Liquid Chromatography Tandem Mass Spectrometry Quantification of 13C-Labeling in Sugars
Abstract
1. Introduction
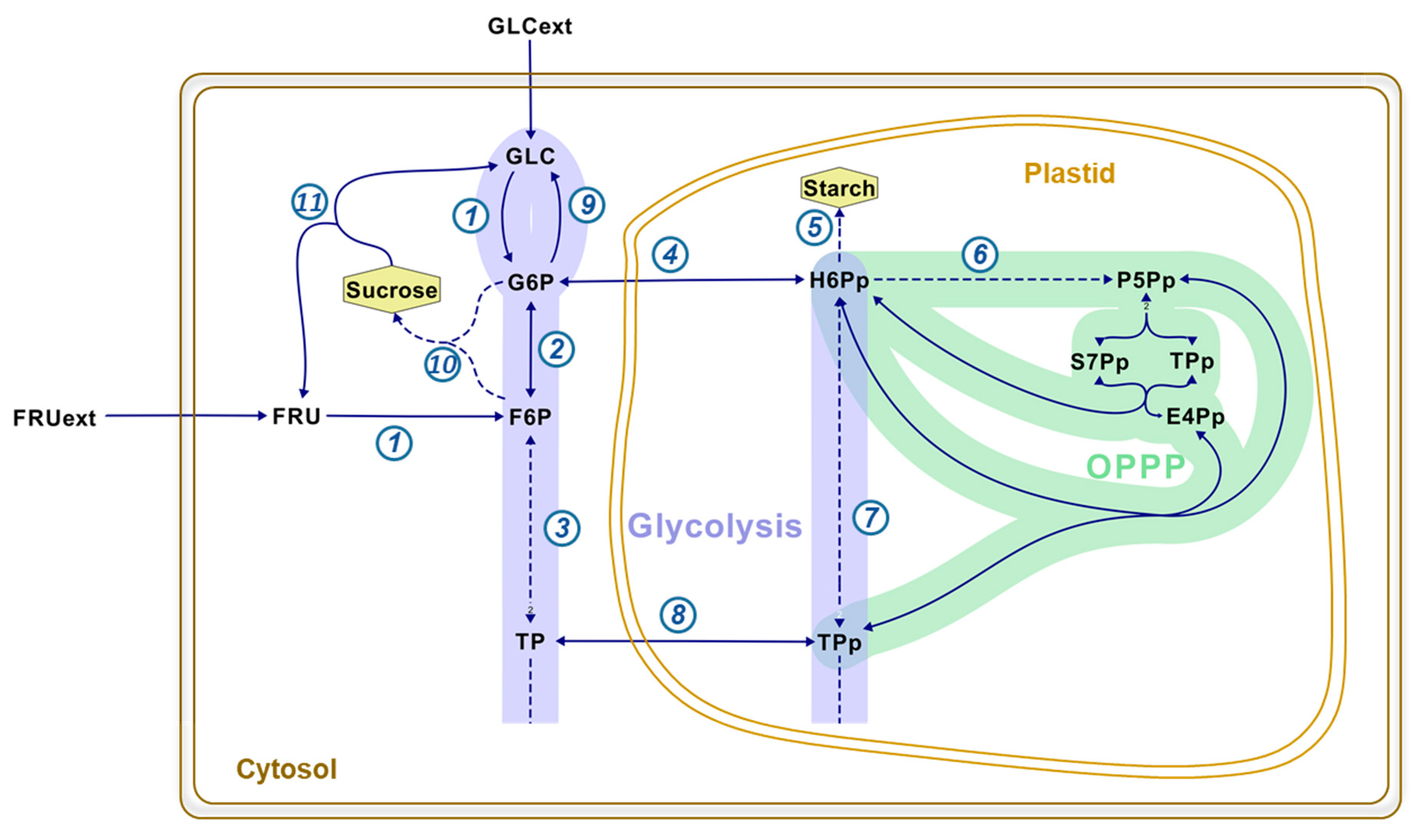
2. Results and Discussion
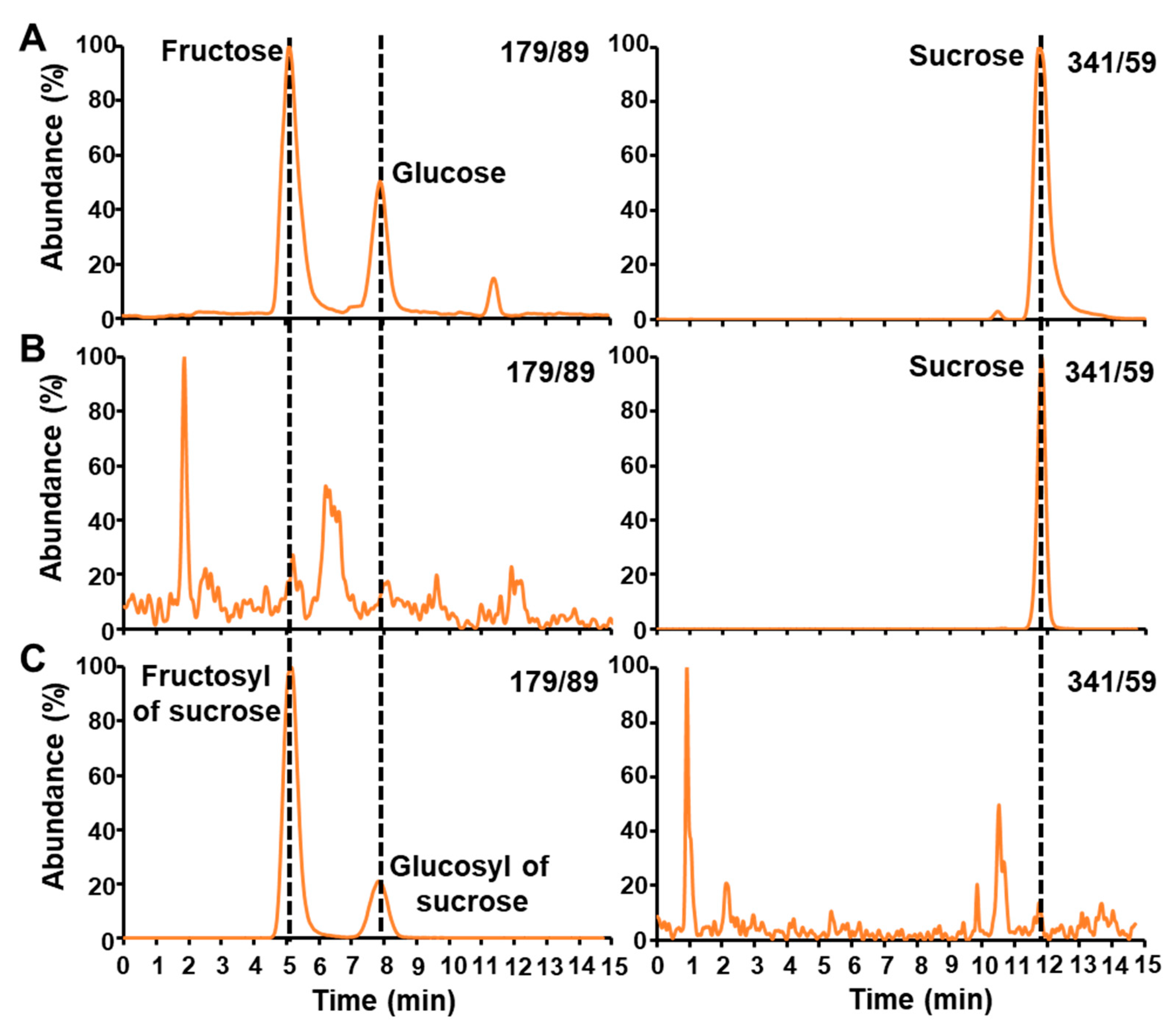
2.1. LC-MS/MS Analysis of Free Sugars
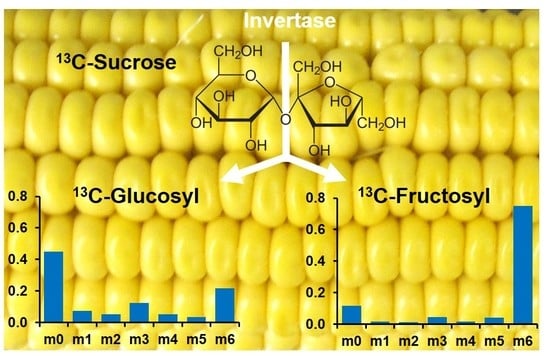
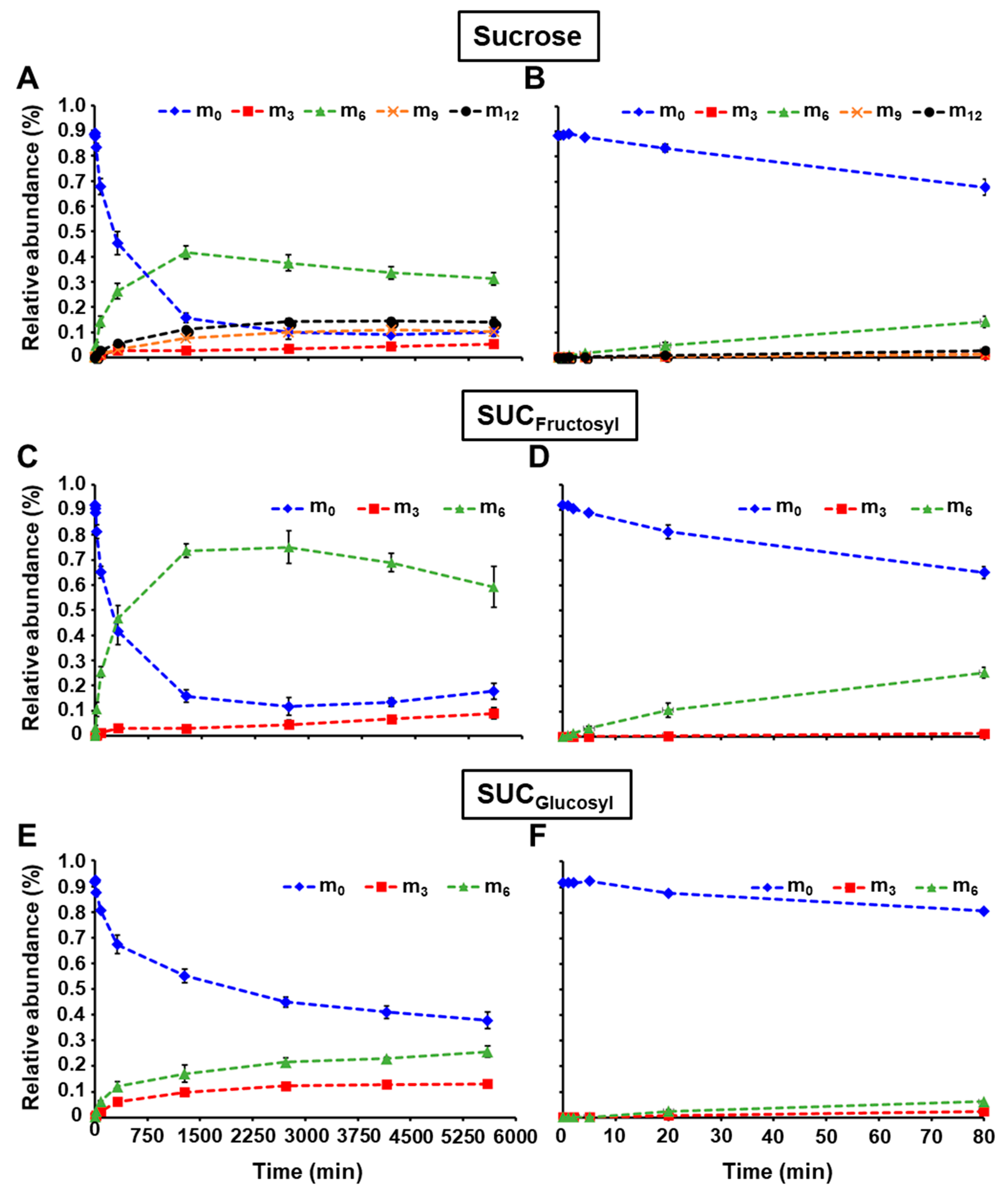
2.2. LC-MS/MS Analysis of the Glucosyl and Fructosyl Moieties of Sucrose
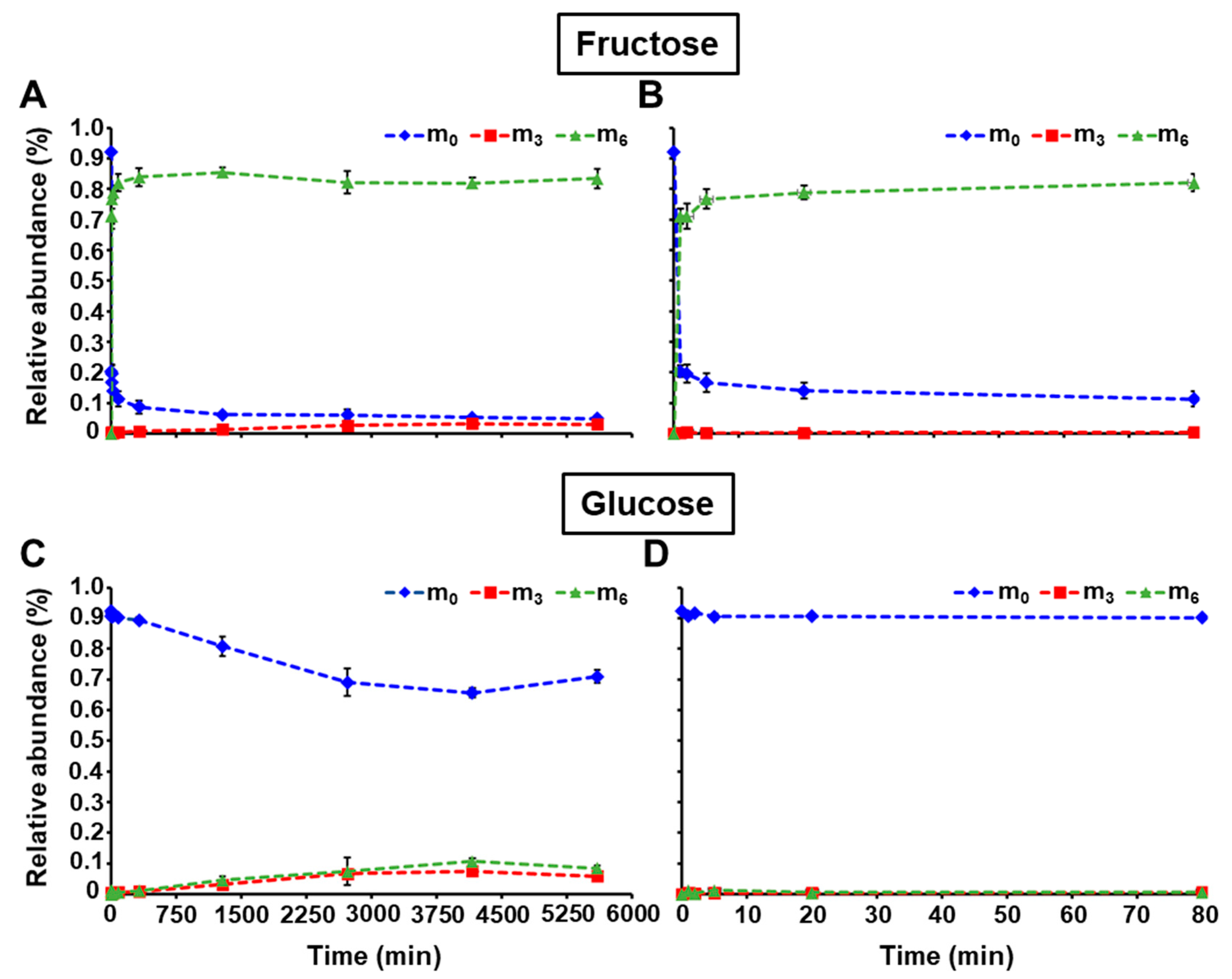
2.3. Application of LC-MS/MS to Monitor 13C-Labeling in Free Sugars
3. Materials and Methods
3.1. Chemicals
3.2. Plant Materials and Growth Conditions
3.3. Extraction of Soluble Sugars
3.4. Analysis of 13C-Labeling in Free Sugars
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ratcliffe, R.G.; Shachar-Hill, Y. Measuring multiple fluxes through plant metabolic networks. Plant J. 2006, 45, 490–511. [Google Scholar] [CrossRef] [PubMed]
- Hatzfeld, W.D.; Stitt, M. A study of the rate of recycling of triose phosphates in heterotrophic Chenopodium rubrum cells, potato tubers, and maize endosperm. Planta 1990, 180, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.A.; ap Rees, T. Fluxes of carbohydrate metabolism in ripening bananas. Planta 1993, 192, 52–60. [Google Scholar] [CrossRef]
