Insights into the Metabolome of the Cyanobacterium Leibleinia gracilis from the Lagoon of Tahiti and First Inspection of Its Variability
Abstract
1. Introduction
2. Results
2.1. Major Metabolites from the Cyanobacterium Leibleinia gracilis
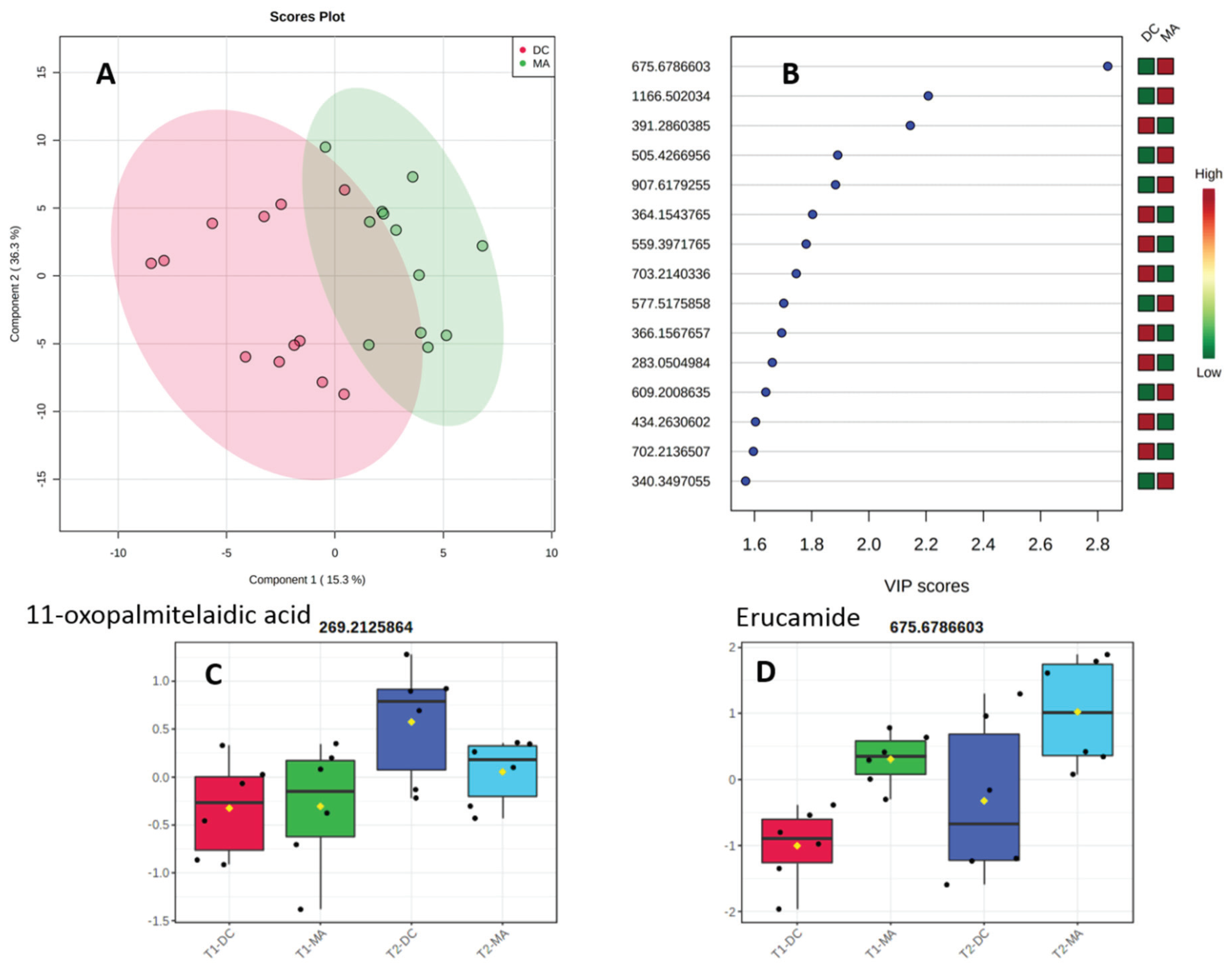
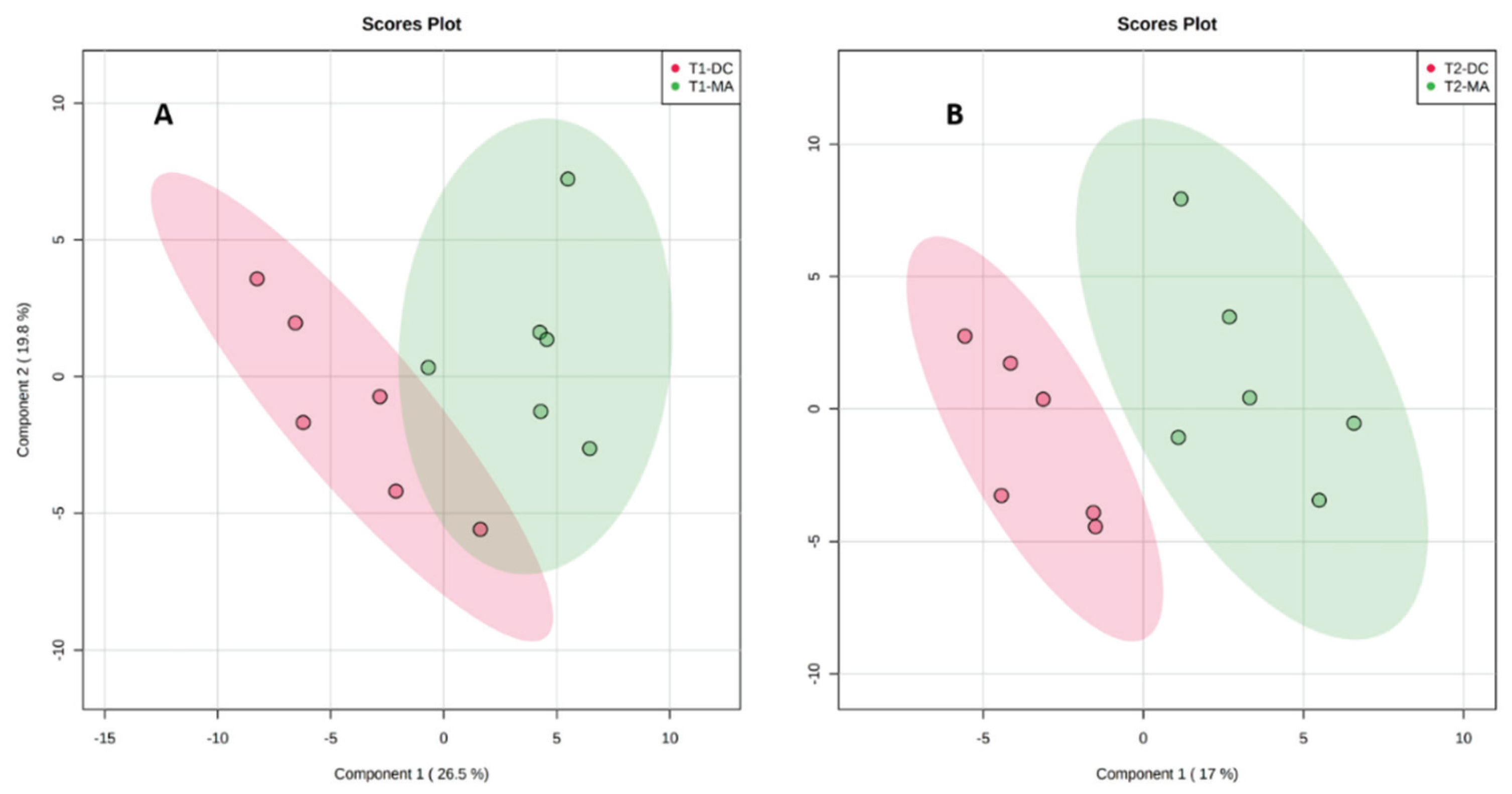
2.2. Comparative Metabolomics Study
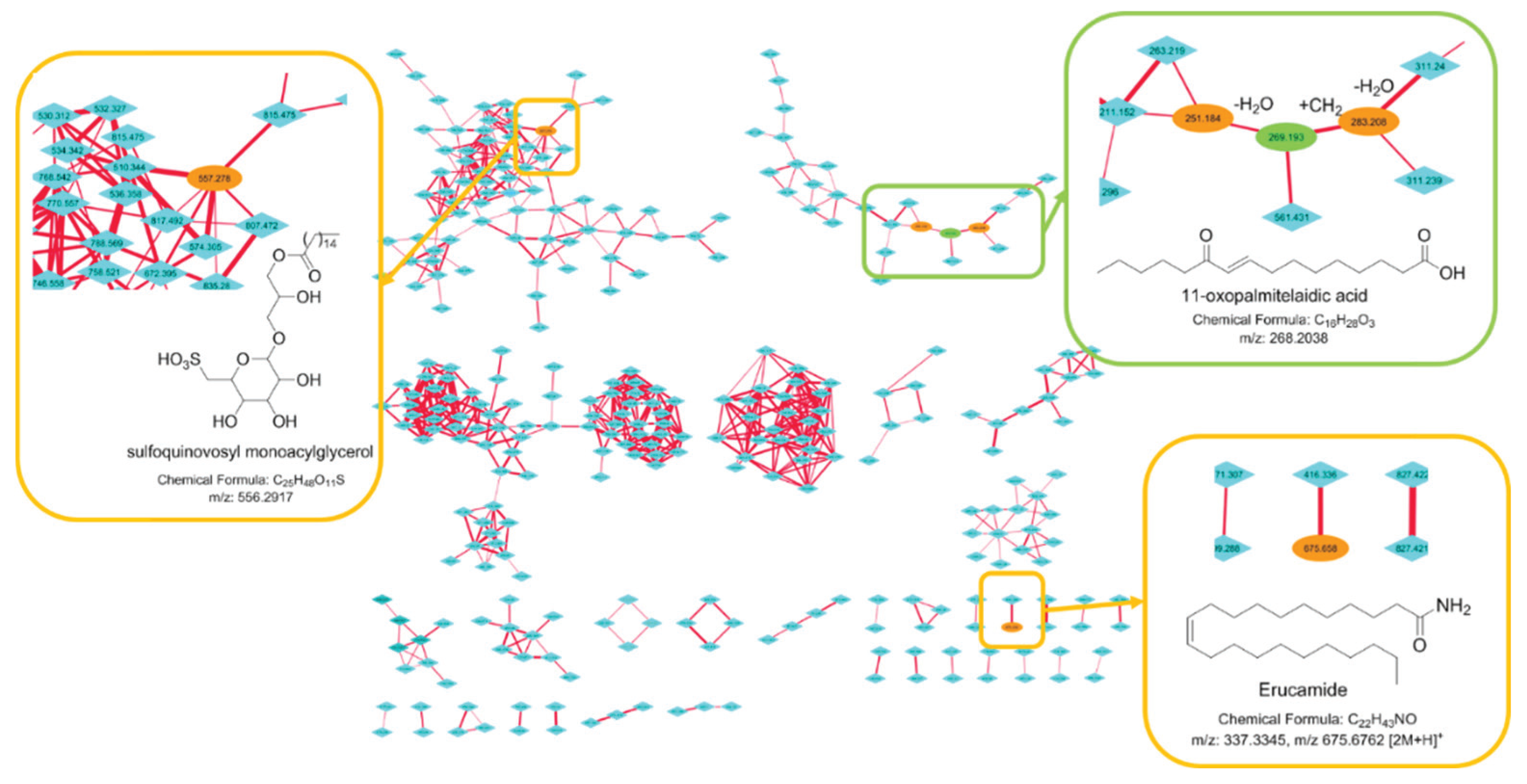
2.3. Molecular Networking
3. Discussion
4. Materials and Methods
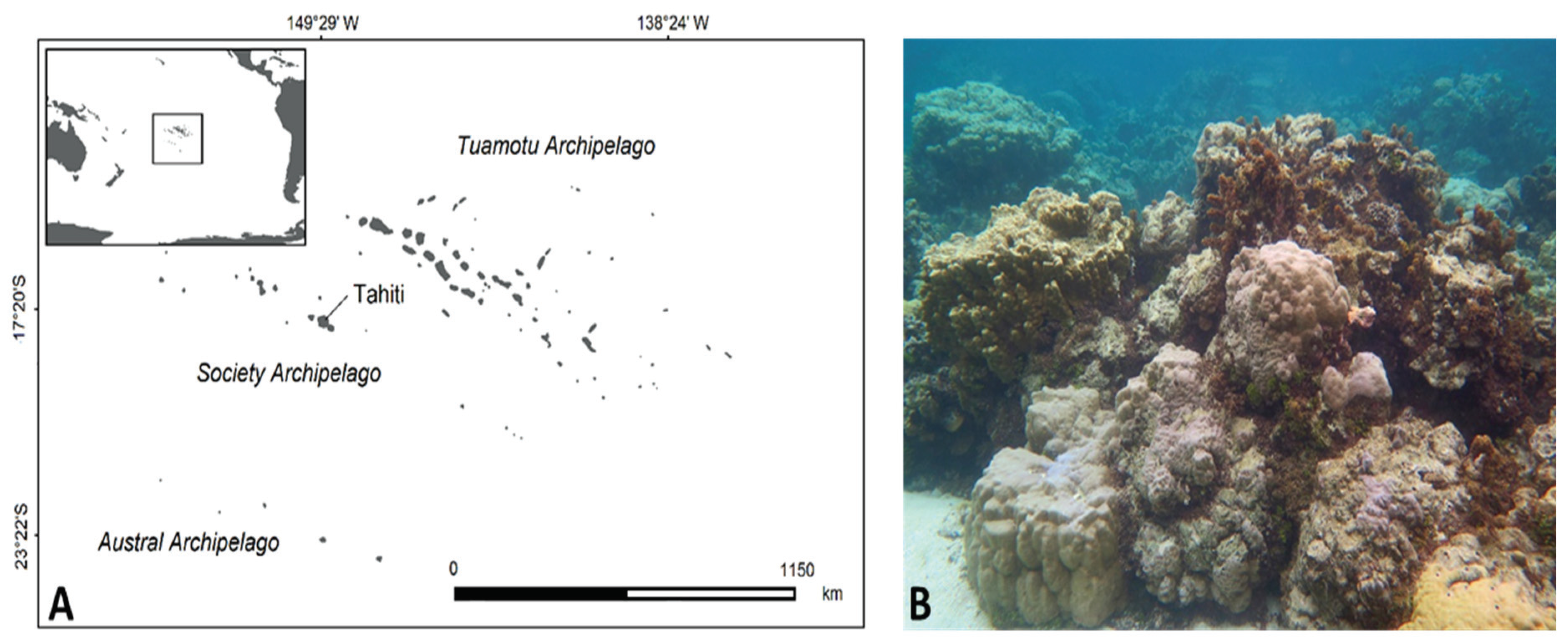
4.1. Study Site
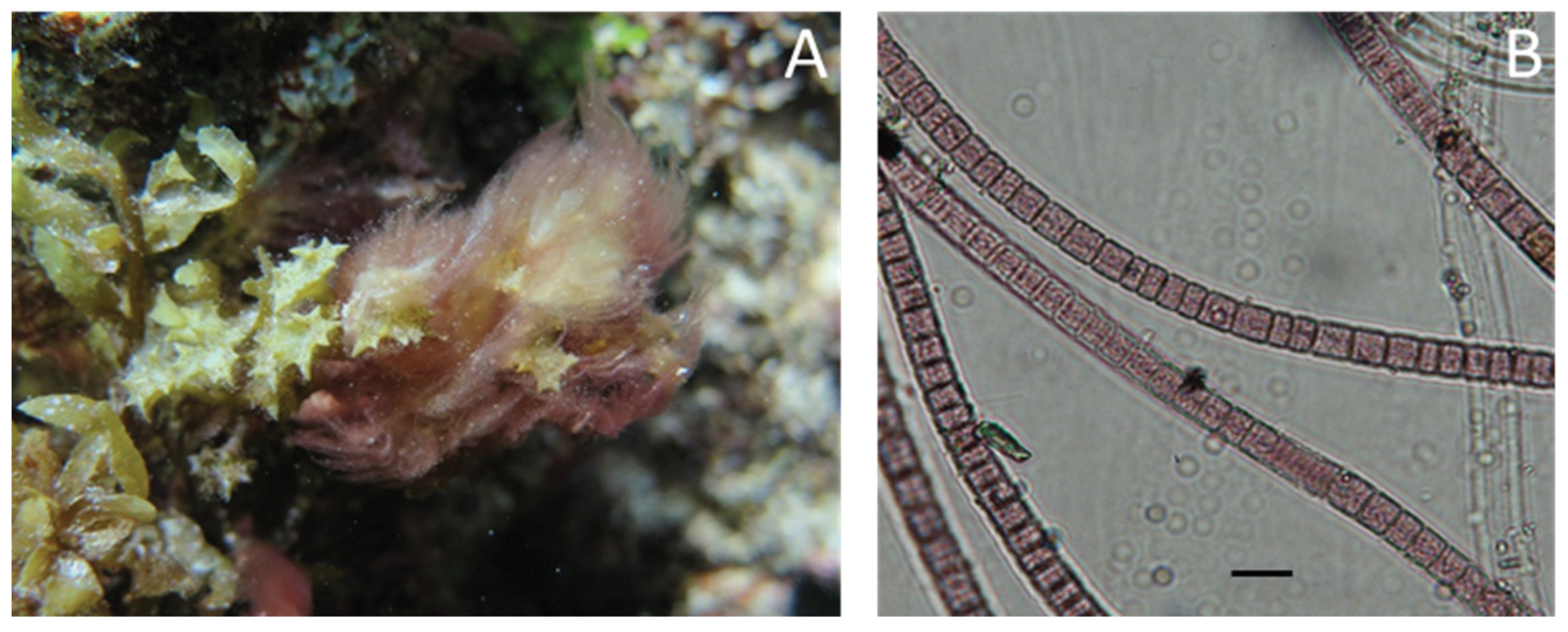
4.2. Biological Material
4.3. Sampling
4.4. Chemical Diversity of L. gracilis
4.5. Comparative Metabolomics and Molecular Metwork
4.5.1. Sample Preparation and Analyses
4.5.2. Data Analyses for the Metabolomic Study
4.5.3. Data Analyses for the Molecular network
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ford, A.; Bejarano, S.; Nugues, M.M.; Visser, P.; Albert, S.; Ferse, S.C.A. Reefs under Siege—the Rise, Putative Drivers, and Consequences of Benthic Cyanobacterial Mats. Front. Mar. Sci. 2018, 5, 18. [Google Scholar] [CrossRef]
- Charpy, L.; Casareto, B.E.; Langlade, M.J.; Suzuki, Y. Cyanobacteria in Coral Reef Ecosystems: A Review. J. Mar. Boil. 2012, 2012, 1–9. [Google Scholar] [CrossRef]
- O’Neil, J.; Davis, T.; Burford, M.A.; Gobler, C. The rise of harmful cyanobacteria blooms: The potential roles of eutrophication and climate change. Harmful Algae 2012, 14, 313–334. [Google Scholar] [CrossRef]
- Paerl, H.W.; Paul, V.J. Climate change: Links to global expansion of harmful cyanobacteria. Water Res. 2012, 46, 1349–1363. [Google Scholar] [CrossRef] [PubMed]
- Birrell, C.; McCook, L.J.; Willis, B.; Villamil-Diaz, L.M. Effects of Benthic Algae on the Replenishment of Corals and the Implications for the Resilience of Coral Reefs. Oceanogr. Mar. Biol. 2008, 46, 25–63. [Google Scholar]
- Kuffner, I.B.; Paul, V.J. Effects of the benthic cyanobacterium Lyngbya majuscula on larval recruitment of the reef corals Acropora surculosa and Pocillopora damicornis. Coral Reefs 2004, 23, 455–458. [Google Scholar] [CrossRef]
