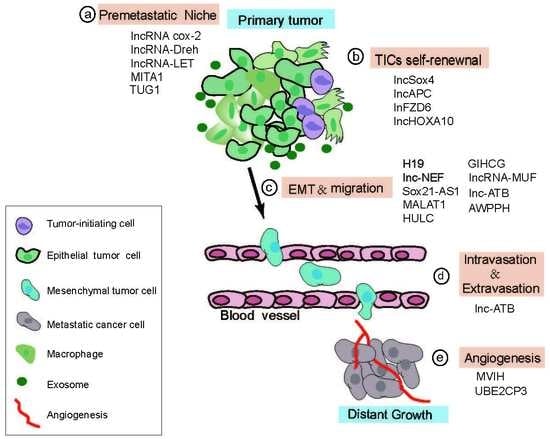
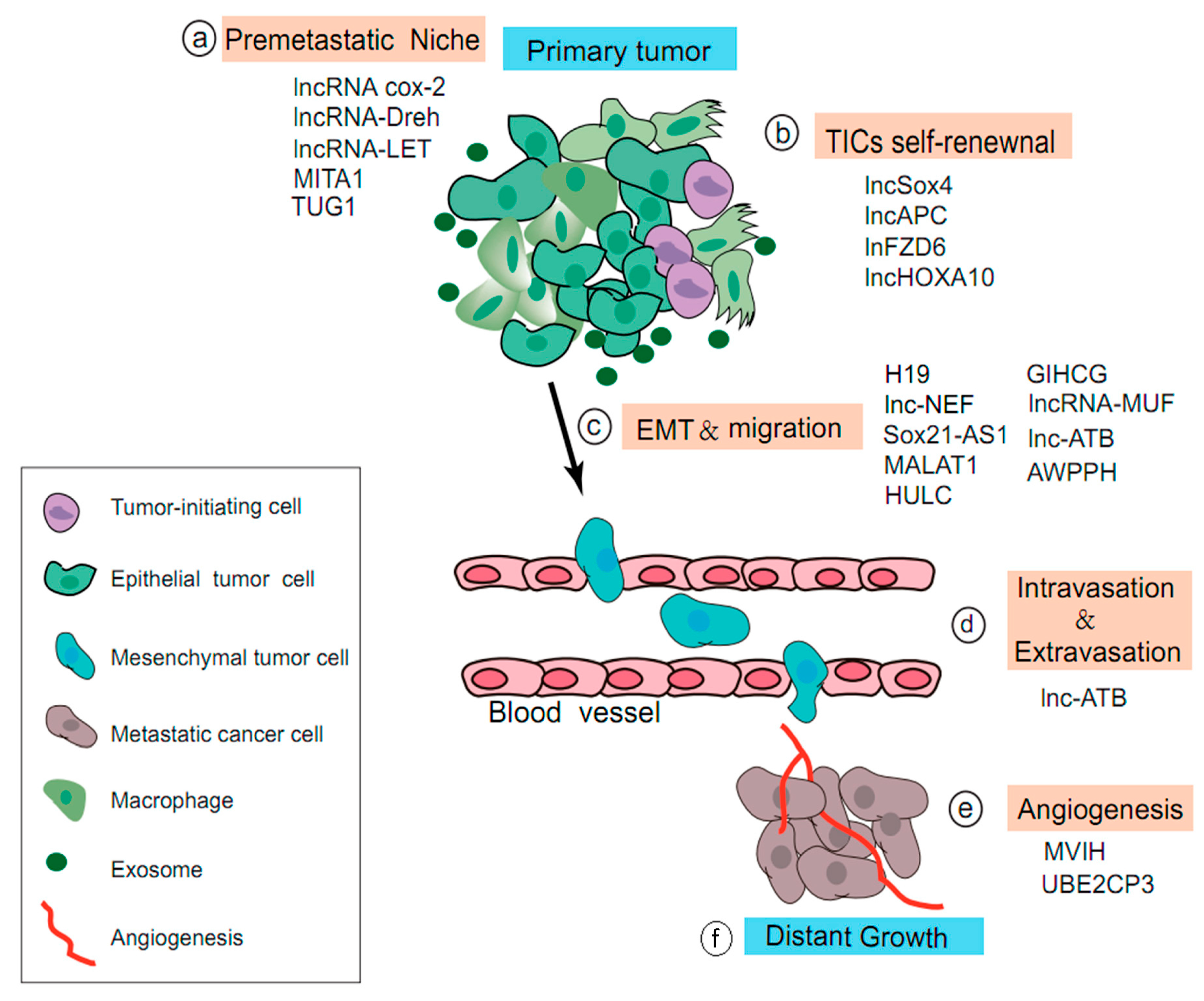
4.2. Role of LncRNAs in the Multi-Step Process of HCC Metastasis
5. Potential Diagnostic and Therapeutic Applications
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Shimokawa, M.; Ohta, Y.; Nishikori, S.; Matano, M.; Takano, A.; Fujii, M.; Date, S.; Sugimoto, S.; Kanai, T.; Sato, T. Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature 2017, 545, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- DiStefano, J.K. Long noncoding RNAs in the initiation, progression, and metastasis of hepatocellular carcinoma. Non-Coding RNA Res. 2017, 2, 129–136. [Google Scholar] [CrossRef]
- Mai, H.; Zhou, B.; Liu, L.; Yang, F.; Conran, C.; Ji, Y.; Hou, J.; Jiang, D. Molecular pattern of lncRNAs in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. CR 2019, 38, 198. [Google Scholar] [CrossRef]
- Huo, X.; Han, S.; Wu, G.; Latchoumanin, O.; Zhou, G.; Hebbard, L.; George, J.; Qiao, L. Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: Implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol. Cancer 2017, 16, 165. [Google Scholar] [CrossRef]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef]
- Liu, C.J.; Gao, C.; Ma, Z.; Cong, R.; Zhang, Q.; Guo, A.Y. lncRInter: A database of experimentally validated long non-coding RNA interaction. J. Genet. Genom. 2017, 44, 265–268. [Google Scholar] [CrossRef]
- Weidle, U.H.; Birzele, F.; Kollmorgen, G.; Ruger, R. Long Non-coding RNAs and their Role in Metastasis. Cancer Genom. Proteom. 2017, 14, 143–160. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Meng, X.M.; Huang, C.; Wu, B.M.; Zhang, L.; Lv, X.W.; Li, J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014, 344, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Lou, W.; Xu, L.; Fan, W. Non-coding RNA in drug resistance of hepatocellular carcinoma. Biosci. Rep. 2018, 38, BSR20180915. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wang, Y.Y.; Xin, H.W.; Wang, L.; Arfuso, F.; Dharmarajan, A.; Kumar, A.P.; Wang, H.; Tang, F.R.; Warrier, S.; et al. The expanding roles of long non-coding RNAs in the regulation of cancer stem cells. Int. J. Biochem. Cell Biol. 2019, 108, 17–20. [Google Scholar] [CrossRef]
- Cheng, J.T.; Wang, L.; Wang, H.; Tang, F.R.; Cai, W.Q.; Sethi, G.; Xin, H.W.; Ma, Z. Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition. Cells 2019, 8, 1178. [Google Scholar] [CrossRef]
- Li, S.P.; Xu, H.X.; Yu, Y.; He, J.D.; Wang, Z.; Xu, Y.J.; Wang, C.Y.; Zhang, H.M.; Zhang, R.X.; Zhang, J.J.; et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget 2016, 7, 42431–42446. [Google Scholar] [CrossRef]
- Yuan, S.X.; Yang, F.; Yang, Y.; Tao, Q.F.; Zhang, J.; Huang, G.; Yang, Y.; Wang, R.Y.; Yang, S.; Huo, X.S.; et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology 2012, 56, 2231–2241. [Google Scholar] [CrossRef]
