Lipidomic Profiles of Lipid Biosynthesis in Oil Palm during Fruit Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Sample Preparation and Extraction
2.3. RNA Extraction and Transcriptomic Analysis
2.4. Lipidomic Analysis Using Liquid Chromatography–Mass Spectrometry
2.5. Cluster Analyses
3. Results
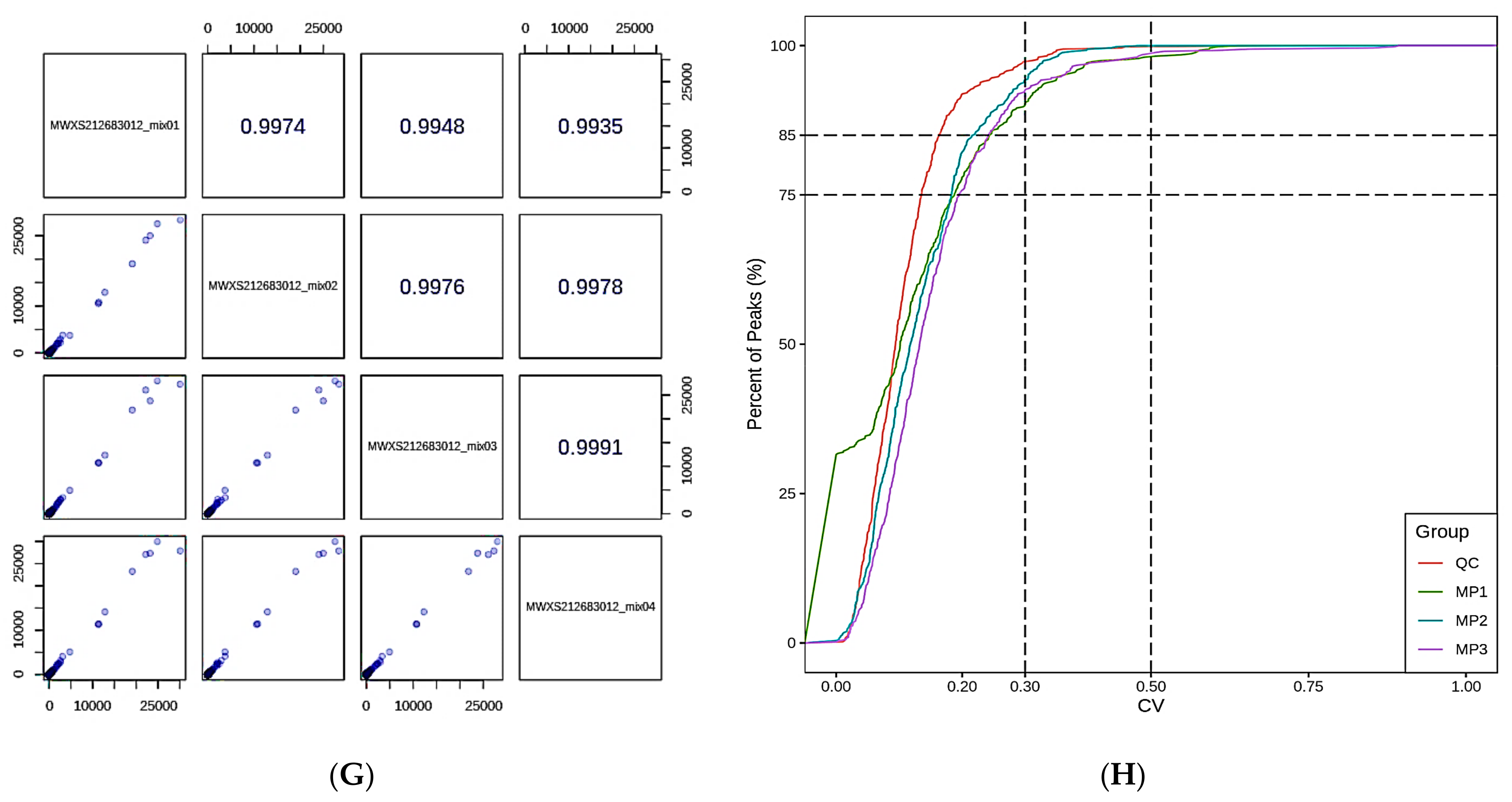
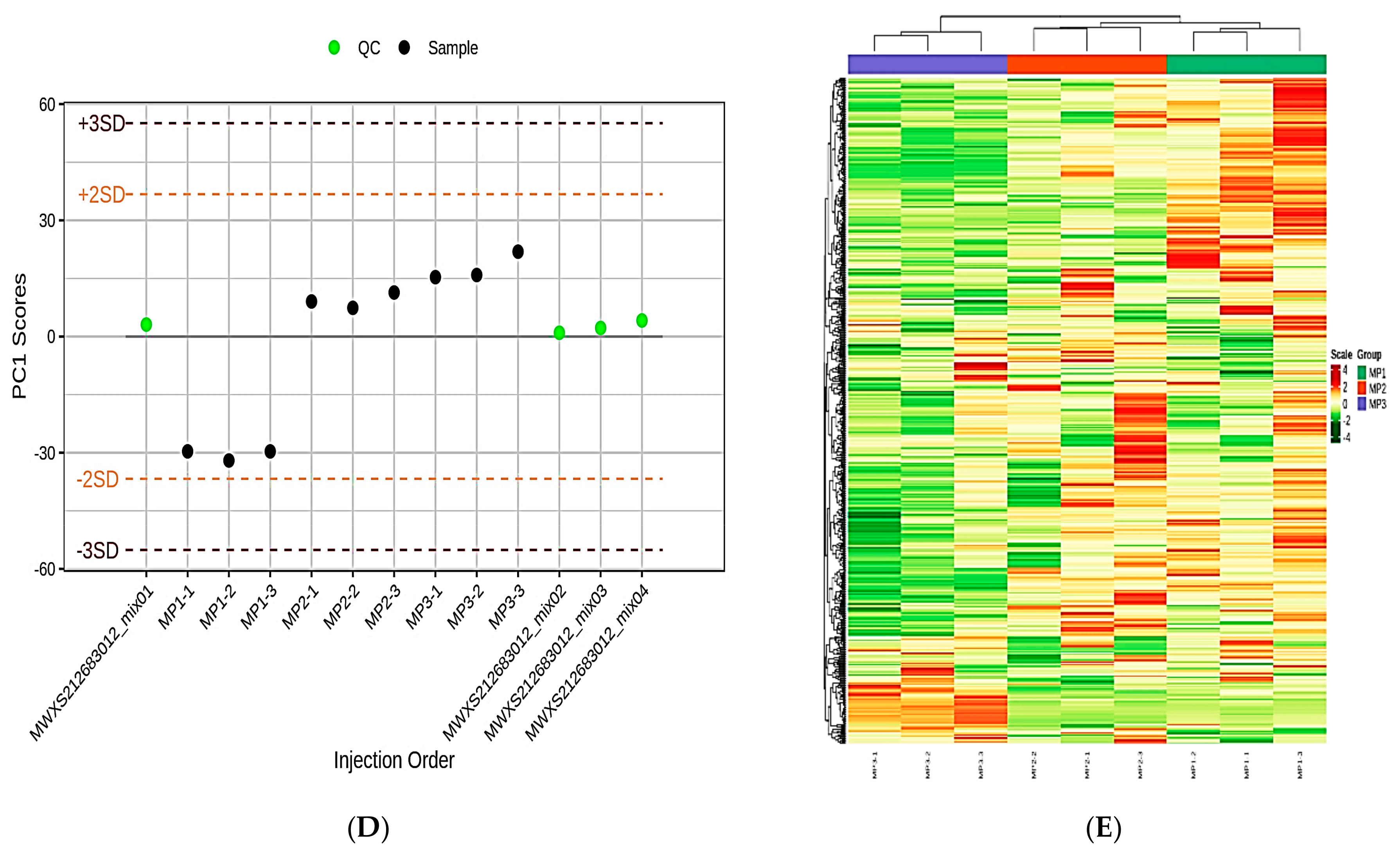
3.1. Data Pre-Processing and Quality Control (QC) of the Lipidomics
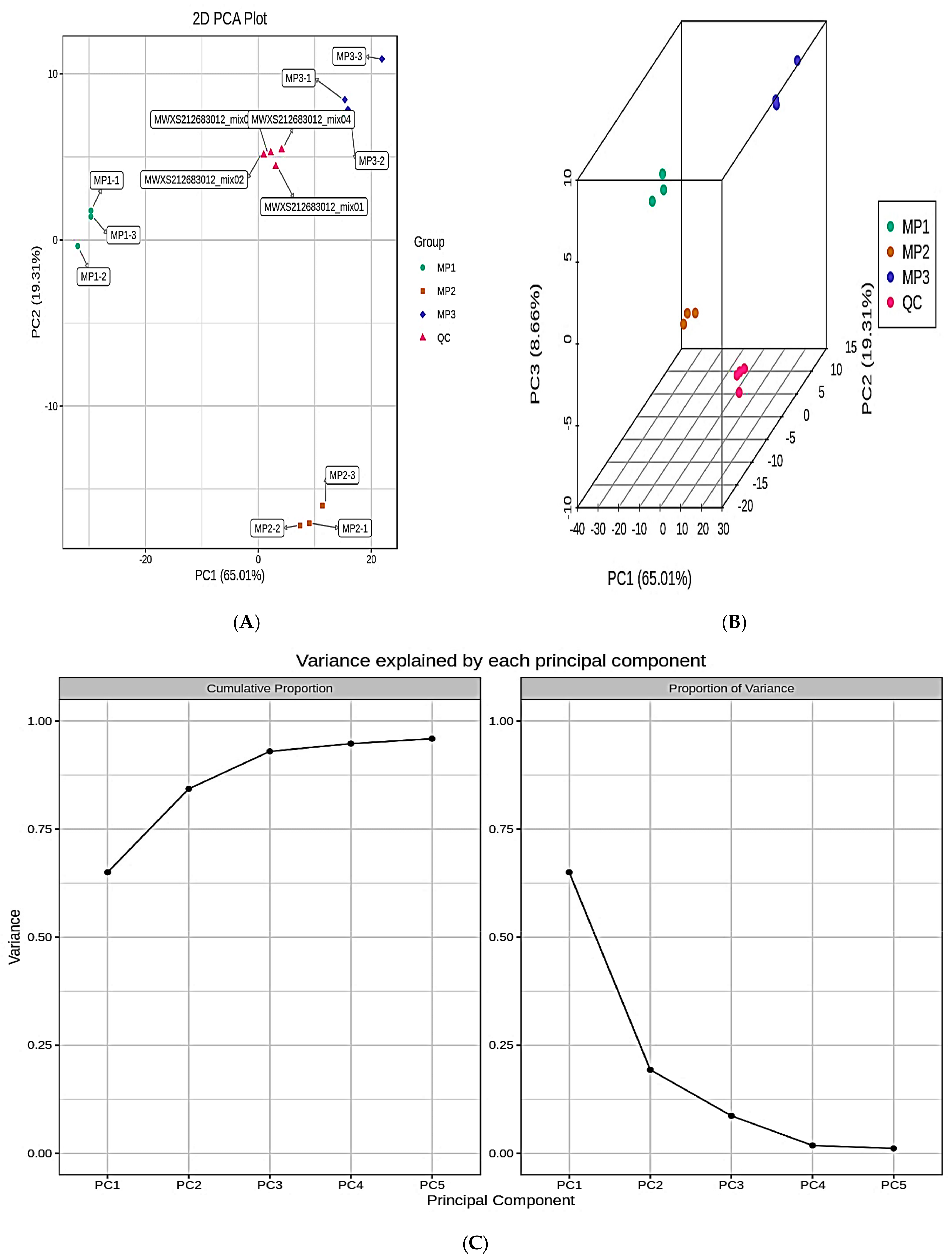
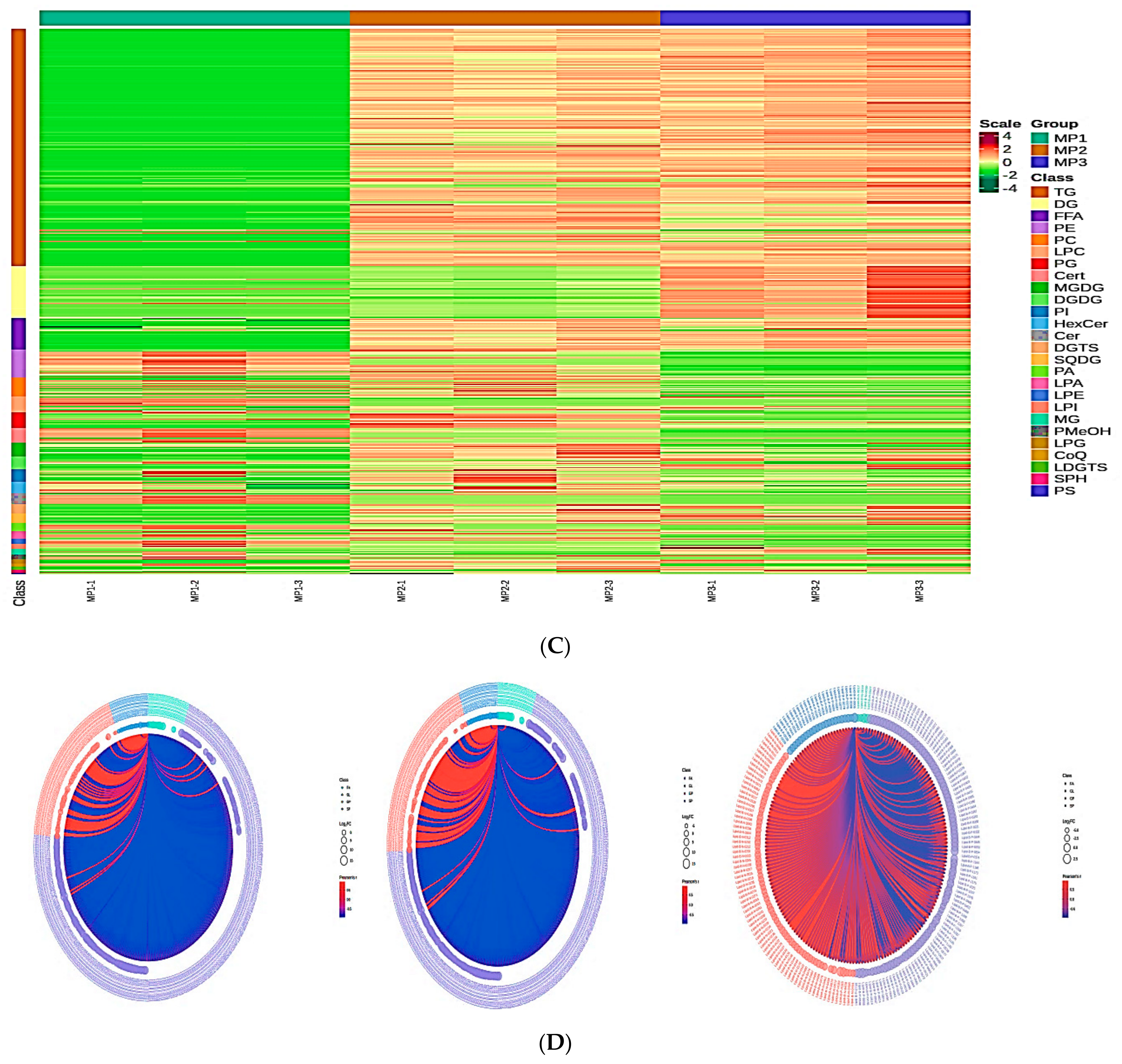
3.2. Principal Component Analysis and Cluster Analysis of the Lipidomics
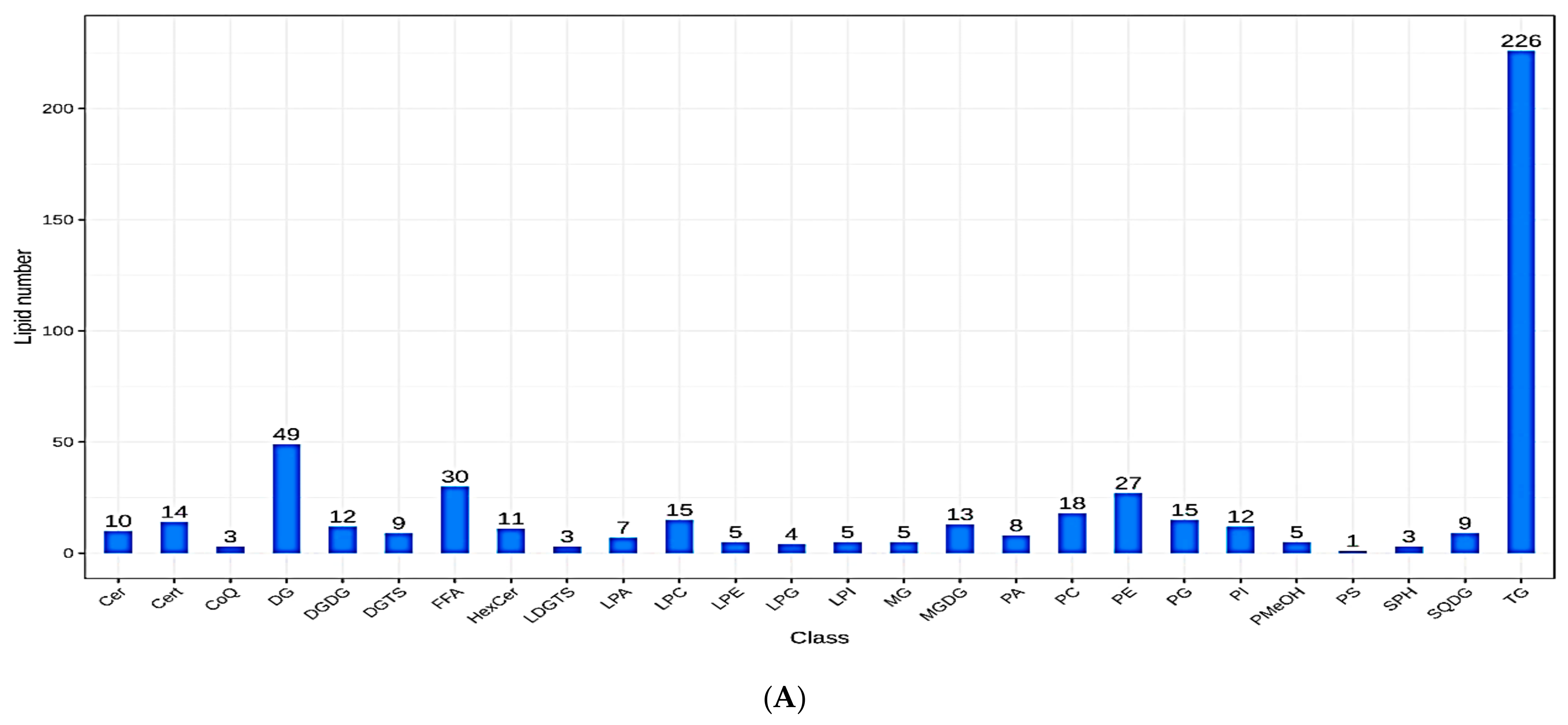
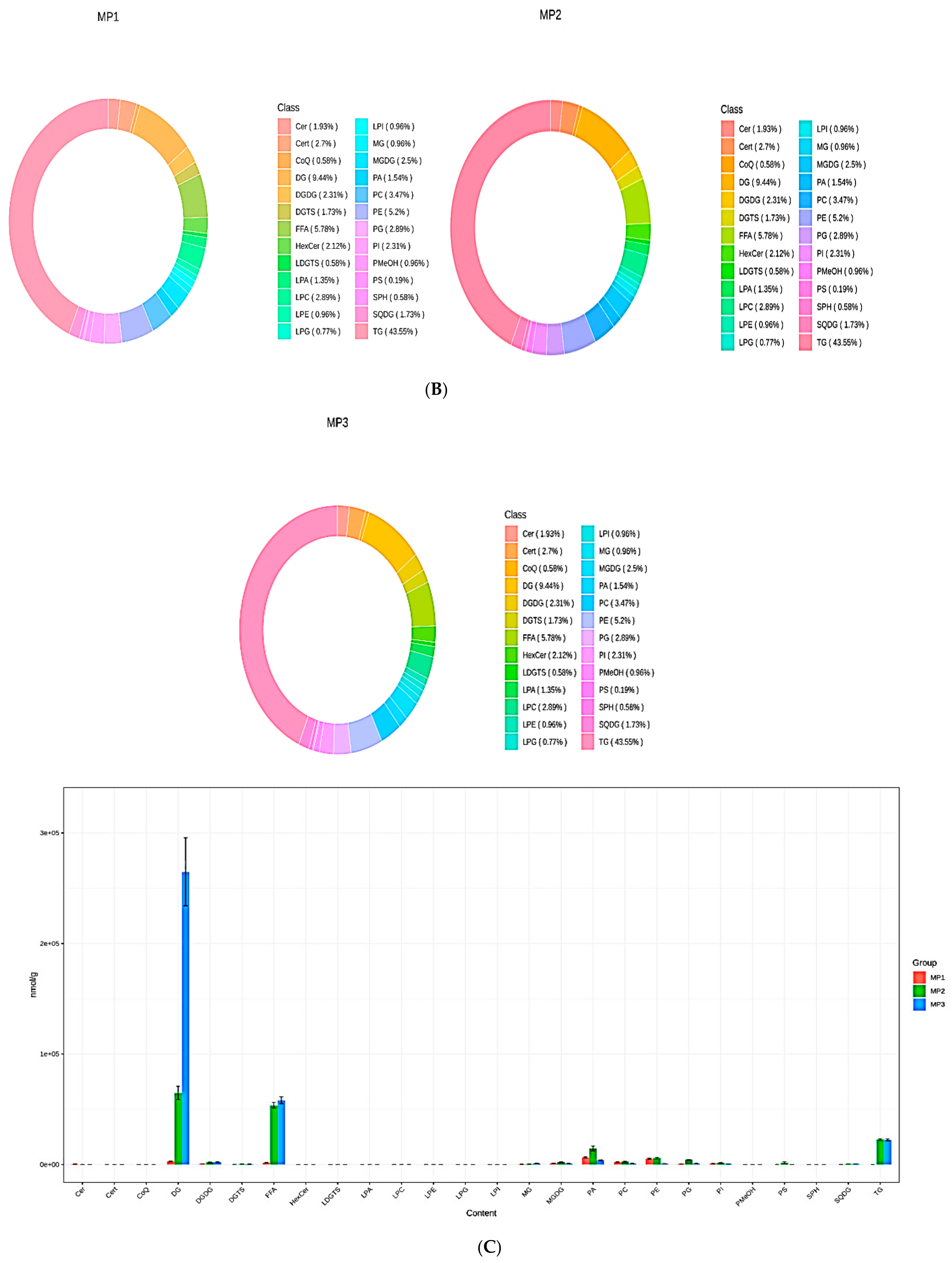
3.3. Lipid Metabolites Composition and Changes in Subclass Content
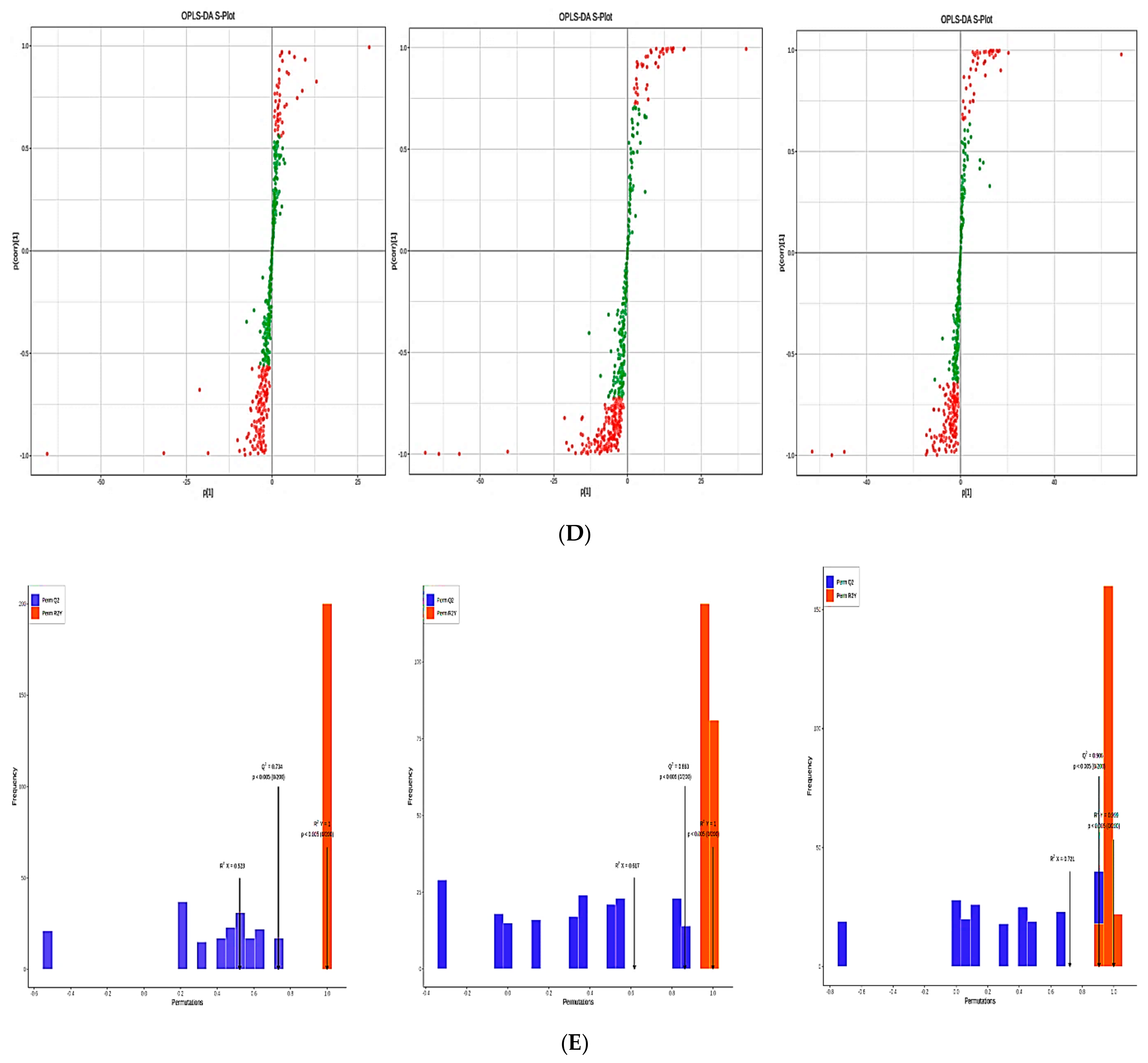
3.4. Discriminant Analysis of the Lipid Metabolites Using Subgroup Principal Component Analysis and Orthogonal Partial Least Squares
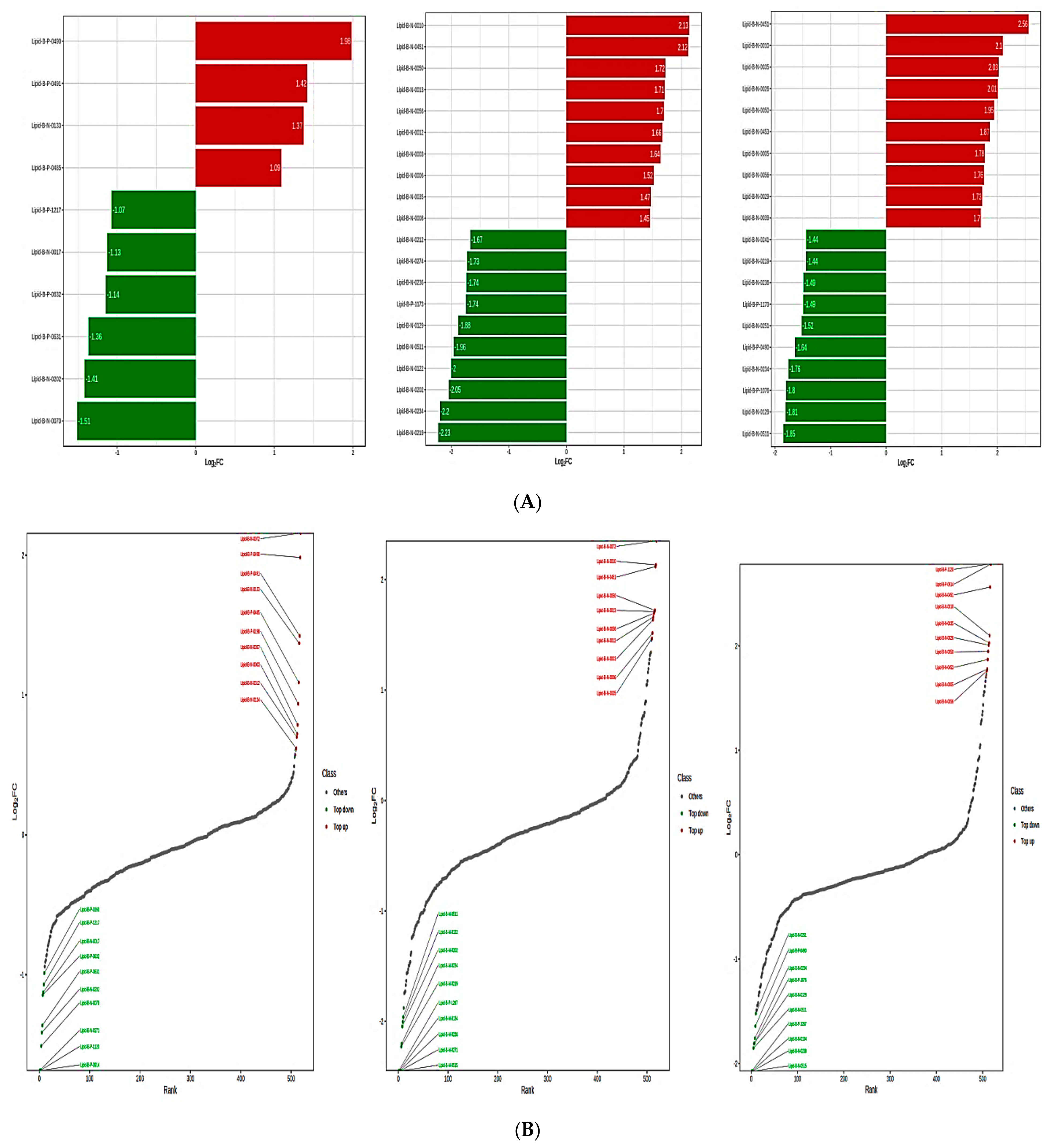
3.5. Identification of the Lipid Metabolites
3.6. Transcriptome Analysis of the Oil Palm
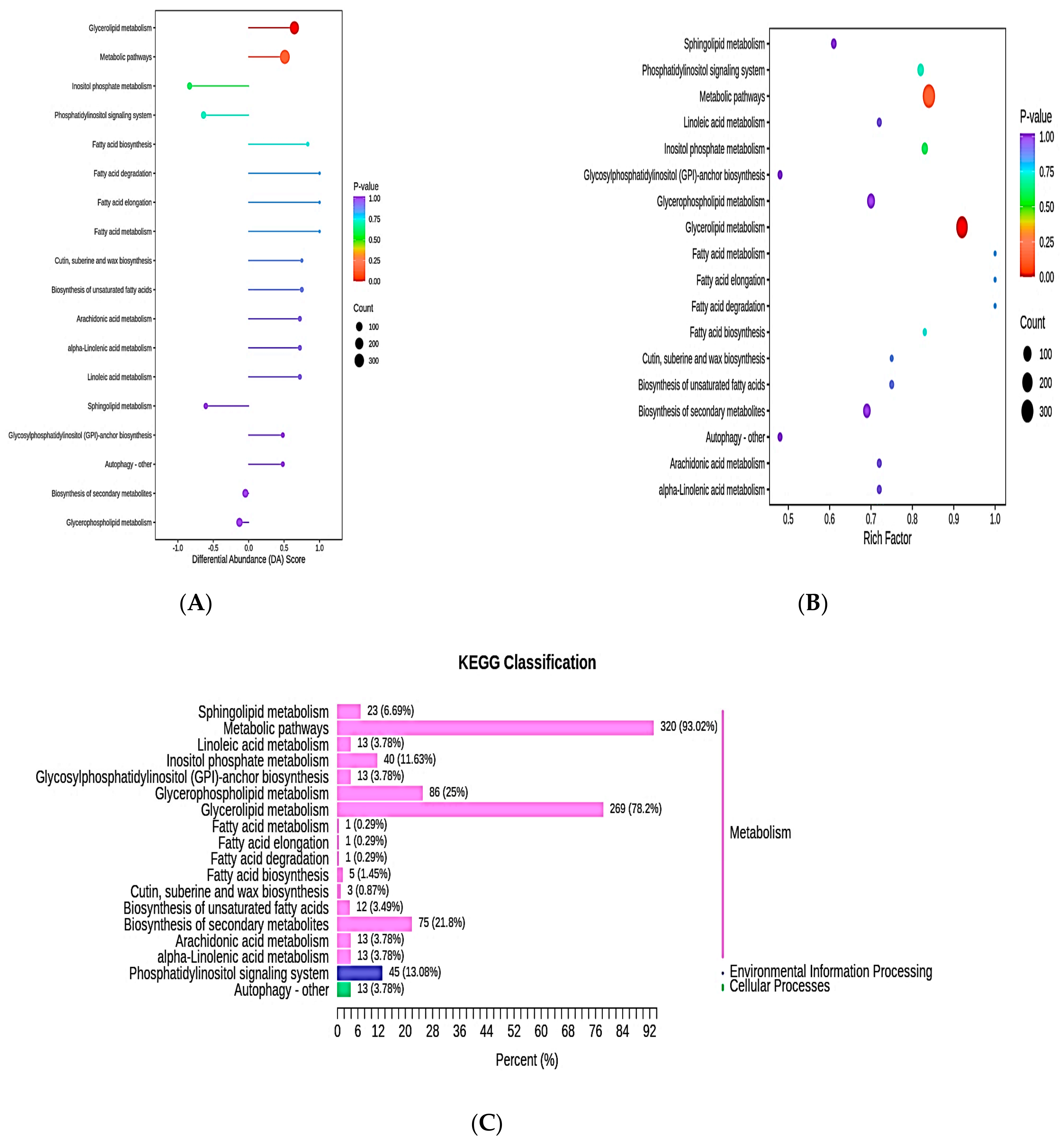