- Frias-Lopez, J.; Bonheyo, G.T.; Jin, Q.; Fouke, B.W. Cyanobacteria Associated with Coral Black Band Disease in Caribbean and Indo-Pacific Reefs. Appl. Environ. Microbiol. 2003, 69, 2409–2413. [Google Scholar] [CrossRef]
- Myers, J.L.; Sekar, R.; Richardson, L.L. Molecular Detection and Ecological Significance of the Cyanobacterial Genera Geitlerinema and Leptolyngbya in Black Band Disease of Corals. Appl. Environ. Microbiol. 2007, 73, 5173–5182. [Google Scholar] [CrossRef]
- Kramarsky-Winter, E.; Arotsker, L.; Rasoulouniriana, D.; Siboni, N.; Loya, Y.; Kushmaro, A. The Possible Role of Cyanobacterial Filaments in Coral Black Band Disease Pathology. Microb. Ecol. 2013, 67, 177–185. [Google Scholar] [CrossRef]
- Jones, A.C.; Monroe, E.A.; Eisman, E.B.; Gerwick, L.; Sherman, D.H.; Gerwick, W.H. The unique mechanistic transformations involved in the biosynthesis of modular natural products from marine cyanobacteria. Nat. Prod. Rep. 2010, 27, 1048–1065. [Google Scholar] [CrossRef]
- Leao, T.; Castelao, G.; Korobeynikov, A.; Monroe, E.A.; Podell, S.; Glukhov, E.; Allen, E.E.; Gerwick, W.H.; Gerwick, L. Comparative genomics uncovers the prolific and distinctive metabolic potential of the cyanobacterial genus Moorea. Proc. Natl. Acad. Sci. USA 2017, 114, 3198–3203. [Google Scholar] [CrossRef] [PubMed]
- Capper, A.; Erickson, A.A.; Ritson-Williams, R.; Becerro, M.A.; Arthur, K.A.; Paul, V.J. Palatability and chemical defences of benthic cyanobacteria to a suite of herbivores. J. Exp. Mar. Boil. Ecol. 2016, 474, 100–108. [Google Scholar] [CrossRef]
- Paul, D.G.N.A.V.J. Production of secondary metabolites by filamentous tropical marine cyanobacteria: Ecological functions of the compounds. J. Phycol. 1999, 35, 1412–1421. [Google Scholar] [CrossRef]
- Pennings, S.C.; Pablo, S.R.; Paul, V.J. Chemical defenses of the tropical, benthic marine cyanobacterium Hormothamnion enteromorphoides: Diverse consumers and synergisms. Limnol. Oceanogr. 1997, 42, 911–917. [Google Scholar] [CrossRef]
- Paul, V.J.; Thacker, R.W.; Banks, K.; Golubic, S. Benthic cyanobacterial bloom impacts the reefs of South Florida (Broward County, USA). Coral Reefs 2005, 24, 693–697. [Google Scholar] [CrossRef]