- Xu, X.; Lou, Y.; Tang, J.; Teng, Y.; Zhang, Z.; Yin, Y.; Zhuo, H.; Tan, Z. The long non-coding RNA Linc-GALH promotes hepatocellular carcinoma metastasis via epigenetically regulating Gankyrin. Cell Death Dis. 2019, 10, 86. [Google Scholar] [CrossRef]
- Sui, C.J.; Zhou, Y.M.; Shen, W.F.; Dai, B.H.; Lu, J.J.; Zhang, M.F.; Yang, J.M. Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429. J. Mol. Med. 2016, 94, 1281–1296. [Google Scholar] [CrossRef]
- Wei, C.; Wang, H.; Xu, F.; Liu, Z.; Jiang, R. LncRNA SOX21-AS1 is associated with progression of hepatocellular carcinoma and predicts prognosis through epigenetically silencing p21. Biomed. Pharmacother. 2018, 104, 137–144. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, Y.; Yao, L.; Liu, Y.; Huang, L.; Yan, Z.; Zhao, W.; Zhu, P.; Weng, H. LncFZD6 initiates Wnt/beta-catenin and liver TIC self-renewal through BRG1-mediated FZD6 transcriptional activation. Oncogene 2018, 37, 3098–3112. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Yang, Q.; Zhu, W.; Jin, H.; Wang, J.; Song, J.; Kong, Y.; Lv, X. LncHOXA10 drives liver TICs self-renewal and tumorigenesis via HOXA10 transcription activation. Mol. Cancer 2018, 17, 173. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Z.; Huang, L.; Wu, Y.H.; Zhai, W.J.; Zhu, P.P.; Gao, Y.F. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat. Commun. 2016, 7, 12598. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Lin, J.; Qin, F.; Yang, Z.; Ding, Y.; Zhang, Y.; Han, L.; Zhu, X.; Zhang, Q. LncAPC drives Wnt/beta-catenin activation and liver TIC self-renewal through EZH2 mediated APC transcriptional inhibition. Mol. Carcinog. 2018, 57, 408–418. [Google Scholar] [CrossRef]
- Chen, B. A novel long noncoding RNA lncWDR26 suppresses the growth and metastasis of hepatocellular carcinoma cells through interaction with SIX3. Am. J. Cancer Res. 2018, 8, 688–698. [Google Scholar]
- Liang, W.C.; Ren, J.L.; Wong, C.W.; Chan, S.O.; Waye, M.M.; Fu, W.M.; Zhang, J.F. LncRNA-NEF antagonized epithelial to mesenchymal transition and cancer metastasis via cis-regulating FOXA2 and inactivating Wnt/beta-catenin signaling. Oncogene 2018, 37, 1445–1456. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, F.; Yuan, J.H.; Yuan, S.X.; Zhou, W.P.; Huo, X.S.; Xu, D.; Bi, H.S.; Wang, F.; Sun, S.H. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis 2013, 34, 577–586. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Zou, X.J.; Cao, C.H.; Zhang, T.; Lei, L.; Qi, X.L.; Liu, L.; Wu, D.H. Identification and Functional Characterization of Long Non-coding RNA MIR22HG as a Tumor Suppressor for Hepatocellular Carcinoma. Theranostics 2018, 8, 3751–3765. [Google Scholar] [CrossRef]
- He, J.; Zuo, Q.; Hu, B.; Jin, H.; Wang, C.; Cheng, Z.; Deng, X.; Yang, C.; Ruan, H.; Yu, C.; et al. A novel, liver-specific long noncoding RNA LINC01093 suppresses HCC progression by interaction with IGF2BP1 to facilitate decay of GLI1 mRNA. Cancer Lett. 2019, 450, 98–109. [Google Scholar] [CrossRef]
