Metagenomic Analysis of the Skin Microbiota of Brazilian Women: How to Develop Anti-Aging Cosmetics Based on This Knowledge?
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Skin Biometrological Measurements
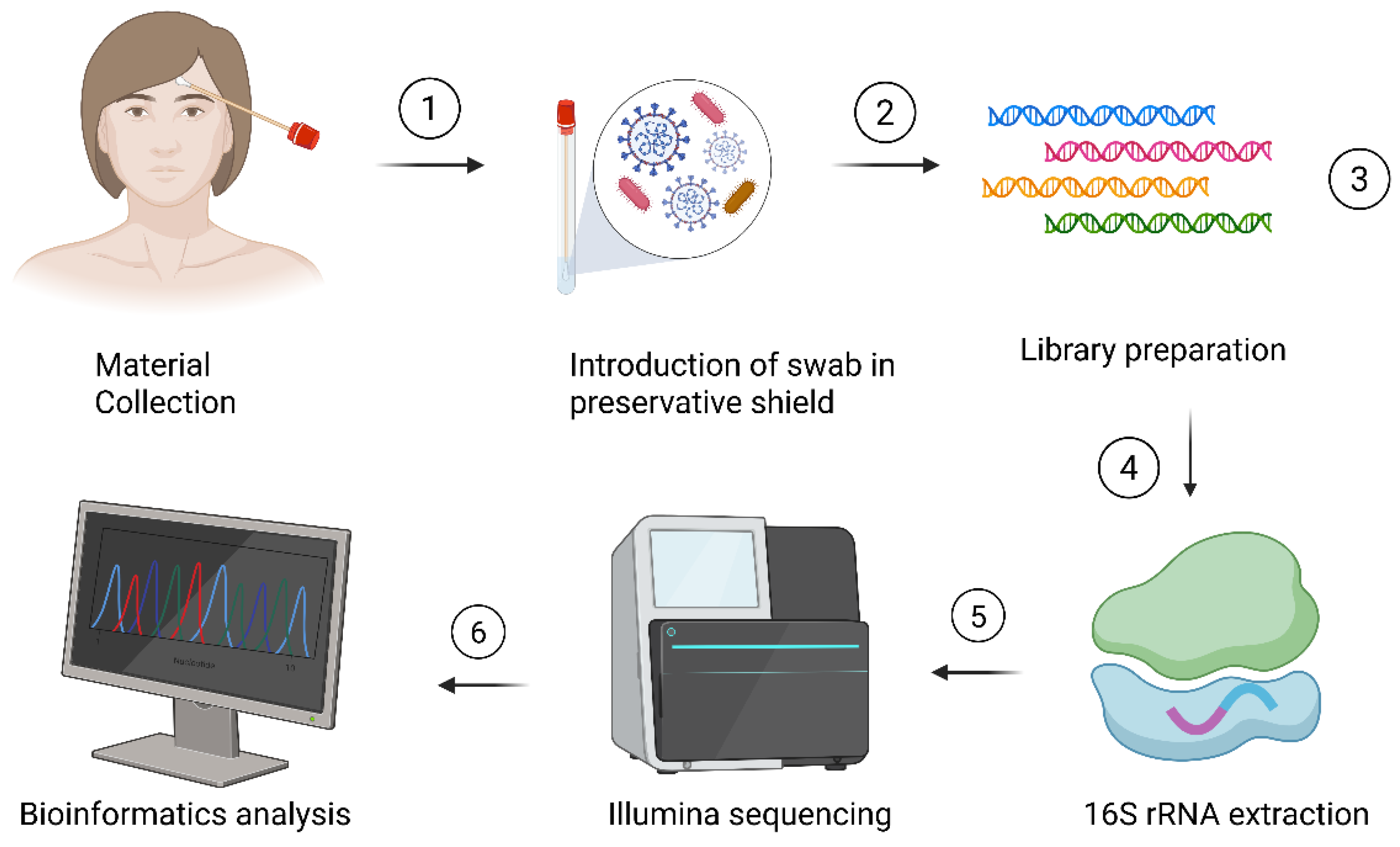