- Thacker, R.; Ginsburg, D.; Paul, V. Effects of herbivore exclusion and nutrient enrichment on coral reef macroalgae and cyanobacteria. Coral Reefs 2001, 19, 318–329. [Google Scholar] [CrossRef]
- Tsuda, R.T.; Kami, H.T. Algal Succession on Artifical Reefs in a Marine Lagoon environment in Guam. J. Phycol. 1973, 9, 260–264. [Google Scholar]
- Morrow, K.M.; Paul, V.J.; Liles, M.R.; Chadwick, N. Allelochemicals produced by Caribbean macroalgae and cyanobacteria have species-specific effects on reef coral microorganisms. Coral Reefs 2011, 30, 309–320. [Google Scholar] [CrossRef]
- Abed, R.M.; Golubić, S.; Garcia-Pichel, F.; Camoin, G.; Lee, S.-J. Identity and speciation in marine benthic cyanobacteria: The Phormidium-complex. Algological Stud. 2003, 109, 35–56. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Golubić, S.; Camoin, G.F.; Sprachta, S.; Garcia-Pichel, F. Characterization of microbialite-forming cyanobacteria in a tropical lagoon: Tikehau atoll, Tuamotu, French Polynesia. J. Phycol. 2003, 39, 862–873. [Google Scholar] [CrossRef]
- Palinska, K.A.; Abed, R.M.M.; Wendt, K.; Charpy, L.; Lotocka, M.; Golubic, S. Opportunistic Cyanobacteria in benthic microbial mats of a tropical lagoon, Tikehau Atoll, Tuamotu Archipelago: Minor in natural populations, major in cultures. Fottea 2012, 12, 127–140. [Google Scholar] [CrossRef]
- Zubia, M.; Vieira, C.; Palinska, K.A.; Roué, M.; Gaertner, J.-C.; Zloch, I.; Grellier, M.; Golubic, S. Benthic cyanobacteria on coral reefs of Moorea Island (French Polynesia): Diversity response to habitat quality. Hydrobiol. 2019, 843, 61–78. [Google Scholar] [CrossRef]
- Roué, M.; Gugger, M.; Golubić, S.; Amzil, Z.; Araoz, R.; Turquet, J.; Chinain, M.; Laurent, M. Marine Cyanotoxins Potentially Harmful to Human Health. In Outstanding Marine Molecules; Wiley: Hoboken, NJ, USA, 2014; pp. 1–22. [Google Scholar]