- Ding, C.H.; Yin, C.; Chen, S.J.; Wen, L.Z.; Ding, K.; Lei, S.J.; Liu, J.P.; Wang, J.; Chen, K.X.; Jiang, H.L.; et al. The HNF1alpha-regulated lncRNA HNF1A-AS1 reverses the malignancy of hepatocellular carcinoma by enhancing the phosphatase activity of SHP-1. Mol. Cancer 2018, 17, 63. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, J.; Liu, X.; Li, S.; Wang, Q.; Di, C.; Hu, Z.; Yu, T.; Ding, J.; Li, J.; et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat. Commun. 2018, 9, 1572. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liang, L.; Dong, Q.; Huan, L.; He, J.; Li, B.; Yang, C.; Jin, H.; Wei, L.; Yu, C.; et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-kappaB pathway in hepatocellular carcinoma. Theranostics 2018, 8, 2814–2829. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Zhang, Y.; Zhan, Z.; Ye, F.; Liang, Y.; Huang, J.; Chen, K.; Chen, L.; Ding, Y. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J. Hematol. Oncol. 2017, 10, 91. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Huo, X.S.; Yuan, S.X.; Zhang, L.; Zhou, W.P.; Wang, F.; Sun, S.H. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol. Cell 2013, 49, 1083–1096. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, Y.; Yu, S. Long noncoding RNA AWPPH promotes hepatocellular carcinoma progression through YBX1 and serves as a prognostic biomarker. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1805–1816. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Z.; Liu, L.; Wang, Q.; Li, S.; Chen, D.; Hu, Z.; Yu, T.; Ding, J.; Li, J.; et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology 2018, 67, 171–187. [Google Scholar] [CrossRef]
- Xie, C.R.; Wang, F.; Zhang, S.; Wang, F.Q.; Zheng, S.; Li, Z.; Lv, J.; Qi, H.Q.; Fang, Q.L.; Wang, X.M.; et al. Long Noncoding RNA HCAL Facilitates the Growth and Metastasis of Hepatocellular Carcinoma by Acting as a ceRNA of LAPTM4B. Mol. Ther. Nucleic Acids 2017, 9, 440–451. [Google Scholar] [CrossRef]
- Hou, Z.; Xu, X.; Zhou, L.; Fu, X.; Tao, S.; Zhou, J.; Tan, D.; Liu, S. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumour Biol. 2017, 39, 1010428317718135. [Google Scholar] [CrossRef]
- Wang, H.; Huo, X.; Yang, X.R.; He, J.; Cheng, L.; Wang, N.; Deng, X.; Jin, H.; Wang, N.; Wang, C.; et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol. Cancer 2017, 16, 136. [Google Scholar] [CrossRef]
- Lu, S.; Zhou, J.; Sun, Y.; Li, N.; Miao, M.; Jiao, B.; Chen, H. The noncoding RNA HOXD-AS1 is a critical regulator of the metastasis and apoptosis phenotype in human hepatocellular carcinoma. Mol. Cancer 2017, 16, 125. [Google Scholar] [CrossRef]
