Mosaic-CLSM Assessment of Bacterial Spatial Distribution in Cosmetic Matrices According to Matrix Viscosity and Bacterial Hydrophobicity
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Chemicals and Materials
2.3. Characterization of Bacterial Surface Properties
2.3.1. Bacterial Surface Hydrophobicity
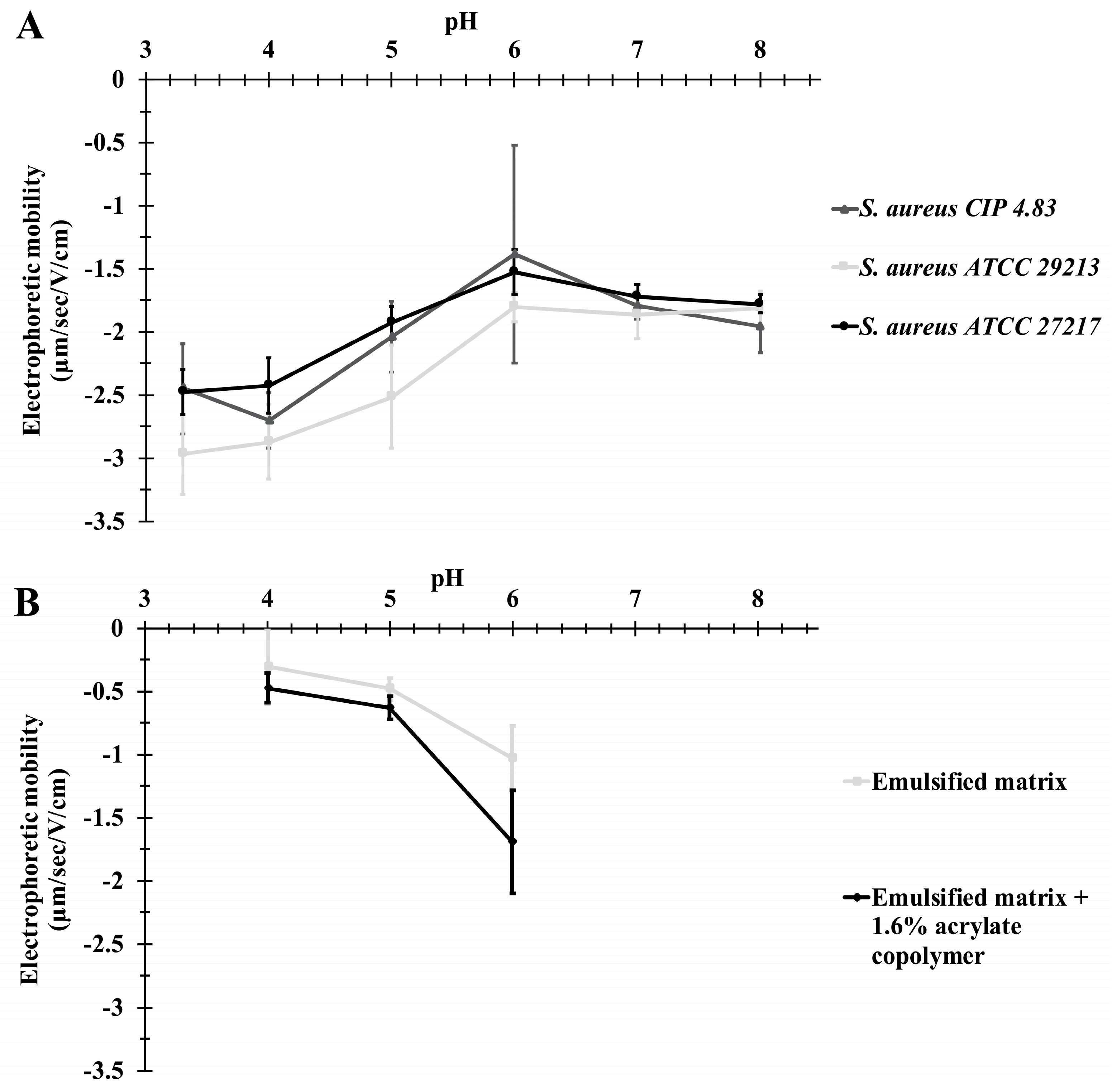
2.3.2. Bacterial Electrophoretic Mobility
2.4. Cosmetic Matrices
2.4.1. Commercial Matrices
2.4.2. Preparation of the Emulsified Model Matrix (35/65 O/W%)
2.5. Characterization of the Emulsified Matrices
2.5.1. Viscosity Measurements
2.5.2. Determination of Lipid Droplet Electrophoretic Mobility
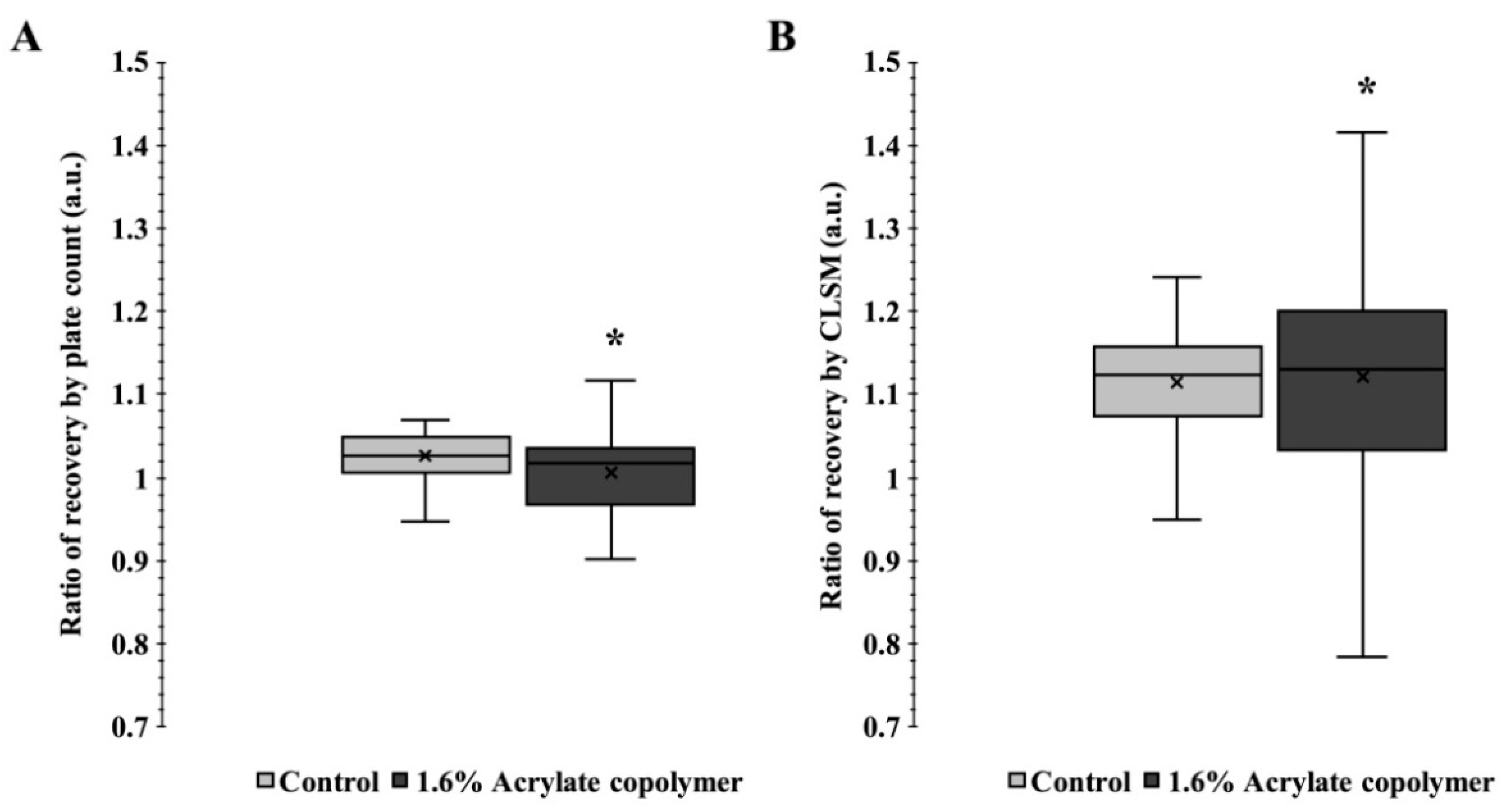
2.6. Enumeration of the Bacterial Population
2.7. Mosaic-Confocal Laser Scanning Microscopy (M-CLSM) and Image Analysis
2.8. Statistical Analysis
3. Results
3.1. Determination of Bacterial Surface Properties
3.1.1. Bacterial Surface Hydrophobicity
3.1.2. Bacterial Surface Charge
3.2. Determination of Matrix Properties
3.2.1. Matrix Viscosity
3.2.2. Matrix Droplet Electrophoretic Mobility
3.3. Bacterial Spatial Distribution in Commercial Emulsions
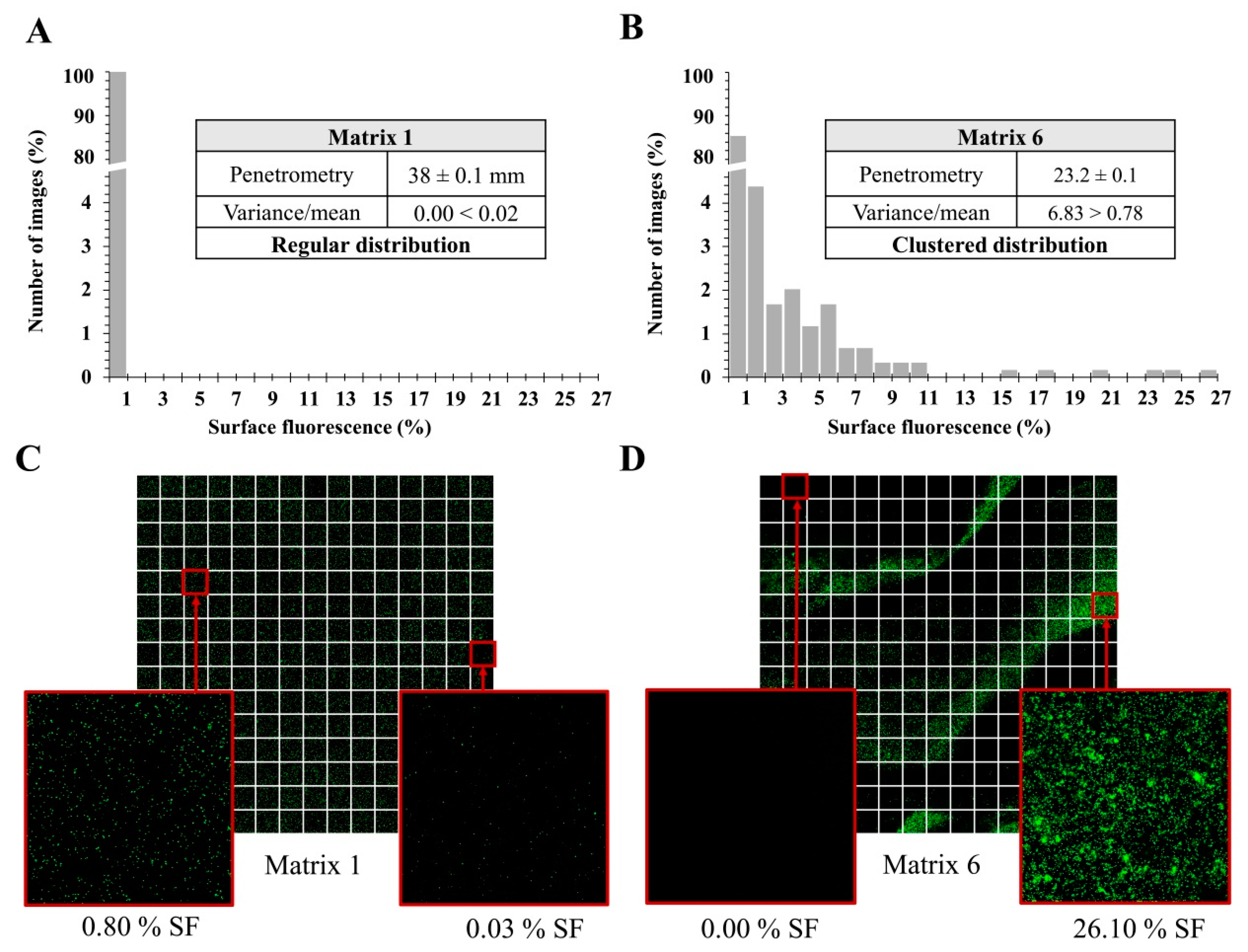
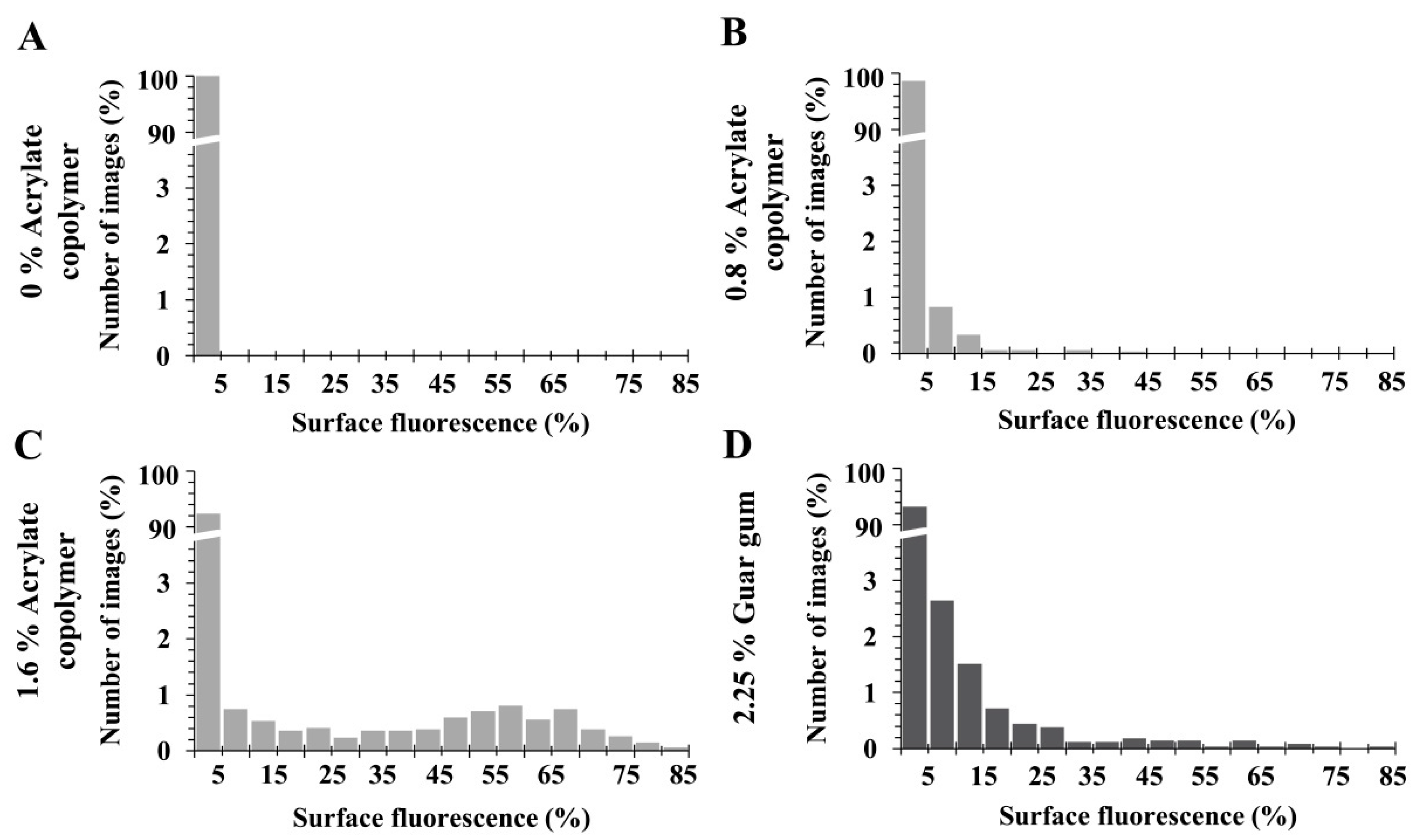
3.4. Bacterial Spatial Distribution and Enumeration in Matrices with Thickener
3.5. Spatial Distribution of Three S. aureus Strains According to Their Level of Hydrophobicity in Matrices with Thickener
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tan, A.S.B.; Tuysuz, M.; Otuk, G. Investigation of preservative efficacy and microbiological content of some cosmetics found on the market. Pak. J. Pharm. Sci. 2013, 26, 153–157. [Google Scholar]
