Association of Neonatal Jaundice with Gut Dysbiosis Characterized by Decreased Bifidobacteriales
Abstract
:1. Introduction
2. Results
2.1. Participants’ Characteristics
2.2. Alpha Diversity
2.3. Beta Diversity
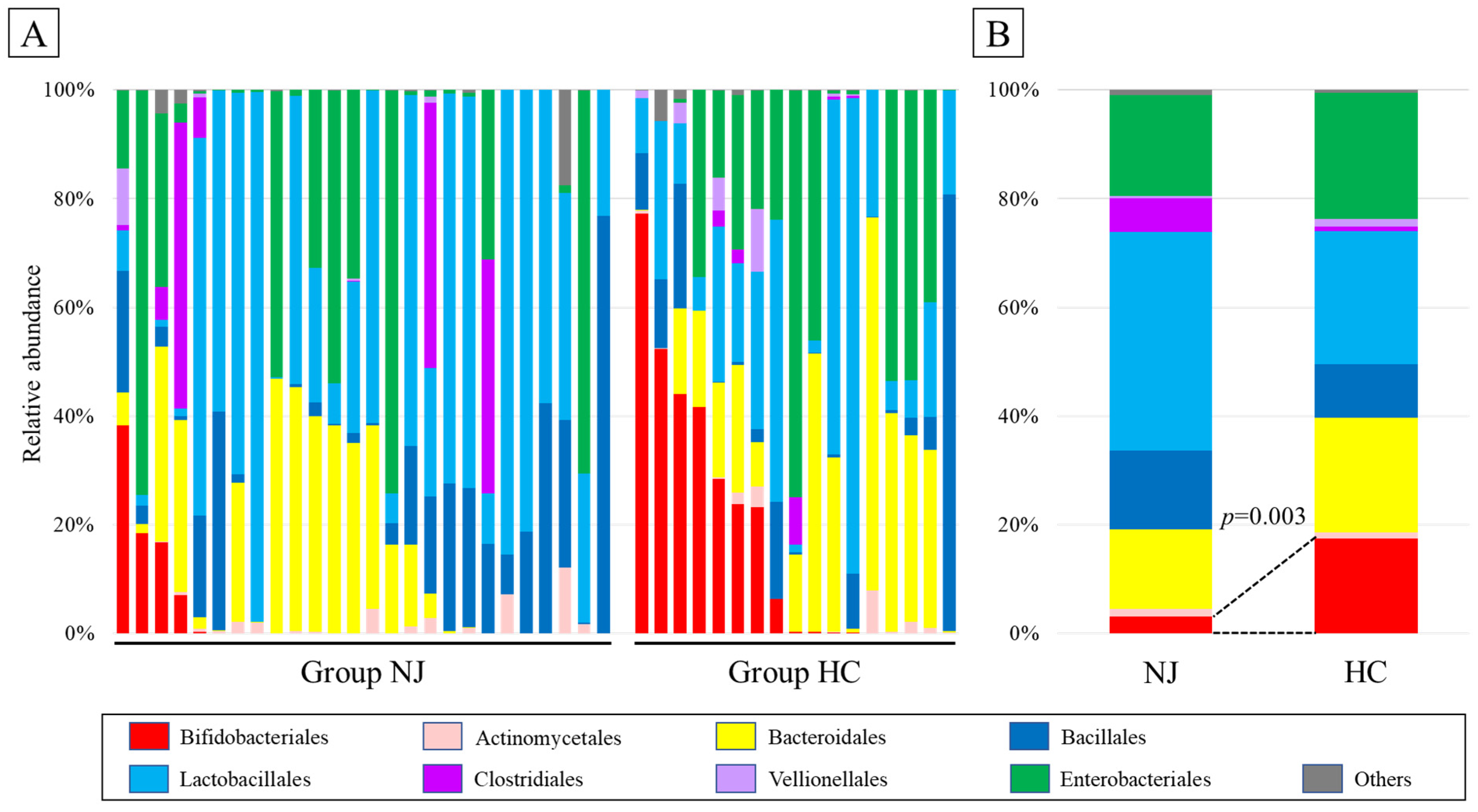
2.4. Taxonomic Composition
2.5. Linear Discriminant Analysis Effect Size (LEfSe)
2.6. Post Hoc Power Analysis
3. Discussion
4. Materials and Methods
4.1. Participants and Study Design
4.2. Blood Sampling and Total Serum Bilirubin Measurements
4.3. Stool Sampling and 16S rRNA Gene Sequencing
4.4. LEfSe
4.5. Statistical Analysis and Sample Size
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhutani, V.K.; Stark, A.R.; Lazzeroni, L.C.; Poland, R.; Gourley, G.R.; Kazmierczak, S.; Meloy, L.; Burgos, A.E.; Hall, J.Y.; Stevenson, D.K.; et al. Predischarge screening for severe neonatal hyperbilirubinemia identifies infants who need phototherapy. J. Pediatr. 2013, 162, 477–482.e1. [Google Scholar] [CrossRef]
- Brits, H.; Adendorff, J.; Huisamen, D.; Beukes, D.; Botha, K.; Herbst, H.; Joubert, G. The prevalence of neonatal jaundice and risk factors in healthy term neonates at National District Hospital in Bloemfontein. Afr. J. Prim. Health Care Fam. Med. 2018, 10, e1–e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olusanya, B.O.; Kaplan, M.; Hansen, T.W.R. Neonatal hyperbilirubinaemia: A global perspective. Lancet Child Adolesc. Health 2018, 2, 610–620. [Google Scholar] [CrossRef] [Green Version]
- Slusher, T.M.; Zamora, T.G.; Appiah, D.; Stanke, J.U.; Strand, M.A.; Lee, B.W.; Richardson, S.B.; Keating, E.M.; Siddappa, A.M.; Olusanya, B.O. Burden of severe neonatal jaundice: A systematic review and meta-analysis. BMJ Paediatr. Open 2017, 1, e000105. [Google Scholar] [CrossRef] [Green Version]
- Ullah, S.; Rahman, K.; Hedayati, M. Hyperbilirubinemia in neonates: Types, causes, clinical examinations, preventive measures and treatments: A narrative review article. Iran. J. Public Health 2016, 45, 558–568. [Google Scholar]
- Duan, M.; Yu, J.; Feng, J.; He, Y.; Xiao, S.; Zhu, D.; Zou, Z. 16S ribosomal RNA-based gut microbiome composition analysis in infants with breast milk jaundice. Open Life Sci. 2018, 13, 208–216. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Shen, N.; Li, J.; Hu, R.; Mo, X.; Xu, L. Changes in intestinal flora and metabolites in neonates with breast milk jaundice. Front. Pediatr. 