- Li, W.; Kang, Y. A new Lnc in metastasis: Long noncoding RNA mediates the prometastatic functions of TGF-beta. Cancer Cell 2014, 25, 557–559. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhang, D.; Wu, W.; Wu, S.; Qian, J.; Hao, Y.; Yan, F.; Zhu, P.; Wu, J.; Huang, G.; et al. Mesenchymal Stem Cells Promote Hepatocarcinogenesis via lncRNA-MUF Interaction with ANXA2 and miR-34a. Cancer Res. 2017, 77, 6704–6716. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lu, L.; Feng, B.; Zhang, K.; Han, S.; Hou, D.; Chen, L.; Chu, X.; Wang, R. The lincRNA-ROR/miR-145 axis promotes invasion and metastasis in hepatocellular carcinoma via induction of epithelial-mesenchymal transition by targeting ZEB2. Sci. Rep. 2017, 7, 4637. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yao, H.; Wang, K.; Liu, X. Long Non-Coding RNA MALAT1 Regulates ZEB1 Expression by Sponging miR-143-3p and Promotes Hepatocellular Carcinoma Progression. J. Cell. Biochem. 2017, 118, 4836–4843. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Liu, S.; Cheng, Y.; Lu, L.; Shi, J.; Xu, G.; Li, N.; Cheng, K.; Wu, M.; Cheng, S.; et al. ICAM-1-Related Noncoding RNA in Cancer Stem Cells Maintains ICAM-1 Expression in Hepatocellular Carcinoma. Clin. Cancer Res. 2016, 22, 2041–2050. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, B.; Zhang, M.; Guo, W.; Wu, Z.; Wang, Y.; Jia, L.; Li, S.; Cancer Genome Atlas Research, N.; Xie, W.; et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell 2018, 33, 706–720. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Lin, A.; Li, C.; Liang, K.; Wang, S.; Liu, Y.; Park, P.K.; Qin, L.; Wei, Y.; Hawke, D.H.; et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 2014, 159, 1110–1125. [Google Scholar] [CrossRef]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2017, 169, 361. [Google Scholar] [CrossRef]
- Guccione, E.; Bassi, C.; Casadio, F.; Martinato, F.; Cesaroni, M.; Schuchlautz, H.; Luscher, B.; Amati, B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 2007, 449, 933–937. [Google Scholar] [CrossRef]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Quirk, M.; Kim, Y.H.; Saab, S.; Lee, E.W. Management of hepatocellular carcinoma with portal vein thrombosis. World J. Gastroenterol. 2015, 21, 3462–3471. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Panda, P.K.; Naik, P.P.; Praharaj, P.P.; Meher, B.R.; Gupta, P.K.; Verma, R.S.; Maiti, T.K.; Shanmugam, M.K.; Chinnathambi, A.; Alharbi, S.A.; et al. Abrus agglutinin stimulates BMP-2-dependent differentiation through autophagic degradation of beta-catenin in colon cancer stem cells. Mol. Carcinog. 2018, 57, 664–677. [Google Scholar] [CrossRef]
- Clevers, H. Wnt/beta-catenin signaling in development and disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef]
- Clevers, H.; Nusse, R. Wnt/beta-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef]
- Monga, S.P. beta-Catenin Signaling and Roles in Liver Homeostasis, Injury, and Tumorigenesis. Gastroenterology 2015, 148, 1294–1310. [Google Scholar] [CrossRef]
- Xu, D.; Yang, F.; Yuan, J.H.; Zhang, L.; Bi, H.S.; Zhou, C.C.; Liu, F.; Wang, F.; Sun, S.H. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta-catenin signaling. Hepatology 2013, 58, 739–751. [Google Scholar] [CrossRef]