- Salon, C.; Raymond, P.; Pradet, A. Quantification of carbon fluxes through the tricarboxylic acid cycle in early germinating lettuce embryos. J. Biol. Chem. 1988, 263, 12278–12287. [Google Scholar] [PubMed]
- Bowsher, C.G.; Tobin, A.K. Compartmentation of metabolism within mitochondria and plastids. J. Exp. Bot. 2001, 52, 513–527. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E. Compartmentation in plant metabolism. J. Exp. Bot. 2007, 58, 35–47. [Google Scholar] [CrossRef]
- Dieuaide-Noubhani, M.; Alonso, A.P. Application of metabolic flux analysis to plants. Methods Mol. Biol. 2014, 1090, 1–18. [Google Scholar]
- Alonso, A.P.; Piasecki, R.J.; Wang, Y.; LaClair, R.W.; Shachar-Hill, Y. Quantifying the Labeling and the Levels of Plant Cell Wall Precursors Using Ion Chromatography Tandem Mass Spectrometry. Plant Physiol. 2010, 153, 915–924. [Google Scholar] [CrossRef]
- Cocuron, J.C.; Alonso, A.P. Liquid chromatography tandem mass spectrometry for measuring 13C-labeling in intermediaries of the glycolysis and pentose-phosphate pathway. Methods Mol. Biol. 2014, 1090, 131–142. [Google Scholar]
- Allen, D.K.; Shachar-Hill, Y.; Ohlrogge, J.B. Compartment-specific labeling information in C-13 metabolic flux analysis of plants. Phytochemistry 2007, 68, 2197–2210. [Google Scholar] [CrossRef]