- Campana, R.; Scesa, C.; Patrone, V.; Vittoria, E.; Baffone, W. Microbiological study of cosmetic products during their use by consumers: Health risk and efficacy of preservative systems. Lett. Appl. Microbiol. 2006, 43, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.; Song, P.; Seo, C.; Cheong, H.; Park, Y. Colonization and infection of the skin by S. aureus: Immune system evasion and the response to cationic antimicrobial peptides. Int. J. Mol. Sci. 2014, 15, 8753. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F.D. Staphylococcus aureus infections. New Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603. [Google Scholar] [CrossRef] [PubMed]
- Berthele, H.; Sella, O.; Lavarde, M.; Mielcarek, C.; Pense-Lheritier, A.M.; Pirnay, S. Determination of the influence of factors (ethanol, pH and aw) on the preservation of cosmetics using experimental design. Int. J. Cosmet. Sci. 2014, 36, 54–61. [Google Scholar] [CrossRef]
- Halla, N.; Fernandes, I.P.; Heleno, S.A.; Costa, P.; Boucherit-Otmani, Z.; Boucherit, K.; Rodrigues, A.E.; Ferreira, I.; Barreiro, M.F. Cosmetics Preservation: A Review on Present Strategies. Molecules 2018, 23, 1571. [Google Scholar] [CrossRef]
- Jongenburger, I.; Reij, M.W.; Boer, E.P.J.; Gorris, L.G.M.; Zwietering, M.H. Random or systematic sampling to detect a localised microbial contamination within a batch of food. Food Control 2011, 22, 1448–1455. [Google Scholar] [CrossRef]
- Jongenburger, I.; Reij, M.W.; Boer, E.P.J.; Zwietering, M.H.; Gorris, L.G.M. Modelling homogeneous and heterogeneous microbial contaminations in a powdered food product. Int. J. Food Microbiol. 2012, 157, 35–44. [Google Scholar] [CrossRef]