3.7. Kyoto Encyclopedia of Genes and Genomes Pathway Analysis of Differential Lipid Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Corley, R.H.V.; Tinker, P.B. The Oil Palm, 4th ed.; Wiley: Hoboken, NJ, USA, 2003; p. 562. [Google Scholar] [CrossRef]
- Barcelos, E.; Rios, S.d.A.; Cunha, R.N.V.; Lopes, R.; Motoike, S.Y.; Babiychuk, E.; Skirycz, A.; Kushnir, S. Oil palm natural diversity and the potential for yield improvement. Front. Plant Sci. 2015, 6, 190. [Google Scholar] [CrossRef] [PubMed]
- Montoya, C.; Cochard, B.; Flori, A.; Cros, D.; Lopes, R.; Cuellar, T.; Espeout, S.; Syaputra, I.; Villeneuve, P.; Pina, M.; et al. Genetic architecture of palm oil fatty acid composition in cultivated oil palm (Elaeis guineensis Jacq.) compared to its wild relative E. oleifera (H.B.K) Cortés. PLoS ONE 2014, 9, e95412. [Google Scholar] [CrossRef] [PubMed]
- Siew, W.-L.; Chong, C.-L.; Tan, Y.-A. Composition of the oil in palm kernel from Elaeis guineensis. J. Am. Oil Chem. Soc. 1995, 72, 1587–1589. [Google Scholar] [CrossRef]
- Hou, Q.; Ufer, G.; Bartels, D. Lipid signalling in plant responses to abiotic stress. Plant Cell Environ. 2016, 39, 1029–1048. [Google Scholar] [CrossRef] [PubMed]