2020, 8, 177. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, Z.; He, F.; Qiu, H.; Wang, Y.; Wang, H.; Zhou, J.; Zhou, J.; Cheng, G.; Zhou, W.; et al. Association of serum bilirubin in newborns affected by jaundice with gut microbiota dysbiosis. J. Nutr. Biochem. 2019, 63, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Yang, R.X.; Wang, A.H.; Zhang, Y.Q. The effect of intestinal flora on the neural development of severe hyperbilirubinemia neonates. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1291–1295. [Google Scholar] [CrossRef]
- Huang, L.; Bao, Y.; Xu, Z.; Lei, X.; Chen, Y.; Zhang, Y.; Zhang, J. Neonatal bilirubin levels and childhood asthma in the US collaborative perinatal project, 1959–1965. Am. J. Epidemiol. 2013, 178, 1691–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzniewicz, M.W.; Niki, H.; Walsh, E.M.; McCulloch, C.E.; Newman, T.B. Hyperbilirubinemia, phototherapy, and childhood asthma. Pediatrics 2018, 142, e20180662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safar, H.; Elsary, A.Y. Neonatal Jaundice: The Other Side of the Coin in the Development of Allergy. Am. J. Perinatol. 2020, 37, 1357–1363. [Google Scholar] [CrossRef] [PubMed]
- Kuniyoshi, Y.; Tsujimoto, Y.; Banno, M.; Taito, S.; Ariie, T. Neonatal jaundice, phototherapy and childhood allergic diseases: An updated systematic review and meta-analysis. Pediatr. Allergy Immunol. 2021, 32, 690–701. [Google Scholar] [CrossRef]
- McNamee, M.B.; Cardwell, C.R.; Patterson, C.C. Neonatal jaundice is associated with a small increase in the risk of childhood type 1 diabetes: A meta-analysis of observational studies. Acta Diabetol. 2012, 49, 83–87. [Google Scholar] [CrossRef]
- Tunç, G.; Uzun Çiçek, A.; Kiliçbay, F. Risk of autism spectrum disorder in children with a history of hospitalization for neonatal jaundice. Turk. J. Med. Sci. 2021, 51, 2657–2665. [Google Scholar] [CrossRef]
- Fazlollahi, M.; Chun, Y.; Grishin, A.; Wood, R.A.; Burks, A.W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.A.; Sicherer, S.H.; et al. Early-life gut microbiome and egg allergy. Allergy 2018, 73, 1515–1524. [Google Scholar] [CrossRef]
- Simonyte Sjodin, K.; Hammarstrom, M.L.; Ryden, P.; Sjodin, A.; Hernell, O.; Engstrand, L.; West, C.E. Temporal and long-term gut microbiota variation in allergic disease: A prospective study from infancy to school age. Allergy 2019, 74, 176–185. [Google Scholar] [CrossRef]
