Identification of Bioactive Phytochemicals in Mulberries
Abstract
:1. Introduction
2. Results
2.1. Identification of Phytochemicals in Morus using LC-PDA-Orbitrap FTMS
2.2. Global Metabolome Differences between Morus Alba and Morus Nigra Fruits
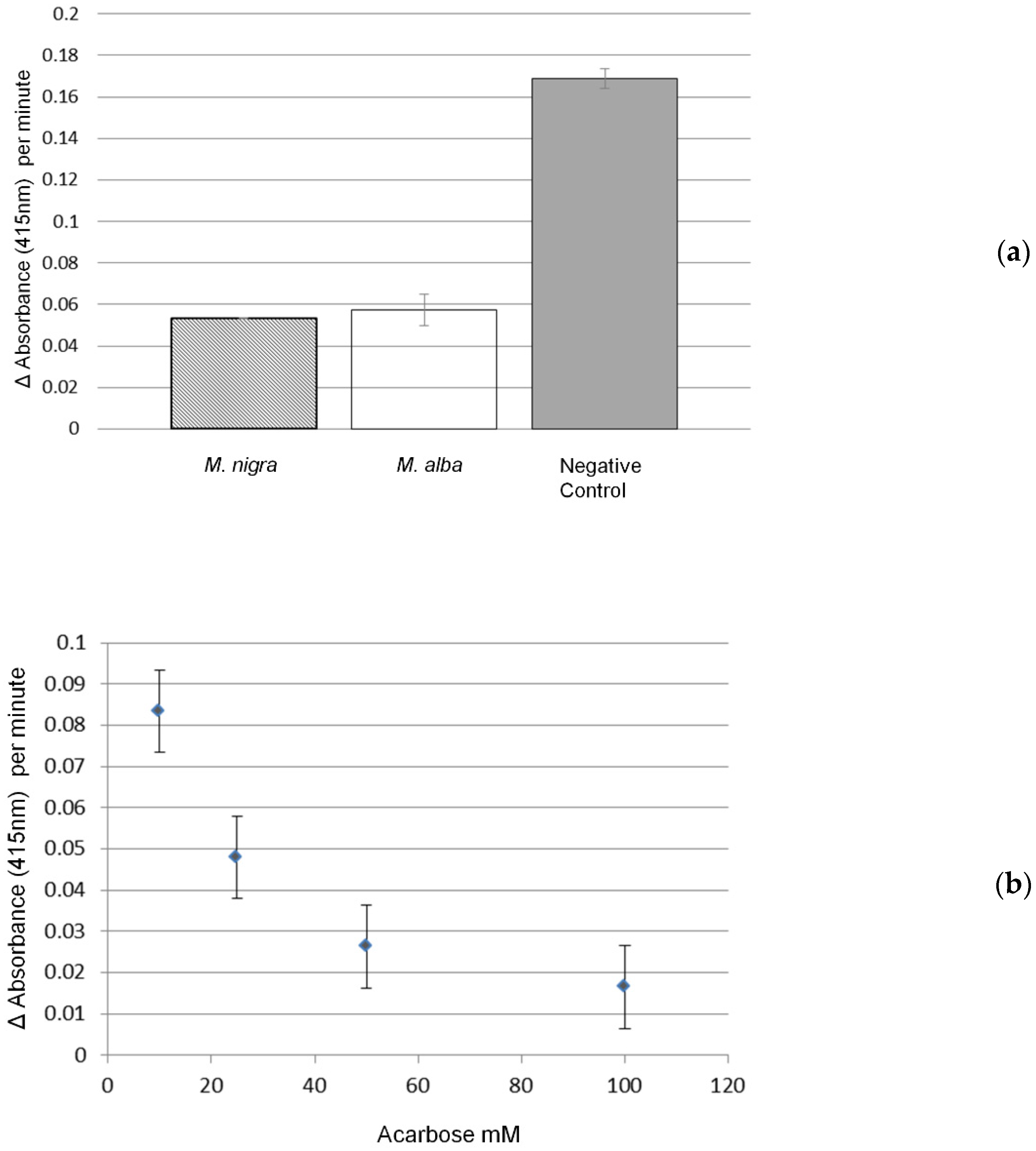
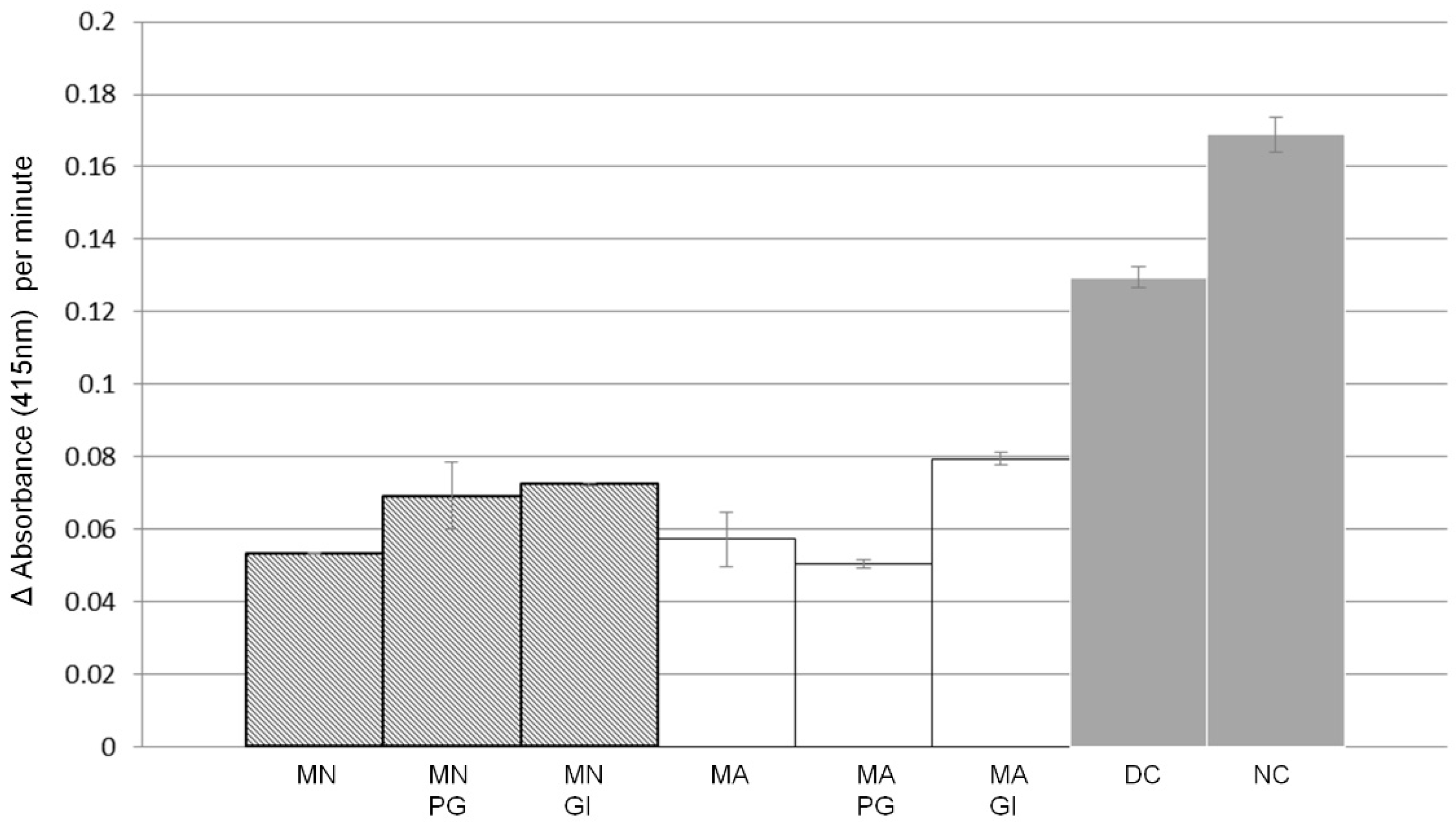
2.3. α-Glucosidase Inhibitory Activity and Effect of In Vitro Gastrointestinal Digestion
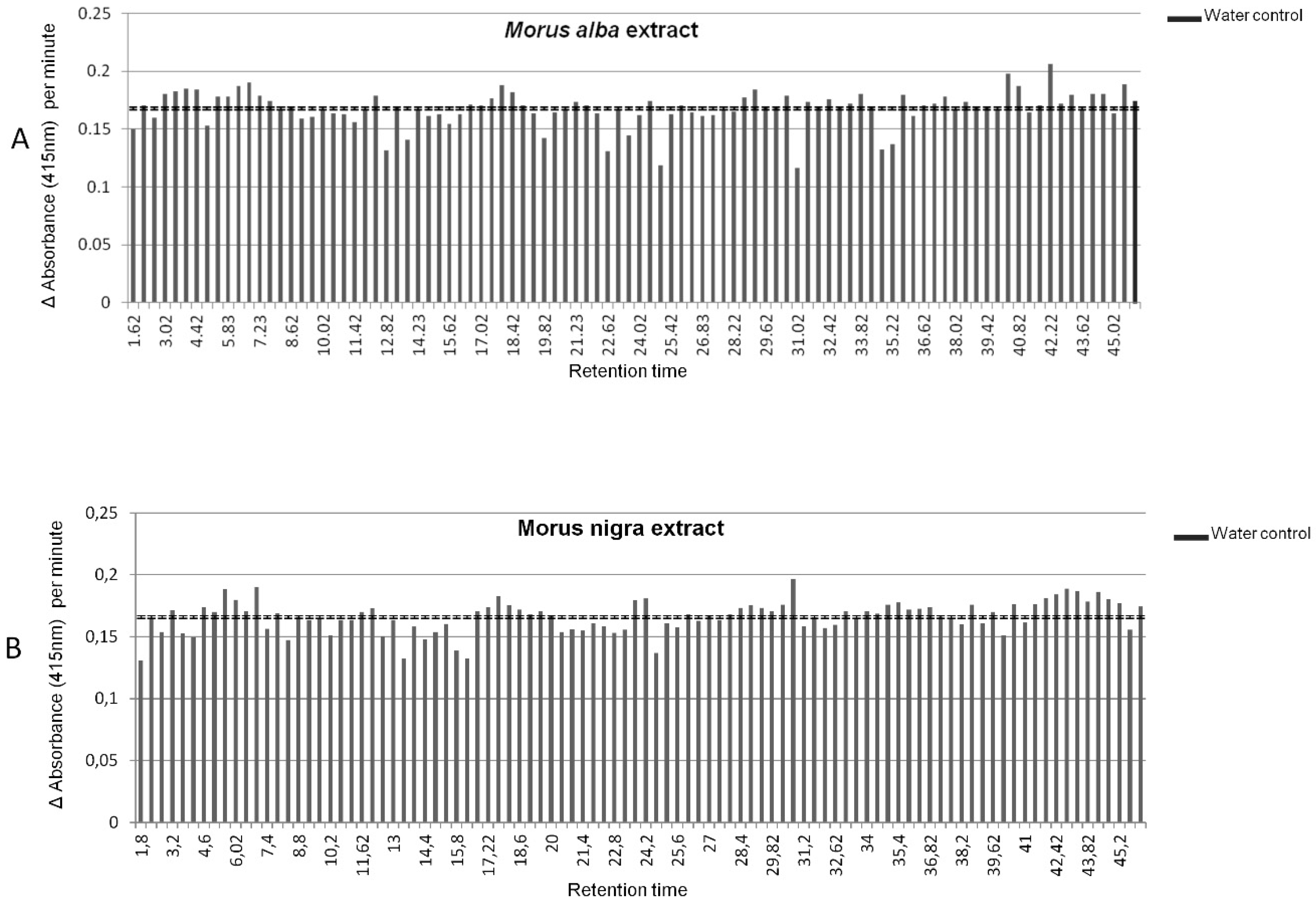
2.4. LCMS Combined with 96-Well Format Fractionation
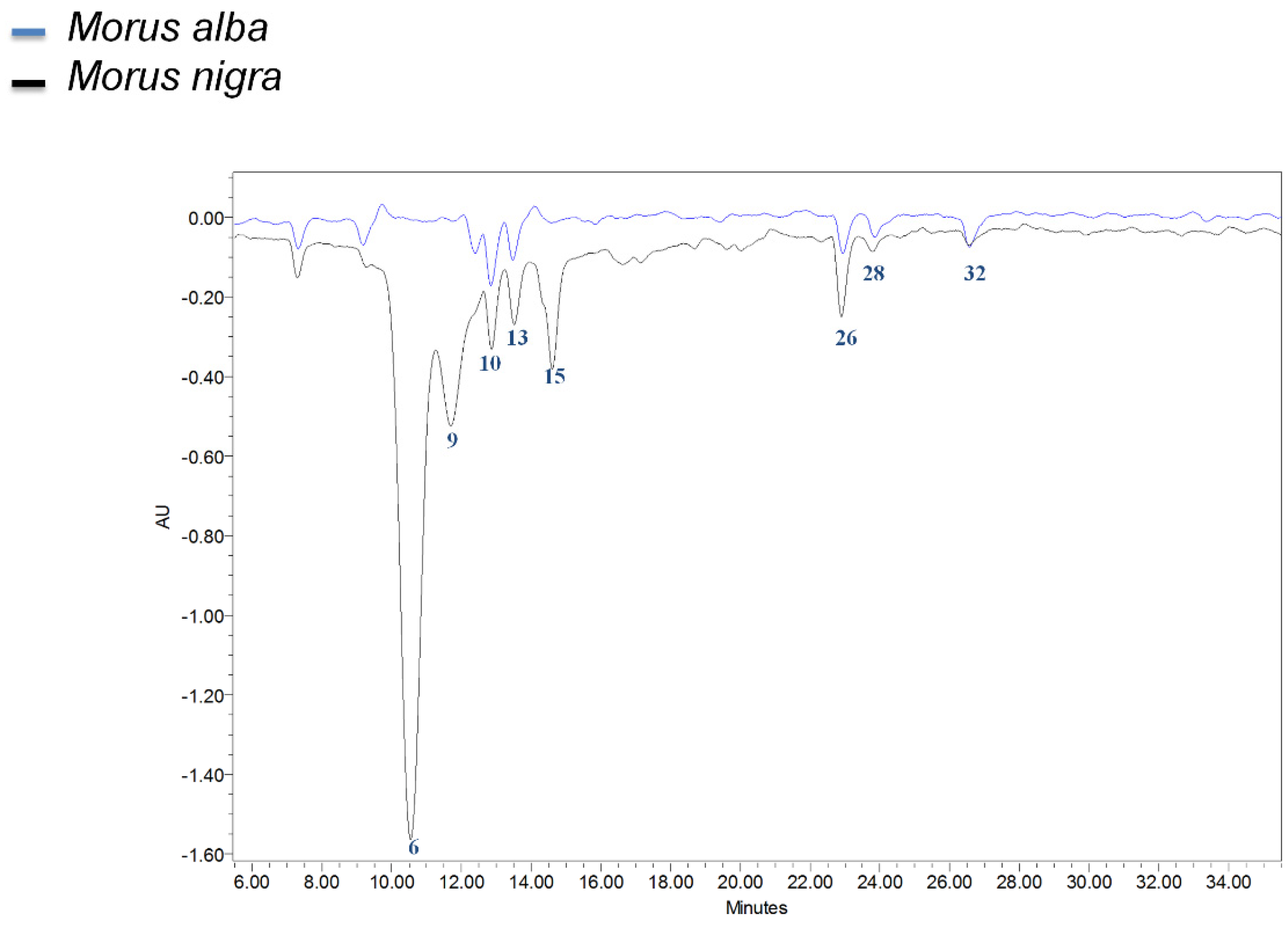
2.5. Total Antioxidant Activity and HPLC with Online Antioxidant Detection
3. Discussion
4. Materials and Methods
4.1. Mulberry Materials
4.2. Extract Preparation
4.3. LC-PDA-Orbitrap FTMS Analysis
4.4. LCMS data Processing and Multivariate Analysis
4.5. In Vitro Simulated Gastrointestinal Digestion
4.6. α-Glucosidase Inhibition Assay
4.7. NanoMate LC-Fractionation of Extracts
4.8. Antioxidant Activity and HPLC Analysis with Online Antioxidant Detection
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ercisli, S.; Orhan, E. Chemical composition of white (Morus alba), red (Morus rubra) and black (Morus nigra) mulberry fruits. Food Chem. 2007, 103, 1380–1384. [Google Scholar] [CrossRef]
- Gundogdu, M.; Muradoglu, F.; Sensoy, R.I.G.; Yilmaz, H. Determination of fruit chemical properties of Morus nigra L., Morus alba L. and Morus rubra L. by HPLC. Sci. Hortic Amst. 2011, 132, 37–41. [Google Scholar] [CrossRef]
- Liang, L.H.; Wu, X.Y.; Zhao, T.; Zhao, J.L.; Li, F.; Zou, Y.; Mao, G.H.; Yang, L.Q. In vitro bioaccessibility and antioxidant activity of anthocyanins from mulberry (Morus atropurpurea Roxb.) following simulated gastro-intestinal digestion. Food Res. Int. 2012, 46, 76–82. [Google Scholar] [CrossRef]
- Banu, S.; Jabir, N.R.; Manjunath, N.C.; Khan, M.S.; Ashraf, G.M.; Kamal, M.A.; Tabrez, S. Reduction of post-prandial hyperglycemia by mulberry tea in type-2 diabetes patients. Saudi J. Biol. Sci. 2015, 22, 32–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.; Lu, Y.H. Comparison of Inhibitory Activities and Mechanisms of Five Mulberry Plant Bioactive Components against alpha-Glucosidase. J. Agric. Food Chem. 2013, 61, 8110–8119. [Google Scholar] [CrossRef]