- Mamode Cassim, A.; Gouguet, P.; Gronnier, J.; Laurent, N.; Germain, V.; Grison, M.; Boutté, Y.; Gerbeau-Pissot, P.; Simon-Plas, F.; Mongrand, S. Plant lipids: Key players of plasma membrane organization and function. Prog. Lipid Res. 2019, 73, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Cheong, W.F.; Wenk, M.R.; Shui, G. Comprehensive Analysis of Lipid Composition in Crude Palm Oil Using Multiple Lipidomic Approaches. J. Genet. Genom. 2014, 41, 293–304. [Google Scholar] [CrossRef]
- Enyoh, E.A.; Ihionu, E.A.; Verla, A.W.; Ebosie, P.N. Physicochemical parameter of palm oil and soil from Ihube community, Okigwe, Imo state Nigeria. Int. Lett. Nat. Sci. 2017, 62, 35–43. [Google Scholar] [CrossRef]
- Pati, S.; Nie, B.; Arnold, R.D.; Cummings, B.S. Extraction, chromatographic and mass spectrometric methods for lipid analysis. Biomed. Chromatogr. 2016, 30, 695–709. [Google Scholar] [CrossRef]
- Bates, P.D.; Stymne, S.; Ohlrogge, J. Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 2013, 16, 358–364. [Google Scholar] [CrossRef]