- Fu, X.; Zhu, X.; Qin, F.; Zhang, Y.; Lin, J.; Ding, Y.; Yang, Z.; Shang, Y.; Wang, L.; Zhang, Q.; et al. Linc00210 drives Wnt/beta-catenin signaling activation and liver tumor progression through CTNNBIP1-dependent manner. Mol. Cancer 2018, 17, 73. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yuan, J.H.; Huang, J.F.; Yang, F.; Wang, T.T.; Ma, J.Z.; Zhang, L.; Zhou, C.C.; Wang, F.; Yu, J.; et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene 2016, 35, 5422–5434. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Ahn, K.S.; Kim, C.; Siveen, K.S.; Ong, T.H.; Shanmugam, M.K.; Li, F.; Shi, J.; Kumar, A.P.; Wang, L.Z.; et al. Ascochlorin, an isoprenoid antibiotic inhibits growth and invasion of hepatocellular carcinoma by targeting STAT3 signaling cascade through the induction of PIAS3. Mol. Oncol. 2015, 9, 818–833. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, P.; Li, F.; Shanmugam, M.K.; Vali, S.; Abbasi, T.; Kapoor, S.; Ahn, K.S.; Kumar, A.P.; Sethi, G. Honokiol inhibits signal transducer and activator of transcription-3 signaling, proliferation, and survival of hepatocellular carcinoma cells via the protein tyrosine phosphatase SHP-1. J. Cell. Physiol. 2012, 227, 2184–2195. [Google Scholar] [CrossRef]
- Huang, J.L.; Cao, S.W.; Ou, Q.S.; Yang, B.; Zheng, S.H.; Tang, J.; Chen, J.; Hu, Y.W.; Zheng, L.; Wang, Q. The long non-coding RNA PTTG3P promotes cell growth and metastasis via up-regulating PTTG1 and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol. Cancer 2018, 17, 93. [Google Scholar] [CrossRef]
- Tang, J.; Zhuo, H.; Zhang, X.; Jiang, R.; Ji, J.; Deng, L.; Qian, X.; Zhang, F.; Sun, B. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis. 2014, 5, e1549. [Google Scholar] [CrossRef]
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Rev. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef]
- Paget, S. The distribution of secondary growths in cancer of the breast. Cancer Metastasis Rev. 1989, 8, 98–101. [Google Scholar]
- Ye, Y.; Xu, Y.; Lai, Y.; He, W.; Li, Y.; Wang, R.; Luo, X.; Chen, R.; Chen, T. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J. Cell. Biochem. 2018, 119, 2951–2963. [Google Scholar] [CrossRef]
- Huang, J.F.; Guo, Y.J.; Zhao, C.X.; Yuan, S.X.; Wang, Y.; Tang, G.N.; Zhou, W.P.; Sun, S.H. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology 2013, 57, 1882–1892. [Google Scholar] [CrossRef]
- Lin, Y.H.; Wu, M.H.; Huang, Y.H.; Yeh, C.T.; Cheng, M.L.; Chi, H.C.; Tsai, C.Y.; Chung, I.H.; Chen, C.Y.; Lin, K.H. Taurine up-regulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology 2018, 67, 188–203. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Xu, H.; Liu, G.; Wu, J.; Li, C.; Wang, X.; Zhang, S.; Xu, H.; Ju, S.; Cheng, W.; et al. Metabolism-induced tumor activator 1 (MITA1), an Energy Stress-Inducible Long Noncoding RNA, Promotes Hepatocellular Carcinoma Metastasis. Hepatology 2019, 70, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Cao, S.; Wang, Y.; Hu, Y.; Liu, H.; Li, J.; Chen, J.; Li, P.; Liu, J.; Wang, Q.; et al. Long non-coding RNA UBE2CP3 enhances HCC cell secretion of VEGFA and promotes angiogenesis by activating ERK1/2/HIF-1alpha/VEGFA signalling in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. CR 2018, 37, 113. [Google Scholar] [CrossRef] [PubMed]