- Gilbert, P.; Caplan, F.; Brown, M.R.W. Centrifugation injury of Gram-negative bacteria. J. Antimicrob. Chemother. 1991, 27, 550–551. [Google Scholar] [CrossRef]
- Pembrey, R.S.; Marshall, K.C.; Schneider, R.P. Cell surface analysis techniques: What do cell preparation protocols do to cell surface properties? Appl. Environ. Microbiol. 1999, 65, 2877–2894. [Google Scholar] [CrossRef] [PubMed]
- Bruinsma, G.M.; Rustema-Abbing, M.; Van der Mei, H.C.; Busscher, H.J. Effects of cell surface damage on surface properties and adhesion of Pseudomonas aeruginosa. J. Microbiol. Methods 2001, 45, 95–101. [Google Scholar] [CrossRef]
- Jeanson, S.; Chadoeuf, J.; Madec, M.N.; Aly, S.; Floury, J.; Brocklehurst, T.F.; Lortal, S. Spatial distribution of bacterial colonies in a model cheese. Appl. Environ. Microbiol. 2011, 77, 1493–1500. [Google Scholar] [CrossRef]
- Hedges, A.J. Estimating the precision of serial dilutions and viable bacterial counts. Int. J. Food Microbiol. 2002, 76, 207–214. [Google Scholar] [CrossRef]
- Jongenburger, I.; Bassett, J.; Jackson, T.; Zwietering, M.H.; Jewell, K. Impact of microbial distributions on food safety I. Factors influencing microbial distributions and modelling aspects. Food Control 2012, 26, 601–609. [Google Scholar] [CrossRef]
- Nauta, M.J. Microbiological risk assessment models for partitioning and mixing during food handling. Int. J. Food Microbiol. 2005, 100, 311–322. [Google Scholar] [CrossRef]
- Lopez, C.; Maillard, M.B.; Briard-Bion, V.; Camier, B.; Hannon, J.A. Lipolysis during ripening of emmental cheese considering organization of fat and preferential localization of bacteria. J. Agric. Food Chem. 2006, 54, 5855–5867. [Google Scholar] [CrossRef]