- Tarazona, P.; Feussner, K.; Feussner, I. An enhanced plant lipidomics method based on multiplexed liquid chromatography-mass spectrometry reveals additional insights into cold- and drought-induced membrane remodeling. Plant J. 2015, 84, 621–633. [Google Scholar] [CrossRef]
- Tranbarger, T.J.; Dussert, S.; Joët, T.; Argout, X.; Summo, M.; Champion, A.; Cros, D.; Omore, A.; Nouy, B.; Morcillo, F. Regulatory mechanisms underlying oil palm fruit mesocarp maturation, ripening, and functional specialization in lipid and carotenoid metabolism. Plant Physiol. 2011, 156, 564–584. [Google Scholar] [CrossRef]
- Ramli, U.S.; Salas, J.J.; Quant, P.A.; Harwood, J.L. Use of metabolic control analysis to give quantitative information on control of lipid biosynthesis in the important oil crop, Elaeis guineensis (oilpalm). New Phytol. 2009, 184, 330–339. [Google Scholar] [CrossRef]
- Saito, K.; Matsuda, F. Metabolomics for Functional Genomics, Systems Biology, and Biotechnology. Ann. Rev. Plant Biol. 2010, 61, 463–489. [Google Scholar] [CrossRef]
- Dreier, D.A.; Bowden, J.A.; Aristizabal-Henao, J.J.; Denslow, N.D.; Martyniuk, C.J. Ecotoxico-lipidomics: An emerging concept to understand chemical-metabolic relationships in comparative fish models. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 36, 100742. [Google Scholar] [CrossRef]
- Manaf, A.M.; Harwood, J.L. Purification and characterization of acyl-CoA: Glycerol 3-phosphate acyltransferase from oil palm (Elaeis guineensis) tissues. Planta 2000, 210, 318–328. [Google Scholar] [CrossRef]
- Lv, Y.; Zhang, X.; Luo, L.; Yang, H.; Li, P.; Zhang, K. Characterization of glycerol-3-phosphate acyltransferase 9 (AhGPAT9) genes, their allelic polymorphism and association with oil content in peanut (Arachis hypogaea L.). Sci. Rep. 2020, 10, 14648. [Google Scholar] [CrossRef]
- Jing, F.; Cantu, D.C.; Tvaruzkova, J.; Chipman, J.P.; Nikolau, B.J.; Yandeau-Nelson, M.D. Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. BMC Biochem. 2011, 12, 44. [Google Scholar] [CrossRef]
- Aymé, L.; Jolivet, P.; Nicaud, J.M.; Chardot, T. Molecular Characterization of the Elaeis guineensis Medium-Chain Fatty Acid Diacylglycerol Acyltransferase DGAT1-1 by Heterologous Expression in Yarrowia lipolytica. PLoS ONE 2015, 10, e0143113. [Google Scholar] [CrossRef]