- Alonso, A.P.; Dale, V.L.; Shachar-Hill, Y. Understanding fatty acid synthesis in developing maize embryos using metabolic flux analysis. Metab. Eng. 2010, 12, 488–497. [Google Scholar] [CrossRef]
- Alonso, A.P.; Goffman, F.D.; Ohlrogge, J.B.; Shachar-Hill, Y. Carbon conversion efficiency and central metabolic fluxes in developing sunflower (Helianthus annuus L.) embryos. Plant J. 2007, 52, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.P.; Raymond, P.; Hernould, M.; Rondeau-Mouro, C.; de Graaf, A.; Chourey, P.; Lahaye, M.; Shachar-Hill, Y.; Rolin, D.; Dieuaide-Noubhani, M. A metabolic flux analysis to study the role of sucrose synthase in the regulation of the carbon partitioning in central metabolism in maize root tips. Metab. Eng. 2007, 9, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.P.; Raymond, P.; Rolin, D.; Dieuaide-Noubhani, M. Substrate cycles in the central metabolism of maize root tips under hypoxia. Phytochemistry 2007, 68, 2222–2231. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.P.; Val, D.L.; Shachar-Hill, Y. Central metabolic fluxes in the endosperm of developing maize seeds and their implications for metabolic engineering. Metab. Eng. 2011, 13, 96–107. [Google Scholar] [CrossRef]