- Oss, C.J.V. Long-range and short-range mechanisms of hydrophobic attraction and hydrophilic repulsion in specific and aspecific interactions. J. Mol. Recognit. 2003, 16, 177–190. [Google Scholar] [CrossRef]
- Burgain, J.; Scher, J.; Francius, G.; Borges, F.; Corgneau, M.; Revol-Junelles, A.M.; Cailliez-Grimal, C.; Gaiani, C. Lactic acid bacteria in dairy food: Surface characterization and interactions with food matrix components. Adv. Colloid Interface Sci. 2014, 213, 21–35. [Google Scholar] [CrossRef]
- Ly, M.H.; Aguedo, M.; Goudot, S.; Le, M.L.; Cayot, P.; Teixeira, J.A.; Le, T.M.; Belin, J.M.; Waché, Y. Interactions between bacterial surfaces and milk proteins, impact on food emulsions stability. Food Hydrocoll. 2008, 22, 742–751. [Google Scholar] [CrossRef]
- Dorobantu, L.S.; Yeung, A.K.C.; Foght, J.M.; Gray, M.R. Stabilization of oil-water emulsions by hydrophobic bacteria. Appl. Environ. Microbiol. 2004, 70, 6333–6336. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; McClements, D.J.; McLandsborough, L.A. Interaction between emulsion droplets and Escherichia coli cells. J. Food Sci. 2001, 66, 570–657. [Google Scholar] [CrossRef]
- Ly, M.H.; Naitali-Bouchez, M.; Meylheuc, T.; Bellon-Fontaine, M.N.; Le, T.M.; Belin, J.M.; Wache, Y. Importance of bacterial surface properties to control the stability of emulsions. Int. J. Food Microbiol. 2006, 112, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Pensé-Lhéritier, A.M. Conception des Produits Cosmétiques—La Formulation; Lavoisier: Paris, France, 2016. [Google Scholar]