- Karaman, B.; Battal, B.; Sari, S.; Verim, S. Hepatocellular carcinoma review: Current treatment, and evidence-based medicine. World J. Gastroenterol. 2014, 20, 18059–18060. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, R.; Lahiri, N. Tissue- and Serum-Associated Biomarkers of Hepatocellular Carcinoma. Biomark. Cancer 2016, 8, 37–55. [Google Scholar] [CrossRef]
- Li, J.; Wang, X.; Tang, J.; Jiang, R.; Zhang, W.; Ji, J.; Sun, B. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2015, 37, 687–696. [Google Scholar] [CrossRef]
- Fujita, K.; Nonomura, N. Urinary biomarkers of prostate cancer. Int. J. Urol. 2018, 25, 770–779. [Google Scholar] [CrossRef]
- Luo, P.; Liang, C.; Zhang, X.; Liu, X.; Wang, Y.; Wu, M.; Feng, X.; Tu, J. Identification of long non-coding RNA ZFAS1 as a novel biomarker for diagnosis of HCC. Biosci. Rep. 2018, 38, BSR20171359. [Google Scholar] [CrossRef]
- Sasaki, R.; Kanda, T.; Yokosuka, O.; Kato, N.; Matsuoka, S.; Moriyama, M. Exosomes and Hepatocellular Carcinoma: From Bench to Bedside. Int. J. Mol. Sci. 2019, 20, 1406. [Google Scholar] [CrossRef]
- Zhang, C.; Ji, Q.; Yang, Y.; Li, Q.; Wang, Z. Exosome: Function and Role in Cancer Metastasis and Drug Resistance. Technol. Cancer Res. Treat. 2018, 17, 1533033818763450. [Google Scholar] [CrossRef]
- Kogure, T.; Yan, I.K.; Lin, W.L.; Patel, T. Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes cancer 2013, 4, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.R.; Hsieh, M.J.; Chiang, W.L.; Hsueh, K.C.; Yang, S.F.; Su, S.C. Association of lncRNA CCAT2 and CASC8 Gene Polymorphisms with Hepatocellular Carcinoma. Int. J. Environ. Res. Public Health 2019, 16, 2833. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Tan, G.; Jiang, X.; Han, P.; Zhai, B.; Dong, X.; Qiao, H.; Jiang, H.; Sun, X. An artificial lncRNA targeting multiple miRNAs overcomes sorafenib resistance in hepatocellular carcinoma cells. Oncotarget 2016, 7, 73257–73269. [Google Scholar] [CrossRef] [PubMed]
- Tonus, C.; Cloquette, K.; Ectors, F.; Piret, J.; Gillet, L.; Antoine, N.; Desmecht, D.; Vanderplasschen, A.; Waroux, O.; Grobet, L. Long term-cultured and cryopreserved primordial germ cells from various chicken breeds retain high proliferative potential and gonadal colonisation competency. Reprod. Fertil. Dev. 2016, 28, 628–639. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Kurzrock, R.; Kim, Y.; Woessner, R.; Younes, A.; Nemunaitis, J.; Fowler, N.; Zhou, T.; Schmidt, J.; Jo, M.; et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 2015, 7, 314ra185. [Google Scholar] [CrossRef] [PubMed]