- Villeneuve, A.; Laurent, D.; Chinain, M.; Gugger, M.; Humbert, J.-F. Molecular Characterization of the Diversity and Potential Toxicity of Cynaobacteria Mats in Two Tropical Lagoons in the South Ocean. J. Phycol. 2012, 48, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Laurent, D.; Kerbrat, A.S.; Darius, H.; Rossi, F.; Yeeting, B.; Haddad, M.; Pauillac, S.; Chinain, M. Ciguatera Shellfish Poisoning (CSP), a new ecotoxicological phenomenon. In Cyanobacteria to Humans via Giant clams; Jensen, M.A., Muller, D.W., Eds.; Nova Science: New York, NY, USA, 2012; pp. 1–44. [Google Scholar]
- Bornancin, L.; Boyaud, F.; Mahiout, Z.; Bonnard, I.; Mills, S.C.; Banaigs, B.; Inguimbert, N. Isolation and Synthesis of Laxaphycin B-Type Peptides: A Case Study and Clues to Their Biosynthesis. Mar. Drugs 2015, 13, 7285–7300. [Google Scholar] [CrossRef]
- Bonnard, I.; Rolland, M.; Salmon, J.-M.; Debiton, E.; Barthomeuf, C.; Banaigs, B. Total Structure and Inhibition of Tumor Cell Proliferation of Laxaphycins. J. Med. Chem. 2007, 50, 1266–1279. [Google Scholar] [CrossRef]
- Bornancin, L.; Alonso, E.; Alvariño, R.; Inguimbert, N.; Bonnard, I.; Botana, L.M.; Banaigs, B. Structure and biological evaluation of new cyclic and acyclic laxaphycin-A type peptides. Bioorganic Med. Chem. 2019, 27, 1966–1980. [Google Scholar] [CrossRef]
- Levert, A.; Alvariño, R.; Bornancin, L.; Mansour, E.A.; Burja, A.M.; Genevière, A.-M.; Bonnard, I.; Alonso, E.; Botana, L.; Banaigs, B. Structures and Activities of Tiahuramides A–C, Cyclic Depsipeptides from a Tahitian Collection of the Marine Cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2018, 81, 1301–1310. [Google Scholar] [CrossRef]
- Mynderse, J.S.; E Moore, R.; Kashiwagi, M.; Norton, T.R. Antileukemia activity in the Osillatoriaceae: Isolation of Debromoaplysiatoxin from Lyngbya. Science 1977, 196, 538–540. [Google Scholar] [CrossRef]
- Higa, T.; Yoshida, W.Y. Louludinium Chloride, an Azabicyclononane Alkaloid from the Marine Cyanophyte Lyngbya gracilis. Heterocycles 1998, 47, 1023. [Google Scholar] [CrossRef]
- Pereira, A.; Cao, Z.; Murray, T.F.; Gerwick, W.H. Hoiamide A, a Sodium Channel Activator of Unusual Architecture from a Consortium of Two Papua New Guinea Cyanobacteria. Chem. Biol. 2009, 16, 893–906. [Google Scholar] [CrossRef]