- Kirk, R.E.; Othmer, D.F. Chemical Technology of Cosmetics; John Wiley & Sons: Hoboken, NJ, USA, 2013; p. 832. [Google Scholar]
- Bridier, A.; Dubois-Brissonnet, F.; Greub, G.; Thomas, V.; Briandet, R. Dynamics of the action of biocides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2011, 55, 2648–2654. [Google Scholar] [CrossRef]
- Bridier, A.; Dubois-Brissonnet, F.; Boubetra, A.; Thomas, V.; Briandet, R. The biofilm architecture of sixty opportunistic pathogens deciphered using a high throughput CLSM method. J. Microbiol. Methods 2010, 82, 64–70. [Google Scholar] [CrossRef]
- Doherty, S.B.; Gee, V.L.; Ross, R.P.; Stanton, C.; Fitzgerald, G.F.; Brodkorb, A. Efficacy of whey protein gel networks as potential viability-enhancing scaffolds for cell immobilization of Lactobacillus rhamnosus GG. J. Microbiol. Methods 2010, 80, 231–241. [Google Scholar] [CrossRef]
- Auty, M.A.; Gardiner, G.E.; McBrearty, S.J.; O’Sullivan, E.O.; Mulvihill, D.M.; Collins, J.K.; Fitzgerald, G.F.; Stanton, C.; Ross, R.P. Direct In Situ viability assessment of bacteria in probiotic dairy products using viability staining in conjunction with confocal scanning laser microscopy. Appl. Environ. Microbiol. 2001, 67, 420–425. [Google Scholar] [CrossRef]
- Rosenberg, M. Bacterial adherence to hydrocarbons: A useful technique for studying cell surface hydrophobicity. FEMS Microbiol. Lett. 1984, 22, 289–295. [Google Scholar] [CrossRef]
- Chen, C.Y.; Nace, G.W.; Irwin, P.L. A 6 × 6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes and Escherichia coli. J. Microbiol. Methods 2003, 55, 475–479. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH image to imageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671. [Google Scholar] [CrossRef]