- Morcillo, F.; Cros, D.; Billotte, N.; Ngando-Ebongue, G.F.; Domonhédo, H.; Pizot, M. Improving palm oil quality through identification and mapping of the lipase gene causing oil deterioration. Nat. Commun. 2013, 4, 2160. [Google Scholar] [CrossRef]
- Choi, Y.; Zaikova, K.; Yeom, S.-J.; Kim, Y.-S.; Lee, D.W. Biogenesis and Lipase-Mediated Mobilization of Lipid Droplets in Plants. Plants 2022, 11, 1243. [Google Scholar] [CrossRef]
- Gidda, S.K.; Watt, S.; Collins-Silva, J.; Kilaru, A.; Arondel, V.; Yurchenko, O.; Horn, P.J.; James, C.N.; Shintani, D.; Ohlrogge, J.B.; et al. Lipid droplet-associated proteins (LDAPs) are involved in the compartmentalization of lipophilic compounds in plant cells. Plant Signal. Behav. 2013, 8, e27141. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.-Y.; Zhang, Y.; Lyu, Y.-P.; Yao, Z.-J.; Hu, Y.-H. Lipidomic profifiling of the developing kernel clarififies the lipid metabolism of Paeonia ostii. Sci. Rep. 2021, 11, 1–12. [Google Scholar]