- Cocuron, J.C.; Koubaa, M.; Kimmelfield, R.; Ross, Z.; Alonso, A.P. A combined metabolomics and fluxomics analysis identifies steps limiting synthesis in maize embryos. Plant Physiol. 2019, 181, 961–975. [Google Scholar] [CrossRef]
- Dieuaidenoubhani, M.; Raffard, G.; Canioni, P.; Pradet, A.; Raymond, P. Quantification of Compartmented Metabolic Fluxes in Maize Root-Tips Using Isotope Distribution from C-13-Labeled or C-14-Labeled Glucose. J. Biol. Chem. 1995, 270, 13147–13159. [Google Scholar] [CrossRef]
- Masakapalli, S.K.; Le Lay, P.; Huddleston, J.E.; Pollock, N.L.; Kruger, N.J.; Ratcliffe, R.G. Subcellular Flux Analysis of Central Metabolism in a Heterotrophic Arabidopsis Cell Suspension Using Steady-State Stable Isotope Labeling. Plant Physiol. 2010, 152, 602–619. [Google Scholar] [CrossRef]
- Rontein, D.; Dieuaide-Noubhani, M.; Dufourc, E.J.; Raymond, P.; Rolin, D. The metabolic architecture of plant cells - Stability of central metabolism and flexibility of anabolic pathways during the growth cycle of tomato cells. J. Biol. Chem. 2002, 277, 43948–43960. [Google Scholar] [CrossRef]