- Hashim, J.; Looney, L.; Hashmi, M.S.J. Particle distribution in cast metal matrix composites—Part I. J. Mater. Process. Technol. 2002, 123, 251–257. [Google Scholar] [CrossRef]
- Wimpenny, J.W.T.; Leistner, L.; Thomas, L.V.; Mitchell, A.J.; Katsaras, K.; Peetz, P. Submerged bacterial colonies within food and model systems: Their growth, distribution and interactions. Int. J. Food Microbiol. 1995, 28, 299–315. [Google Scholar] [CrossRef]
- Brocklehurst, T.F.; Parker, M.L.; Gunning, P.A.; Coleman, H.P.; Robins, M.M. Growth of food-borne pathogenic bacteria in oil-in-water emulsions: II-Effect of emulsion structure on growth parameters and form of growth. J. Appl. Bacteriol. 1995, 78, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Ly, M.H.; Vo, N.H.; Le, T.M.; Belin, J.M.; Wache, Y. Diversity of the surface properties of Lactococci and consequences on adhesion to food components. Colloids Surf. B Biointerfaces 2006, 52, 149–153. [Google Scholar] [CrossRef]
| Polymer | Acrylate Copolymer | Guar Gum | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0% | 0.4% | 0.8% | 1.6% | 2.5% | ||||||
| Penetrometry (mm) | 31.5 ± 0.4 | 28.5 ± 0.3 | 27.7 ± 0.4 | 26.6 ± 0.3 | 25.5 ± 0.4 | |||||
| Surface Fluorescence% | Mean | Variance | Mean | Variance | Mean | Variance | Mean | Variance | Mean | Variance |
| S. aureus CIP 4.83 | 0.11 | 0.01 | 0.18 | 0.02 | 0.66 | 2.79 | 3.58 | 176.26 | 1.55 | 37.88 |
| Distribution | Regular | Regular | Clustered | Clustered | Clustered | |||||
| S. aureus ATCC 29213 | 0.45 | 0.09 | 0.66 | 0.06 | 0.50 | 0.73 | 1.16 | 11.32 | 0.94 | 6.23 |