- Rodriguez-Sanchez, S.; Quintanilla-Lopez, J.E.; Soria, A.C.; Sanz, M.L. Evaluation of different hydrophilic stationary phases for the simultaneous determination of iminosugars and other low molecular weight carbohydrates in vegetable extracts by liquid chromatography tandem mass spectrometry. J. Chromatogr. A 2014, 1372, 81–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kongstad, K.T.; Ozdemir, C.; Barzak, A.; Wubshet, S.G.; Staerk, D. Combined Use of High-Resolution a-Glucosidase inhibition Profiling and High-Performance Liquid Chromatography High-Resolution. Mass Spectrometry Solid-Phase Extraction Nuclear Magnetic Resonance Spectroscopy for Investigation of Antidiabetic Principles in Crude Plant Extracts. J. Agric. Food Chem. 2015, 63, 2257–2263. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Salcedo, E.M.; Mena, P.; Garcia-Viguera, C.; Hernandez, F.; Martinez, J.J. (Poly)phenolic compounds and antioxidant activity of white (Morus alba) and black (Morus nigra) mulberry leaves: Their potential for new products rich in phytochemicals. J. Funct. Foods 2015, 18, 1039–1046. [Google Scholar] [CrossRef]
- Sanchez-Salcedo, E.M.; Mena, P.; Garcia-Viguera, C.; Martinez, J.J.; Hernandez, F. Phytochemical evaluation of white (Morus alba L.) and black (Morus nigra L.) mulberry fruits, a starting point for the assessment of their beneficial properties. J. Funct. Foods 2015, 12, 399–408. [Google Scholar] [CrossRef]
- Bao, T.; Xu, Y.; Gowd, V.; Zhao, J.C.; Xie, J.H.; Liang, W.K.; Chen, W. Systematic study on phytochemicals and antioxidant activity of some new and common mulberry cultivars in China. J. Funct. Foods 2016, 25, 537–547. [Google Scholar] [CrossRef]