- Yamagishi, M.; Akagawa, S.; Akagawa, Y.; Nakai, Y.; Yamanouchi, S.; Kimata, T.; Hashiyada, M.; Akane, A.; Tsuji, S.; Kaneko, K. Decreased butyric acid-producing bacteria in gut microbiota of children with egg allergy. Allergy 2021, 76, 2279–2282. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Kaser, A. Gut microbiome, obesity, and metabolic dysfunction. J. Clin. Investig. 2011, 121, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuji, H.; Matsuda, K.; Nomoto, K. Counting the countless: Bacterial quantification by targeting rRNA molecules to explore the human gut microbiota in health and disease. Front. Microbiol. 2018, 9, 1417. [Google Scholar] [CrossRef] [Green Version]
- Akagawa, S.; Tsuji, S.; Onuma, C.; Akagawa, Y.; Yamaguchi, T.; Yamagishi, M.; Yamanouchi, S.; Kimata, T.; Sekiya, S.I.; Ohashi, A.; et al. Effect of delivery mode and nutrition on gut microbiota in neonates. Ann. Nutr. Metab. 2019, 74, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019, 574, 117–121. [Google Scholar] [CrossRef]
- Skar, V.; Skar, A.G.; Strømme, J.H. Beta-glucuronidase activity related to bacterial growth in common bile duct bile in gallstone patients. Scand. J. Gastroenterol. 1988, 23, 83–90. [Google Scholar] [CrossRef]
- An, H.M.; Park, S.Y.; Lee, D.K.; Kim, J.R.; Cha, M.K.; Lee, S.W.; Lim, H.T.; Kim, K.J.; Ha, N.J. Antiobesity and lipid-lowering effects of Bifidobacterium spp. in high fat diet-induced obese rats. Lipids Health Dis. 2011, 10, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobles, C.L.; Green, S.I.; Maresso, A.W. A product of heme catabolism modulates bacterial function and survival. PLoS Pathog. 2013, 9, e1003507. [Google Scholar] [CrossRef] [Green Version]
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; Levan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191. [Google Scholar] [CrossRef] [Green Version]
- Nagpal, R.; Kurakawa, T.; Tsuji, H.; Takahashi, T.; Kawashima, K.; Nagata, S.; Nomoto, K.; Yamashiro, Y. Evolution of gut Bifidobacterium population in healthy Japanese infants over the first three years of life: A quantitative assessment. Sci. Rep. 2017, 7, 10097. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faul, F.; Erdfelder, E.; Lang, A.G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]


| Characteristics | Group NJ (n = 26) | Group HC (n = 17) | p-Value |