2.3. Sample Collection and DNA Extraction
2.4. 16S rRNA Sequencing
2.5. Statistical Analysis
3. Results
3.1. In Vivo Cutaneous Biometrology
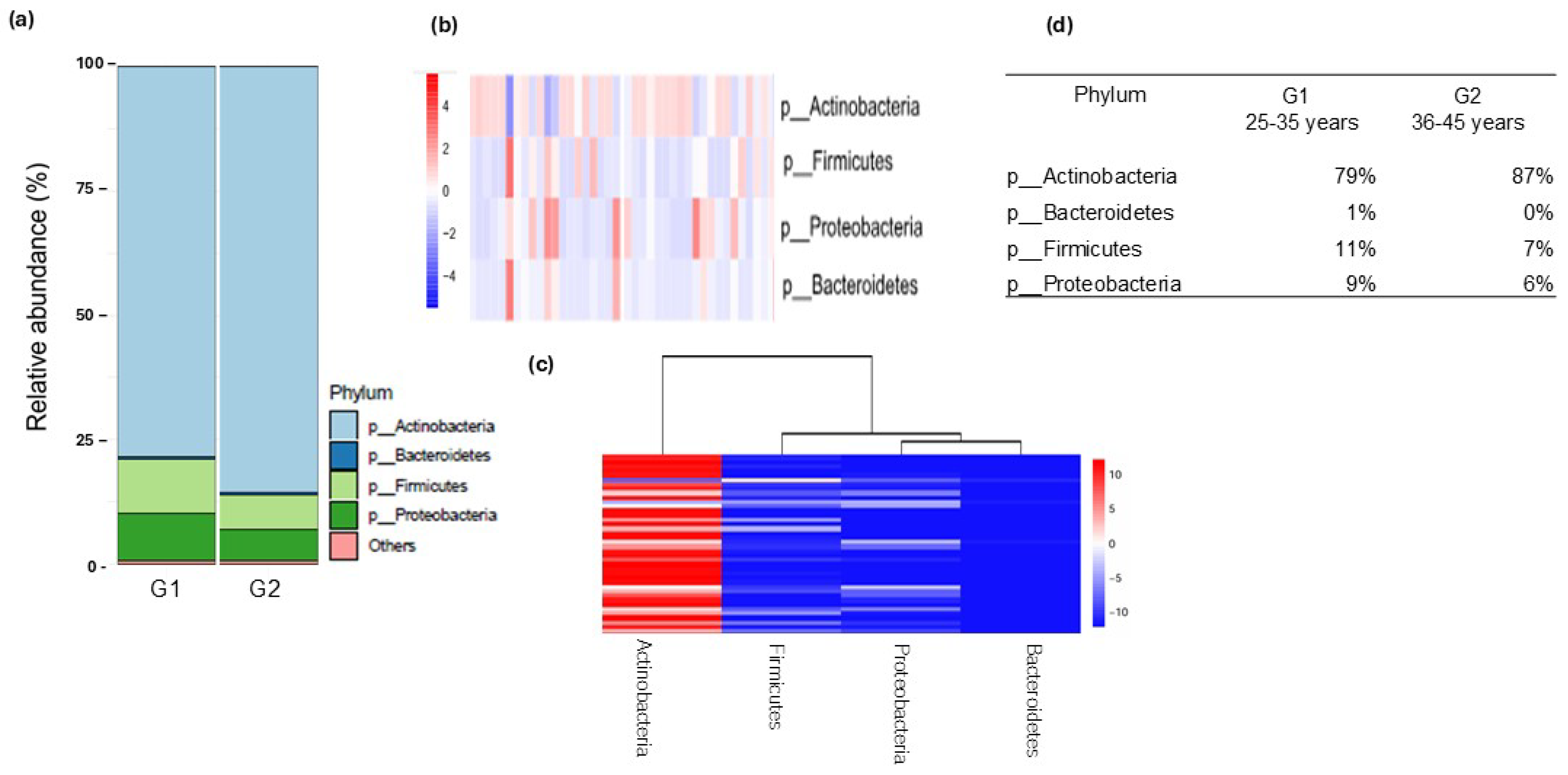
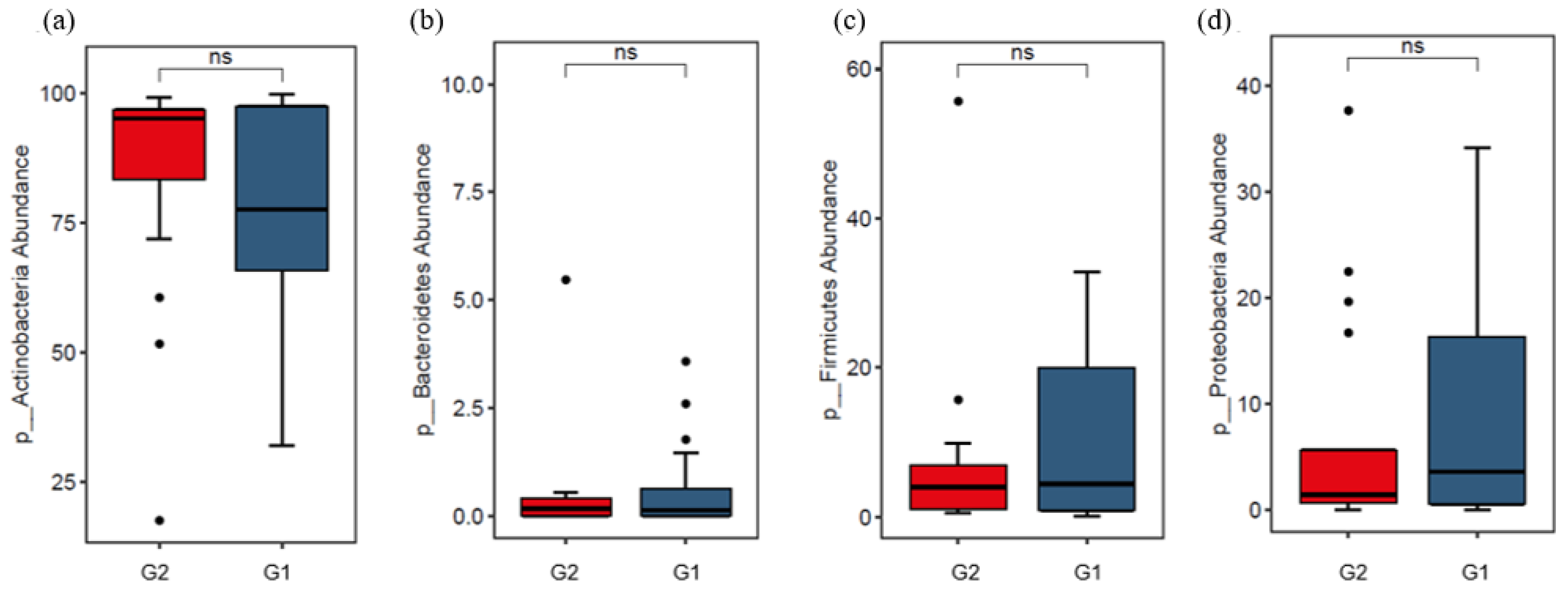