- Woodfield, H.K.; Cazenave-Gassiot, A.; Haslam, R.P.; Guschina, I.A.; Wenk, M.R.; Harwood, J.L. Using lipidomics to reveal details of lipid accumulation in developing seeds from oilseed rape (Brassica napus L.). Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Volk, G.M.; Crane, J.; Caspersen, A.M.; Hill, L.M.; Gardner, C.; Walters, C. Massive cellular disruption occurs during early imbibition of Cuphea seeds containing crystallized triacylglycerols. Planta 2006, 224, 1415–1426. [Google Scholar] [CrossRef]
- Dong, W.; Lv, H.; Xia, G.; Wang, M. Does diacylglycerol serve as a signaling molecule in plants. Plant Signal. Behav 2012, 7, 472–475. [Google Scholar] [CrossRef]
- Fatiha, A.I.D. Plant Lipid Metabolism. In Advances in Lipid Metabolism; Rodrigo Valenzuela, B., Ed.; IntechOpen: Rijeka, Croatia, 2019. [Google Scholar]
- Mancini, A.; Imperlini, E.; Nigro, E.; Montagnese, C.; Daniele, A.; Orrù, S.; Buono, P. Biological and Nutritional Properties of Palm Oil and Palmitic Acid: Effects on Health. Molecules 2015, 20, 17339–17361. [Google Scholar] [CrossRef]