- Kim, S.B.; Chang, B.Y.; Jo, Y.H.; Lee, S.H.; Han, S.B.; Hwang, B.Y.; Kim, S.Y.; Lee, M.K. Macrophage activating activity of pyrrole alkaloids from Morus alba fruits. J. Ethnopharmacol. 2013, 145, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.K.; Chou, F.P.; Chen, Y.C.; Chyau, C.C.; Ho, H.H.; Wang, C.J. Effects of Mulberry (Morus alba L.) Extracts on Lipid Homeostasis In Vitro and In Vivo. J. Agric. Food Chem. 2009, 57, 7605–7611. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Yang, L.; Zheng, H.Y. Hypolipidemic and antioxidant effects of mulberry (Morus alba L.) fruit in hyperlipidaemia rats. Food Chem. Toxicol. 2010, 48, 2374–2379. [Google Scholar] [CrossRef] [PubMed]
- Krishna, P.G.A.; Sivakumar, T.R.; Jin, C.; Li, S.H.; Weng, Y.J.; Yin, J.; Jia, J.Q.; Wang, C.Y.; Gui, Z.Z. Antioxidant and Hemolysis Protective Effects of Polyphenol-Rich Extract from Mulberry Fruits. Pharmacogn. Mag. 2018, 14, 103–109. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Asano, N.; Oseki, K.; Tomioka, E.; Kizu, H.; Matsui, K. N-Containing Sugars from Morus alba and Their Glycosidase Inhibitory Activities. Carbohydr. Res. 1994, 259, 243–255. [Google Scholar] [CrossRef]
- Natic, M.M.; Dabic, D.C.; Papetti, A.; Aksic, M.M.F.; Ognjanov, V.; Ljubojevic, M.; Tesic, Z.L. Analysis and characterisation of phytochemicals in mulberry (Morus alba L.) fruits grown in Vojvodina, North Serbia. Food Chem. 2015, 171, 128–136. [Google Scholar] [CrossRef]
- Kusano, G.; Orihara, S.; Tsukamoto, D.; Shibano, M.; Coskun, M.; Guvenc, A.; Erdurak, C.S. Five new nortropane alkaloids and six new amino acids from the fruit of Morus alba LINNE growing in Turkey. Chem. Pharm. Bull. 2002, 50, 185–192. [Google Scholar] [CrossRef] [Green Version]
- Van Der Hooft, J.J.J.; Vervoort, J.; Bino, R.J.; De Vos, R.C.H. Spectral trees as a robust annotation tool in LC-MS based metabolomics. Metabolomics 2012, 8, 691–703. [Google Scholar] [CrossRef]
- Yang, J.F.; Wen, H.C.; Zhang, L.; Zhang, X.X.; Fu, Z.; Li, J.M. The influence of ripening stage and region on the chemical compounds in mulberry fruits (Morus atropurpurea Roxb.) based on UPLC-QTOF-MS. Food Res. Int. 2017, 100, 159–165. [Google Scholar] [CrossRef]