- Alonso, A.P.; Vigeolas, H.; Raymond, P.; Rolin, D.; Dieuaide-Noubhani, M. A new substrate cycle in plants. evidence for a high glucose-phosphate-to-glucose turnover from in vivo steady-state and pulse-labeling experiments with [13C] glucose and [14C] glucose. Plant Physiol. 2005, 138, 2220–2232. [Google Scholar] [CrossRef]
- Fell, D. Understanding the Control of Metabolism; Portland Press: London, UK, 1997; pp. 213–225. [Google Scholar]
- Acket, S.; Degournay, A.; Merlier, F.; Thomasset, B. 13C labeling analysis of sugars by high resolution-mass spectrometry for metabolic flux analysis. Anal. Biochem. 2017, 527, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Viola, R.; Davies, H.V.; Chudeck, A.R. Pathways of starch and sucrose biosynthesis in developing tubers of potato (Solanum tuberosum L.) and seeds of faba bean (Vicia faba L.): Elucidation by 13C-nuclear-magnetic-resonance spectroscopy. Planta 1991, 183, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Cocuron, J.C.; Anderson, B.; Boyd, A.; Alonso, A.P. Targeted metabolomics of Physaria fendleri, an industrial crop producing hydroxy fatty acids. Plant Cell Physiol. 2014, 55, 620–633. [Google Scholar] [CrossRef]
- Calvano, C.D.; Cataldi, T.R.I.; Kogel, J.F.; Monopoli, A.; Palmisano, F.; Sundermeyer, J. Structural Characterization of Neutral Saccharides by Negative Ion MALDI Mass Spectrometry Using a Superbasic Proton Sponge as Deprotonating Matrix. J. Am. Soc. Mass Spectrom. 2017, 28, 1666–1675. [Google Scholar] [CrossRef] [PubMed]