- Arun, G.; Diermeier, S.; Akerman, M.; Chang, K.C.; Wilkinson, J.E.; Hearn, S.; Kim, Y.; MacLeod, A.R.; Krainer, A.R.; Norton, L.; et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016, 30, 34–51. [Google Scholar] [CrossRef]
| LncRNA | Interaction Class | Interaction Partner | Expression of LncRNA | Pathway | Function | Mechanism | Reference |
|---|---|---|---|---|---|---|---|
| linc-GALH | RNA-TFs | DNMT1 | Upregulated | AKT signaling | Promote metastasis | Epigenetically regulates Gankyrin by adjusting the ubiquitination status of DNMT1 | [18] |
| lncRNA GIHCG | RNA-TFs | EZH2 DNMT1 | Upregulated | Promote metastasis | Inhibits miR200b/a/429 transcript by recruiting DNMT1 and EZH2 to miR-200b/a/429 promoter | [19] | |
| lncRNA SOX21-AS1 | RNA-TFs | EZH2 | Downregulated | Inhibit metastasis | Epigenetically silenced p21 via recruiting EZH2 to the promoter of p21 | [20] | |
| lncFZD6 | RNA-TFs | FZD6 | Upregulated | Wnt/β-catenin signaling | Promote metastasis | Interacts with FZD6 promoter and recruits BRG1 to initiate transcription | [21] |
| lncHOXA10 | RNA-TFs | EZH2 | Upregulated | Promote metastasis | Recruits SNF2L to the promoter to initiate the expression of HOXA10 | [22] | |
| lncSox4 | RNA-TFs | Stat3 | Upregulated | Promote metastasis | Drives Sox4 expression by recruiting Stat3 to be a Sox4 promoter | [23] | |
| lncAPC | RNA-TFs | EZH2 | Upregulated | Wnt/β-catenin signaling | Promote metastasis | Inhibits APC transcription by recruiting EZH2 to be a APC promoter | [24] |
| lncWDR26 | RNA-TFs | SIX3 | Downregulated | Inhibit metastasis | Inhibits WDR26 transcription by binding with SIX3 | [25] | |
| lncRNA-NEF | RNA-TFs | FOXA2 | Upregulated | Promote metastasis | Interacts with β-catenin to increase the binding of GSK3β with β-catenin and inhibits phosphorylation of β-catenin | [26] | |
| H19 | RNA-protein | hnRNP U/PCAF/RNA Pol II | Downregulated | Inhibit metastasis | Associates with hnRNP U/PCAF/RNA Pol II and activates miR-200 family by increasing histone acetylation | [27] | |
| MIR22HG | RNA-protein | HuR | Downregulated | Inhibit metastasis | Interacted with HuR to increase its stability | [28] | |
| LINC01093 | RNA-protein | IGF2BP1 | Downregulated | Inhibit metastasis | Recruits IGF2BP1, preventing GLI1 binding to IGF2BP1 | [29] | |
| HNF1A-AS1 | RNA-protein | SHP-1 C-terminal | Downregulated | Inhibit metastasis | Acts as phosphatase activator through interacting with SHP1 | [30] | |
| LINC01138 | RNA-protein | PRMT5 | Upregulated | Promote metastasis | Interacts with PRMT5 and enhances its protein stability | [31] | |
| miR503HG | RNA-protein | HNRNPA2B1 | Downregulated | NF-κB signaling | Inhibit metastasis | Interacts with the HNRNPA2B1 and modulates the ubiquitination status of HNRNPA2B1 | [32] |
| lncRNA uc.134 | RNA-protein | CUL4A | Downregulated | Hippo kinase signaling | Inhibit metastasis | Inhibits the translocation of CUL4A from the nucleus to the cytoplasm | [33] |