- Llorach, R.; Martinez-Sanchez, A.; Tomas-Barberan, F.A.; Gil, M.I.; Ferreres, F. Characterisation of polyphenols and antioxidant properties of five lettuce varieties and escarole. Food Chem. 2008, 108, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Li, H.X.; Jo, E.; Myung, C.S.; Kim, Y.H.; Yang, S.Y. Lipolytic effect of compounds isolated from leaves of mulberry (Morus alba L.) in 3T3-L1 adipocytes. Nat. Prod. Res. 2018, 32, 1963–1966. [Google Scholar] [CrossRef] [PubMed]
- Paudel, L.; Wyzgoski, F.J.; Scheerens, J.C.; Chanon, A.M.; Reese, R.N.; Smiljanic, D.; Wesdemiotis, C.; Blakeslee, J.J.; Riedl, K.M.; Rinaldi, P.L. Nonanthocyanin Secondary Metabolites of Black Raspberry (Rubus occidentalis L.) Fruits: Identification by HPLC-DAD, NMR, HPLC-ESI-MS, and ESI-MS/MS Analyses. J. Agric. Food Chem. 2013, 61, 12032–12043. [Google Scholar] [CrossRef] [PubMed]
- Carazzone, C.; Mascherpa, D.; Gazzani, G.; Papetti, A. Identification of phenolic constituents in red chicory salads (Cichorium intybus) by high-performance liquid chromatography with diode array detection and electrospray ionisation tandem mass spectrometry. Food Chem. 2013, 138, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.C.; Park, C.H.; Yang, J.L.; Yeo, H.J.; Kim, T.J.; Kim, J.K.; Park, S.U. Molecular characterization of anthocyanin and betulinic acid biosynthesis in red and white mulberry fruits using high-throughput sequencing. Food Chem. 2019, 279, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Mena, P.; Sanchez-Salcedo, E.M.; Tassotti, M.; Martinez, J.J.; Hernandez, F.; Del Rio, D. Phytochemical evaluation of eight white (Morus alba L.) and black (Morus nigra L.) mulberry clones grown in Spain based on UHPLC-ESI-MSn metabolomic profiles. Food Res. Int. 2016, 89, 1116–1122. [Google Scholar] [CrossRef]
- Dugo, P.; Donato, P.; Cacciola, F.; Germano, M.P.; Rapisarda, A.; Mondello, L. Characterization of the polyphenolic fraction of Morus alba leaves extracts by HPLC coupled to a hybrid IT-TOF MS system. J. Sep. Sci. 2009, 32, 3627–3634. [Google Scholar] [CrossRef]