- Koubaa, M.; Cocuron, J.-C.; Thomasset, B.; Alonso, A.P. Highlighting the tricarboxilic acid cycle: Liquid and gas chromatography-mass spectrometry analyses of 13C-labeled organic acids. Anal. Biochem. 2013, 436, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Cocuron, J.C.; Tsogtbaatar, E.; Alonso, A.P. High-throughput quantification of the levels and labeling abundance of free amino acids by liquid chromatography tandem mass spectrometry. J. Chromatogr. A 2017, 1490, 148–155. [Google Scholar] [CrossRef] [PubMed]
| Sugars | Parent Ion Formula | Daughter Ion Formula | Parent/Daughter Transitions | DP (V) | EP (V) | CE (V) | CXP (V) |
|---|---|---|---|---|---|---|---|
| Hexoses | C6H11O6- | C3H6O3- | 179/89 | −30 | −10 | −12 | −11 |
| Disaccharides | C12H21O11- | C2H4O2- | 341/59 | −240 | −10 | −50 | −9 |
| Sugars | RT (min) | Mass Isotopomers | Parent/Daughter Transitions |
|---|---|---|---|
| Fructose | 5.11 | m0 | 179/89 |
| Glucose | 7.90 | m1 | 180/89; 180/90 |
| m2 | 181/89; 181/90; 181/91 | ||
| m3 | 182/90; 182/91; 182/92 | ||
| m4 | 183/90; 183/91; 183/92 | ||
| m5 | 184/91; 184/92 | ||
| m6 | 185/92 | ||
| Sucrose | 11.95 | m0 | 341/59 |