- Dixson, D.; Abrego, D.; Hay, M. Chemically mediated behavior of recruiting corals and fishes: A tipping point that may limit reef recovery. Science 2014, 345, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Vieira, C.; Thomas, O.P.; Culioli, G.; Genta-Jouve, G.; Houlbreque, F.; Gaubert, J.; De Clerck, O.; Payri, C. Allelopathic interactions between the brown algal genus Lobophora (Dictyotales, Phaeophyceae) and scleractinian corals. Sci. Rep. 2016, 6, 18637. [Google Scholar] [CrossRef] [PubMed]
- Greff, S.; Aires, T.; Serrão, E.Á.; Engelen, A.; Thomas, O.P.; Pérez, T. The interaction between the proliferating macroalga Asparagopsis taxiformis and the coral Astroides calycularis induces changes in microbiome and metabolomic fingerprints. Sci. Rep. 2017, 7, 42625. [Google Scholar] [CrossRef] [PubMed]
- Stiger-Pouvreau, V.; Payri, C. Spatial and temporal patterns of settlement of the brown macroalgae Turbinaria ornata and Sargassum mangarevense in a coral reef on Tahiti. Mar. Ecol. Prog. Ser. 1999, 191, 91–100. [Google Scholar] [CrossRef]
- Stiger-Pouvreau, V.; Payri, C. Natural settlement dynamics of a young population of Turbinaria ornata and phenological comparisons with older populations. Aquat. Bot. 2005, 81, 225–243. [Google Scholar] [CrossRef]
- Boada, L.D.; Zumbado, M.; Luzardo, O.P.; González, M.A.; Plakas, S.M.; Granade, H.R.; Abraham, A.; Jester, E.L.; Dickey, R.W. Ciguatera fish poisoning on the West Africa Coast: An emerging risk in the Canary Islands (Spain). Toxicon 2010, 56, 1516–1519. [Google Scholar] [CrossRef] [PubMed]
- Via, C.W.; Glukhov, E.; Costa, S.; Zimba, P.V.; Moeller, P.D.R.; Gerwick, W.H.; Bertin, M.J. The Metabolome of a Cyanobacterial Bloom Visualized by MS/MS-Based Molecular Networking Reveals New Neurotoxic Smenamide Analogs (C, D, and E). Front. Chem. 2018, 6. [Google Scholar] [CrossRef]
- Ding, C.Y.G.; Pang, L.M.; Liang, Z.; Goh, K.K.K.; Glukhov, E.; Gerwick, W.H.; Tan, L.T. MS/MS-Based Molecular Networking Approach for the Detection of Aplysiatoxin-Related Compounds in Environmental Marine Cyanobacteria. Mar. Drugs 2018, 16, 505. [Google Scholar] [CrossRef]