- Thabti, I.; Elfalleh, W.; Hannachi, H.; Ferchichi, A.; Campos, M.D. Identification and quantification of phenolic acids and flavonol glycosides in Tunisian Morus species by HPLC-DAD and HPLC-MS. J. Funct. Foods 2012, 4, 367–374. [Google Scholar] [CrossRef]
- Qi, X.W.; Shuai, Q.; Chen, H.; Fan, L.; Zeng, Q.W.; He, N.J. Cloning and expression analyses of the anthocyanin biosynthetic genes in mulberry plants. Mol. Genet. Genom. 2014, 289, 783–793. [Google Scholar] [CrossRef]
- Wang, H.; Cao, G.H.; Prior, R.L. Total antioxidant capacity of fruits. J. Agric. Food Chem. 1996, 44, 701–705. [Google Scholar] [CrossRef]
- De Vos, R.C.H.; Moco, S.; Lommen, A.; Keurentjes, J.J.B.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.W.; Xu, Y.; Xie, J.H.; Sun, C.D.; Zheng, X.D.; Chen, W. Systematic evaluation of phenolic compounds and protective capacity of a new mulberry cultivar J33 against palmitic acid-induced lipotoxicity using a simulated digestion method. Food Chem. 2018, 258, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Van Der Hooft, J.J.J.; De Vos, R.C.H.; Ridder, L.; Vervoort, J.; Bino, R.J. Structural elucidation of low abundant metabolites in complex sample matrices. Metabolomics 2013, 9, 1009–1018. [Google Scholar] [CrossRef]
- Lommen, A. MetAlign: Interface-Driven, Versatile Metabolomics Tool for Hyphenated Full-Scan Mass Spectrometry Data Preprocessing. Anal. Chem. 2009, 81, 3079–3086. [Google Scholar] [CrossRef] [PubMed]
- Tikunov, Y.M.; Laptenok, S.; Hall, R.D.; Bovy, A.; De Vos, R.C.H. MSClust: A tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics 2012, 8, 714–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDougall, G.J.; Dobson, P.; Smith, P.; Blake, A.; Stewart, D. Assessing potential bioavallability of raspberry anthocyanins using an in vitro digestion system. J. Agric. Food Chem. 2005, 53, 5896–5904. [Google Scholar] [CrossRef] [PubMed]