| lncRNA-LET | RNA-protein | NF90 | Downregulated | Inhibit metastasis | Associates with NF90 to enhance the degradation of NF90 | [34] | |
| AWPPH | RNA-protein | YBX1 | Upregulated | Promote metastasis | Promotes YBX1-mediated activation of SNAIL1 translation and PIK3CA transcription | [35] | |
| TSLNC8 | RNA-protein RNA-TFs | TKT STAT3 | Downregulated | STAT signaling | Inhibit metastasis | Interacts with TKT and STAT3, and inhibits STAT3 phosphorylation and transcriptional activity | [36] |
| lncRNA HCAL | RNA-RNA | miR-15a miR-196a miR-196b | Upregulated | Promote metastasis | Binds to miR-15a, miR-196a, or miR-196b, and by increasing LAPTM4B expression | [37] | |
| MALAT1 | RNA-RNA | miR-204 | Downregulated | Inhibit metastasis | Sponges miR-204 and release SIRT1. | [38] | |
| HOXD-AS1 | RNA-RNA | miR-130a-3p | Upregulated | MEK/ERK signaling | Promote metastasis | Binds to miR-130a-3p that prevented SOX4 degradation, activates the expression of EZH2 and MMP2 | [39] |
| HOXD-AS1 | RNA-RNA | miR19a | Upregulated | Promote metastasis | Upregulates the ARHGAP11A via bind to miR19a | [40] | |
| lnc-ATB | RNA-RNA | miR-200 and IL-11 mRNA | Upregulated | TGF-β signaling | Promote metastasis | Binds with the miR-200 family and sequestrates the repression effect of the miR-200s on ZEB1/2; binds with IL-11 mRNA to promote organ colonization | [41] |
| lnc-MUF | RNA-RNA | miR-34a | Upregulated | Wnt/β-catenin signaling | Promote metastasis | Upregulate SNAIL1 expression | [42] |
| linc-ROR | RNA-RNA | miR-145 | Upregulated | Promote metastasis | Sponges miR-145 to de-repress the expression of target gene ZEB2 | [43] | |
| MALAT1 | RNA-RNA | miR-143-3p | Upregulated | Promote metastasis | Regulates the expression of ZEB1 by sponging miR-143-3p | [44] | |
| lncRNA ICR | RNA-RNA | ICAM-1 mRNA | Upregulated | Promote metastasis | Regulates ICAM-1 expression by increasing the stability of ICAM-1 mRNA through RNA duplex formation | [45] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Tang, F.-R.; Arfuso, F.; Cai, W.-Q.; Ma, Z.; Yang, J.; Sethi, G. The Emerging Role of Long Non-Coding RNAs in the Metastasis of Hepatocellular Carcinoma. Biomolecules 2020, 10, 66. https://doi.org/10.3390/biom10010066
Chen X, Tang F-R, Arfuso F, Cai W-Q, Ma Z, Yang J, Sethi G. The Emerging Role of Long Non-Coding RNAs in the Metastasis of Hepatocellular Carcinoma. Biomolecules. 2020; 10(1):66. https://doi.org/10.3390/biom10010066
Chicago/Turabian StyleChen, Xuejiao, Feng-Ru Tang, Frank Arfuso, Wen-Qi Cai, Zhaowu Ma, Jiyuan Yang, and Gautam Sethi. 2020. "The Emerging Role of Long Non-Coding RNAs in the Metastasis of Hepatocellular Carcinoma" Biomolecules 10, no. 1: 66. https://doi.org/10.3390/biom10010066
APA StyleChen, X., Tang, F.-R., Arfuso, F., Cai, W.-Q., Ma, Z., Yang, J., & Sethi, G. (2020). The Emerging Role of Long Non-Coding RNAs in the Metastasis of Hepatocellular Carcinoma. Biomolecules, 10(1), 66. https://doi.org/10.3390/biom10010066