- Winnikoff, J.R.; Glukhov, E.; Watrous, J.; Dorrestein, P.C.; Gerwick, W.H. Quantitative molecular networking to profile marine cyanobacterial metabolomes. J. Antibiot. 2013, 67, 105–112. [Google Scholar] [CrossRef]
- Le Manach, S.; Duval, C.; Marie, A.; Djediat, C.; Catherine, A.; Edery, M.; Bernard, C.; Marie, B. Global Metabolomic Characterizations of Microcystis spp. Highlights Clonal Diversity in Natural Bloom-Forming Populations and Expands Metabolite Structural Diversity. Front. Microbiol. 2019, 10, 791. [Google Scholar] [CrossRef]
- Jones, M.R.; Pinto, E.; Torres, M.A.; Doerr, F.; Mazur-Marzec, H.; Szubert, K.; Tartaglione, L.; Dell’Aversano, C.; Miles, C.O.; Beach, D.G.; et al. Comprehensive database of secondary metabolites from cyanobacteria. Int. J. Mol. Sci. 2020, 21, 656. [Google Scholar] [CrossRef]
- Zhu, Z.-J.; Schultz, A.W.; Wang, J.; Johnson, C.H.; Yannone, S.M.; Patti, G.J.; Siuzdak, G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 2013, 8, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Getachew, P.; Joo, J.; Choi, Y.S.; Hwang, D.S.; Hong, Y.-K. The slip agents oleamide and erucamide reduce biofouling by marine benthic organisms (diatoms, biofilms and abalones). Toxicol. Environ. Heal. Sci. 2016, 8, 341–348. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Moore, B.S. Lessons from the Past and Charting the Future of Marine Natural Products Drug Discovery and Chemical Biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, N.D.; Watrous, J.; A Kapono, C.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Bai, L.; Cui, X.; Cheng, N.; Cao, W.; Wu, Y.; Guo, S.; Zhang, L.; Ho, C.-T.; Bai, N. Hepatoprotective standardized EtOH–water extract of the leaves of Ziziphus jujuba. Food Funct. 2017, 8, 816–822. [Google Scholar] [CrossRef]
- Los, D.A.; Mironov, K.S. Modes of Fatty Acid Desaturation in Cyanobacteria: An Update. Life 2015, 5, 554–567. [Google Scholar] [CrossRef]
- Roulland, E.; Solanki, H.; Calabro, K.; Zubia, M.; Genta-Jouve, G.; Thomas, O.P. Stereochemical Study of Puna’auic Acid, an Allenic Fatty Acid from the Eastern Indo-Pacific Cyanobacterium Pseudanabaena sp. Org. Lett. 2018, 20, 2311–2314. [Google Scholar] [CrossRef]