- Capanoglu, E.; Beekwilder, J.; Boyacioglu, D.; Hall, R.; De Vos, R. Changes in antioxidant and metabolite profiles during production of tomato paste. J. Agric. Food Chem. 2008, 56, 964–973. [Google Scholar] [CrossRef]
- Beekwilder, J.; Jonker, H.; Meesters, P.; Hall, R.D.; Van Der Meer, I.M.; De Vos, C.H.R. Antioxidants in raspberry: On-line analysis links antioxidant activity to a diversity of individual metabolites. J. Agric. Food Chem. 2005, 53, 3313–3320. [Google Scholar] [CrossRef]
| N° | RT | Accurate Mass | Molecular Ion [M+H]+ | Molecular Formula | Putative ID | Fragment Ions [M+H]+ | L. I. | References |
|---|---|---|---|---|---|---|---|---|
| 1 | 1.58 | 163.0844 | 164.0923 | C6H13NO4 | 1-deoxynojirimycin | - | 3 | [16] |
| 2 | 1.70 | 289.2253 | 290.2332 | C15H31NO4 | n-nonil-deoxynojirimycin | 206.8857/122.9243 | 3 | [16] |
| 3 | 2.14 | 147.0895 | 148.0974 | C6H13NO3 | fagomine | - | 3 | [16] |
| 4 | 2.19 | 181.0738 | 182.0817 | C9H11NO3 | 2-formyl-1H-pyrrole-1-butanoic acid | 165.0544/136.0755 | 3 | [11] |
| 5 | 8.16 | 354.0951 | 355.1024 | C16H18O9 | caffeoylquinic acid isomer I | 163.0386 | 2 | [17] |
| 6 | 9.30 | 449.1084 | 449.1084 | C21H21O11+ | cyanidin hexoside | 287.0546 | 2 | [17] |
| 7 | 9.35 | 507.3043 | 508.3122 | C24H45NO10 | morusimic acid E | 346.2587/284.2579 | 3 | [18] |
| 8 | 10.21 | 595.1662 | 595.1662 | C27H30O15+ | cyanidin hexose deoxyhexose | 449.1058/287.0546 | 2 | [17] |
| 9 | 10.93 | 433.1135 | 433.1135 | C21H21O10+ | pelargonidin hexoside | 271.0596 | 2 | [17] |
| 10 | 11.52 | 354.0951 | 355.1024 | C16H18O9 | caffeoylquinic acid isomer II | 163.0386 | 2 | [17] |
| 11 | 11.92 | 579.1714 | 579.1714 | C27H31O14+ | pelargonidin hexose deoxyhexose | 433.1115/271.0596 | 2 | [17] |
| 12 | 12.24 | 626.1483 | 627.1542 | C27H30O17 | quercetin hexose hexose | 465.1023/303.0489 | 2 | [17] |
| 13 | 12.38 | 354.0951 | 355.1024 | C16H18O9 | caffeoylquinic acid isomer III | 163.0386 | 2 | [17] |
| 14 | 12.42 | 772.2062 | 773.2135 | C33H40O21 | quercetin-3-O-rutinoside-7-O-glucoside | 303.0496/465.0995/611.1576 | 1 | [19] |