| m1 | 342/59; 342/60 | ||
| m2 | 343/59; 343/60; 343/61 | ||
| m3 | 344/59; 344/60; 344/61 | ||
| m4 | 345/59; 345/60; 345/61 | ||
| m5 | 346/59; 346/60; 346/61 | ||
| m6 | 347/59; 347/60; 347/61 | ||
| m7 | 348/59; 348/60; 348/61 | ||
| m8 | 349/59; 349/60; 349/61 | ||
| m9 | 350/59; 350/60; 350/61 | ||
| m10 | 351/59; 351/60; 351/61 | ||
| m11 | 352/60; 352/61 | ||
| m12 | 353/61 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cocuron, J.-C.; Ross, Z.; Alonso, A.P. Liquid Chromatography Tandem Mass Spectrometry Quantification of 13C-Labeling in Sugars. Metabolites 2020, 10, 30. https://doi.org/10.3390/metabo10010030
Cocuron J-C, Ross Z, Alonso AP. Liquid Chromatography Tandem Mass Spectrometry Quantification of 13C-Labeling in Sugars. Metabolites. 2020; 10(1):30. https://doi.org/10.3390/metabo10010030
Chicago/Turabian StyleCocuron, Jean-Christophe, Zacchary Ross, and Ana P. Alonso. 2020. "Liquid Chromatography Tandem Mass Spectrometry Quantification of 13C-Labeling in Sugars" Metabolites 10, no. 1: 30. https://doi.org/10.3390/metabo10010030
APA StyleCocuron, J.-C., Ross, Z., & Alonso, A. P. (2020). Liquid Chromatography Tandem Mass Spectrometry Quantification of 13C-Labeling in Sugars. Metabolites, 10(1), 30. https://doi.org/10.3390/metabo10010030