|---|---|---|---|
| Sex, male (%) | 14 (54%) | 10 (59%) | 0.75 |
| Gestational age (days) | 272 (267–275) | 275 (269–281) | 0.22 |
| Birth weight (g) | 3118 (2773–3385) | 3255 (3094–3410) | 0.16 |
| Delivery mode, vaginal delivery (%) | 26 (100%) | 17 (100%) | 1.00 |
| Nutrition, breastfed (%) | 26 (100%) | 17 (100%) | 1.00 |
| Use of antibiotics after birth (%) | 0 (0%) | 0 (%) | 1.00 |
| Apgar score 1 min | 8 (8–9) | 8 (8–9) | 0.51 |
| Apgar score 5 min | 9 (9–9) | 9 (9–9) | 0.06 |
| Maternal age (years) | 34 (29–38) | 32 (30–35) | 0.67 |
| Gravidity | 1 (1–2) | 2 (1–2) | 0.11 |
| Parity | 0 (0–1) | 1 (0–1) | 0.07 |
| Premature rupture of the membrane | 0 (0%) | 0 (%) | 1.00 |
| Use of antibiotics by mothers during the four weeks prior to delivery | 0 (0%) | 0 (%) | 1.00 |
| Maternal blood type | |||
| A | 17 (65%) | 11 (65%) | 0.96 |
| B | 6 (23%) | 5 (29%) | 0.70 |
| AB | 3 (12%) | 1 (6%) | 0.53 |
| O | 0 (0%) | 0 (0%) | 1.00 |
| Day 4 serum bilirubin (mg/dL) | 16.0 (15.5–16.8) | 7.4 (6.8–8.3) | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akagawa, S.; Akagawa, Y.; Yamanouchi, S.; Teramoto, Y.; Yasuda, M.; Fujishiro, S.; Kino, J.; Hirabayashi, M.; Mine, K.; Kimata, T.; et al. Association of Neonatal Jaundice with Gut Dysbiosis Characterized by Decreased Bifidobacteriales. Metabolites 2021, 11, 887. https://doi.org/10.3390/metabo11120887
Akagawa S, Akagawa Y, Yamanouchi S, Teramoto Y, Yasuda M, Fujishiro S, Kino J, Hirabayashi M, Mine K, Kimata T, et al. Association of Neonatal Jaundice with Gut Dysbiosis Characterized by Decreased Bifidobacteriales. Metabolites. 2021; 11(12):887. https://doi.org/10.3390/metabo11120887
Chicago/Turabian StyleAkagawa, Shohei, Yuko Akagawa, Sohsaku Yamanouchi, Yoshiki Teramoto, Masahiro Yasuda, Sadayuki Fujishiro, Jiro Kino, Masato Hirabayashi, Kenji Mine, Takahisa Kimata, and et al. 2021. "Association of Neonatal Jaundice with Gut Dysbiosis Characterized by Decreased Bifidobacteriales" Metabolites 11, no. 12: 887. https://doi.org/10.3390/metabo11120887
APA StyleAkagawa, S., Akagawa, Y., Yamanouchi, S., Teramoto, Y., Yasuda, M., Fujishiro, S., Kino, J., Hirabayashi, M., Mine, K., Kimata, T., Hashiyada, M., Akane, A., Tsuji, S., & Kaneko, K. (2021). Association of Neonatal Jaundice with Gut Dysbiosis Characterized by Decreased Bifidobacteriales. Metabolites, 11(12), 887. https://doi.org/10.3390/metabo11120887