| 15 | 13.25 | 466.1111 | 467.1190 | C21H22O12 | dihydroquercetin hexoside/taxifolin hexoside | 449.1069/305.0650 | 3 | [20] |
| 16 | 14.41 | 712.1487 | 713.1544 | C30H32O20 | quercetin hexoside malonyl hexoside | 551.1015/463.1021/303.0496 | 2 | [21] |
| 17 | 14.71 | 756.2112 | 757.2192 | C33H40O21 | kaempferol-3-O-rutinoside-7-O-glucoside | 611.1576/449.1065/287.547 | 1 | [19] |
| 18 | 15.35 | 386.1940 | 387.2020 | C19H30O8 | roseoside | 370.1118/208.0599 | 3 | [22] |
| 19 | 16.37 | 772.2062 | 773.2135 | C33H40O21 | quercetin-hexose-hexose-deoxyhexose | 303.0496/465.0995/611.1576 | 3 | [19] |
| 20 | 16.52 | 450.1162 | 451.1235 | C21H22O11 | dihydrokaempferol-hexoside | 289.0703 | 3 | [23] |
| 21 | 17.02 | 696.1517 | 697.1597 | C30H32O19 | kaempferol hexoside malonyl hexoside | 287.0545/449.1065/535.1076 | 2 | [24] |
| 22 | 17.22 | 772.2062 | 773.2135 | C34H40O31 | quercetin-hexose-hexose-deoxyhexose | 303.0496/465.0995/611.1576 | 2 | [19] |
| 23 | 18.28 | 756.2112 | 757.2192 | C33H40O20 | kaempferol hexose-hexose deoxyhexose | 611.1576/449.4065/287.0547 | 2 | [19] |
| 24 | 18.63 | 756.2112 | 757.2192 | C33H40O20 | kaempferol hexose hexose deoxyhexose | 611.1576/449.4065/287.0547 | 2 | [19] |
| 25 | 19.59 | 491.3094 | 492.3173 | C24H46O9N | morusimic acid C | 330.2640 | 3 | [18] |
| 26 | 21.39 | 610.1534 | 611.1606 | C27H30O16 | quercetin-3-O-rutinoside | 303.0495/465.1019 | 1 | [25] |
| 27 | 22.06 | 329.2566 | 330.2645 | C18H35NO4 | morusimic acid B | 312.2529/268.2630/250.2525 | 3 | [18] |
| 28 | 22.42 | 464.0954 | 465.1027 | C21H20O12 | quercetin-hexoside | 303.0498 | 2 | [17] |
| 29 | 24.39 | 594.1584 | 595.1664 | C27H30O15 | kaempferol-3-O-rutinoside | 449.1066/287.0546 | 1 | [17] |
| 30 | 24.88 | 550.0958 | 551.1038 | C24H22O15 | quercetin-malonylhexoside | 303.0499 | 2 | [26] |
| 31 | 25.10 | 516.1268 | 517.1341 | C25H24O12 | dicaffeoylquinic acid I | 163.0387 | 2 | [27] |
| 32 | 25.59 | 448.1006 | 449.1078 | C21H20O11 | kaempferol-hexoside | 287.0546 | 2 | [17] |