| Distribution | Regular | Regular | Clustered | Clustered | Clustered | |||||
| S. aureus ATCC 27217 | 0.60 | 0.24 | 0.62 | 0.11 | 0.31 | 0.57 | 1.19 | 8.43 | 0.91 | 8.99 |
| Distribution | Regular | Regular | Clustered | Clustered | Clustered | |||||
| Beads | 1.79 | 0.24 | 2.28 | 0.38 | 1.74 | 1.90 | 1.71 | 4.21 | 2.23 | 13.21 |
| Distribution | Regular | Regular | Clustered | Clustered | Clustered | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almoughrabie, S.; Ngari, C.; Briandet, R.; Poulet, V.; Dubois-Brissonnet, F. Mosaic-CLSM Assessment of Bacterial Spatial Distribution in Cosmetic Matrices According to Matrix Viscosity and Bacterial Hydrophobicity. Cosmetics 2020, 7, 32. https://doi.org/10.3390/cosmetics7020032
Almoughrabie S, Ngari C, Briandet R, Poulet V, Dubois-Brissonnet F. Mosaic-CLSM Assessment of Bacterial Spatial Distribution in Cosmetic Matrices According to Matrix Viscosity and Bacterial Hydrophobicity. Cosmetics. 2020; 7(2):32. https://doi.org/10.3390/cosmetics7020032
Chicago/Turabian StyleAlmoughrabie, Samia, Chrisse Ngari, Romain Briandet, Valérie Poulet, and Florence Dubois-Brissonnet. 2020. "Mosaic-CLSM Assessment of Bacterial Spatial Distribution in Cosmetic Matrices According to Matrix Viscosity and Bacterial Hydrophobicity" Cosmetics 7, no. 2: 32. https://doi.org/10.3390/cosmetics7020032
APA StyleAlmoughrabie, S., Ngari, C., Briandet, R., Poulet, V., & Dubois-Brissonnet, F. (2020). Mosaic-CLSM Assessment of Bacterial Spatial Distribution in Cosmetic Matrices According to Matrix Viscosity and Bacterial Hydrophobicity. Cosmetics, 7(2), 32. https://doi.org/10.3390/cosmetics7020032