- Kwan, J.C.; Meickle, T.; Ladwa, D.; Teplitski, M.; Paul, V.; Luesch, H. Lyngbyoic acid, a "tagged" fatty acid from a marine cyanobacterium, disrupts quorum sensing in Pseudomonas aeruginosa. Mol. BioSyst. 2011, 7, 1205–1216. [Google Scholar] [CrossRef]
- Schwarz, D.; Orf, I.; Kopka, J.; Hagemann, M. Recent Applications of Metabolomics Toward Cyanobacteria. Metabolites 2013, 3, 72–100. [Google Scholar] [CrossRef]
- Tiam, S.K.; Gugger, M.; DeMay, J.; Le Manach, S.; Duval, C.; Bernard, C.; Marie, B. Insights into the Diversity of Secondary Metabolites of Planktothrix Using a Biphasic Approach Combining Global Genomics and Metabolomics. Toxins 2019, 11, 498. [Google Scholar] [CrossRef] [PubMed]
- Baran, R.; Ivanova, N.N.; Jose, N.; Garcia-Pichel, F.; Kyrpides, N.C.; Gugger, M.; Northen, T.R. Functional Genomics of Novel Secondary Metabolites from Diverse Cyanobacteria Using Untargeted Metabolomics. Mar. Drugs 2013, 11, 3617–3631. [Google Scholar] [CrossRef] [PubMed]
- Kleigrewe, K.; Almaliti, J.; Tian, I.Y.; Kinnel, R.B.; Korobeynikov, A.; Monroe, E.A.; Duggan, B.M.; Di Marzo, V.; Sherman, D.H.; Dorrestein, P.C.; et al. Combining Mass Spectrometric Metabolic Profiling with Genomic Analysis: A Powerful Approach for Discovering Natural Products from Cyanobacteria. J. Nat. Prod. 2015, 78, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Thacker, R.W.; Paul, V.J. Are benthic cyanobacteria indicators of nutrient enrichment? Relationships between cyanobacterial abundance and environmental factors on the reef flats of guam. Bull. Mar. Sci. 2001, 69, 497–508. [Google Scholar]
- Cai, W.; Matthews, J.H.; Paul, V.J.; Luesch, H. Pitiamides A and B, Multifunctional Fatty Acid Amides from Marine Cyanobacteria. Planta Medica 2016, 82, 897–902. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Solanki, H. Metabolomic Study on the cyanobacterium Leibleinia gracilis. Available online: https://www.ebi.ac.uk/metabolights/editor/www.ebi.ac.uk/metabolights/MTBLS1462 (accessed on 20 April 2020).