| 33 | 25.66 | 516.1268 | 517.1341 | C25H24O12 | dicaffeoylquinic acid II | 325.0913/163.0387 | 2 | [27] |
| 34 | 27.83 | 516.1268 | 517.1341 | C25H24O12 | dicaffeoylquinic acid III | 325.0913/163.0387 | 2 | [27] |
| 35 | 28.53 | 534.1009 | 535.1088 | C24H22O14 | kaempferol malonyl hexoside | 287.0546 | 2 | [28] |
| M. alba Retention Time (min) | Bioactive Metabolite | M. nigra Retention Time (min) | Bioactive Metabolite |
|---|---|---|---|
| 1.6–2.08 | 1-2-3 | 1.8–2.27 | 1-2-3 |
| 4.88–5.35 | n.i. | 4.13–4.6 | n.i. |
| 12.82–13.28 | 15 | 8.33–8.8 | 6 |
| 13.75–14.23 | 16/17 | 10.2–10.67 | 9 |
| 19.82–20.3 | 25 | 10.2–10.67 | 9 |
| 22.62–23.08 | 28 | 12.53–13 | 15 |
| 23.55–24.02 | 29 | 13.4–13.93 | 16 |
| 24.95–25.42 | 32 | 14.4–14.87 | 17 |
| 31.02–31.5 | n.i. | 15.8–16.27 | 19 |
| 34.7–35.2 | n.i. | 16.27–16.73 | 20 |
| 35.22–35.68 | n.i. | 24.67–25.12 | 31/32 |
| - | 40.07–40.53 | n.i. |
| Extracts | TEAC mg/g FW |
|---|---|
| Morus alba (White mulberry) | 39.40 ± 0.02 |
| Morus nigra (Black mulberry) | 49.42 ± 0.01 |
| Fragaria vesca (Wild strawberry) | 50.61 ± 0.01 |
| Fragaria ananassa (Strawberry) | 51.31 ± 0.01 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Urso, G.; Mes, J.J.; Montoro, P.; Hall, R.D.; de Vos, R.C.H. Identification of Bioactive Phytochemicals in Mulberries. Metabolites 2020, 10, 7. https://doi.org/10.3390/metabo10010007
D’Urso G, Mes JJ, Montoro P, Hall RD, de Vos RCH. Identification of Bioactive Phytochemicals in Mulberries. Metabolites. 2020; 10(1):7. https://doi.org/10.3390/metabo10010007
Chicago/Turabian StyleD’Urso, Gilda, Jurriaan J. Mes, Paola Montoro, Robert D. Hall, and Ric C.H. de Vos. 2020. "Identification of Bioactive Phytochemicals in Mulberries" Metabolites 10, no. 1: 7. https://doi.org/10.3390/metabo10010007
APA StyleD’Urso, G., Mes, J. J., Montoro, P., Hall, R. D., & de Vos, R. C. H. (2020). Identification of Bioactive Phytochemicals in Mulberries. Metabolites, 10(1), 7. https://doi.org/10.3390/metabo10010007