- Putri, S.P.; Fukusaki, E. Mass Spectrometry-Based Metabolomics: A Practical Guide; CRC Press: Boca Raton, FL, USA, 2014; ISBN 978-1-4822-2376-7. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin, J.J.J.; Wu, Q.; Feng, M.; Li, R.; Zhou, L.; Zhang, S.; Yang, C.; Cao, H. Lipidomic Profiles of Lipid Biosynthesis in Oil Palm during Fruit Development. Metabolites 2023, 13, 727. https://doi.org/10.3390/metabo13060727
Martin JJJ, Wu Q, Feng M, Li R, Zhou L, Zhang S, Yang C, Cao H. Lipidomic Profiles of Lipid Biosynthesis in Oil Palm during Fruit Development. Metabolites. 2023; 13(6):727. https://doi.org/10.3390/metabo13060727
Chicago/Turabian StyleMartin, Jerome Jeyakumar John, Qiufei Wu, Meili Feng, Rui Li, Lixia Zhou, Shuyan Zhang, Cheng Yang, and Hongxing Cao. 2023. "Lipidomic Profiles of Lipid Biosynthesis in Oil Palm during Fruit Development" Metabolites 13, no. 6: 727. https://doi.org/10.3390/metabo13060727
APA StyleMartin, J. J. J., Wu, Q., Feng, M., Li, R., Zhou, L., Zhang, S., Yang, C., & Cao, H. (2023). Lipidomic Profiles of Lipid Biosynthesis in Oil Palm during Fruit Development. Metabolites, 13(6), 727. https://doi.org/10.3390/metabo13060727