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
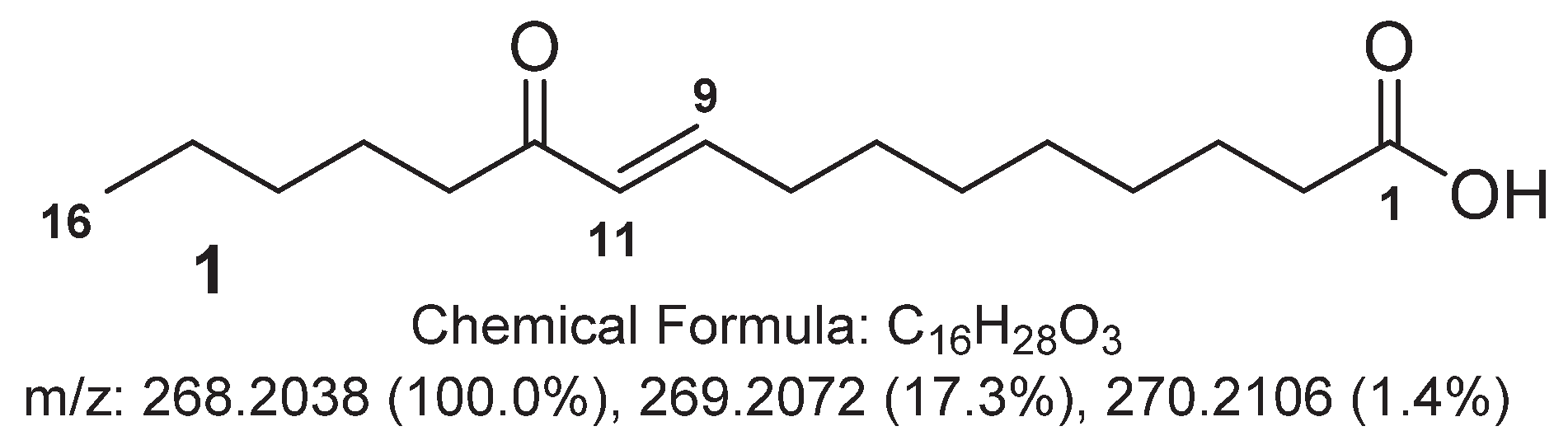

| Position | δC, mult. | δH, mult. (J in Hz) |
|---|---|---|
| 1 | 177.8, qC | - |
| 2 | 34.8, CH2 | 2.29, t (7.0) |
| 3 | 25.8, CH2 | 1.61, m |
| 4 | 29.9, CH2 | 1.38, m |
| 5 | 30.0, CH2 | 1.31, m |
| 6 | 29.9, CH2 | 1.38, m |
| 7 | 29.0, CH2 | 1.51, quint (7.0) |
| 8 | 33.4, CH2 | 2.26, q (7.0) |
| 9 | 149.7, CH | 6.91, dt (16.0, 7.0) |
| 10 | 131.3, CH | 6.11, d (16.0) |
| 11 | 203.8, qC | - |
| 12 | 40.6, CH2 | 2.59, t (7.0) |
| 13 | 25.5, CH2 | 1.58, m |
| 14 | 32.8, CH2 | 1.31, m |
| 15 | 23.5, CH2 | 1.32, m |
| 16 | 14.3, CH3 | 0.91, t (7.0) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solanki, H.; Pierdet, M.; Thomas, O.P.; Zubia, M. Insights into the Metabolome of the Cyanobacterium Leibleinia gracilis from the Lagoon of Tahiti and First Inspection of Its Variability. Metabolites 2020, 10, 215. https://doi.org/10.3390/metabo10050215
Solanki H, Pierdet M, Thomas OP, Zubia M. Insights into the Metabolome of the Cyanobacterium Leibleinia gracilis from the Lagoon of Tahiti and First Inspection of Its Variability. Metabolites. 2020; 10(5):215. https://doi.org/10.3390/metabo10050215
Chicago/Turabian StyleSolanki, Hiren, Manon Pierdet, Olivier P. Thomas, and Mayalen Zubia. 2020. "Insights into the Metabolome of the Cyanobacterium Leibleinia gracilis from the Lagoon of Tahiti and First Inspection of Its Variability" Metabolites 10, no. 5: 215. https://doi.org/10.3390/metabo10050215
APA StyleSolanki, H., Pierdet, M., Thomas, O. P., & Zubia, M. (2020). Insights into the Metabolome of the Cyanobacterium Leibleinia gracilis from the Lagoon of Tahiti and First Inspection of Its Variability. Metabolites, 10(5), 215. https://doi.org/10.3390/metabo10050215