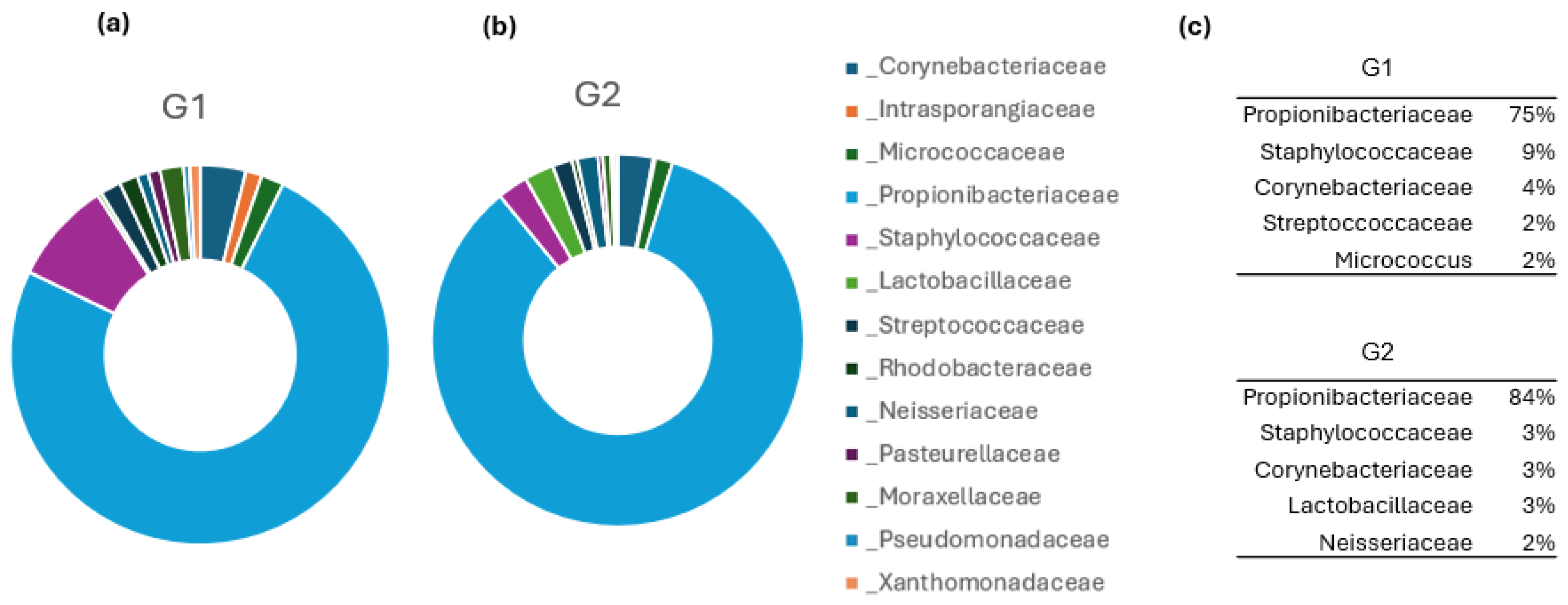
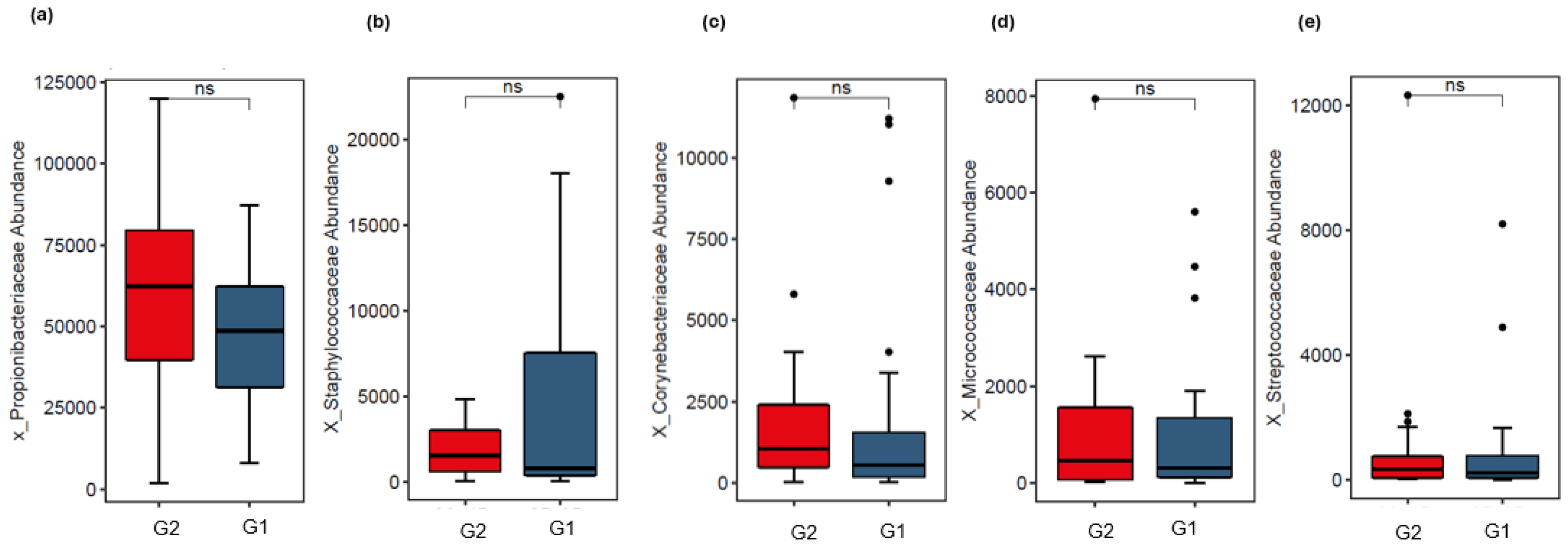
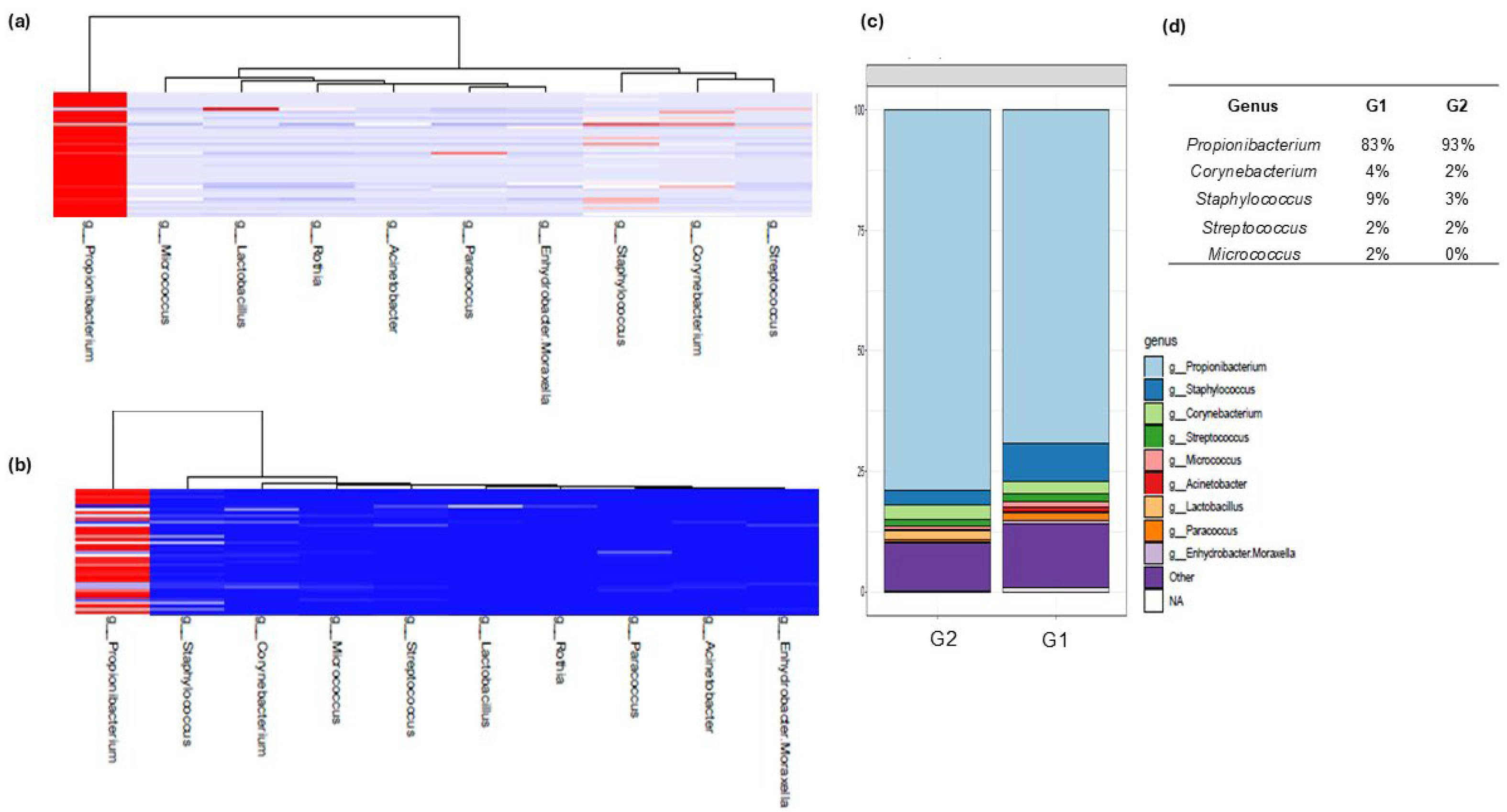
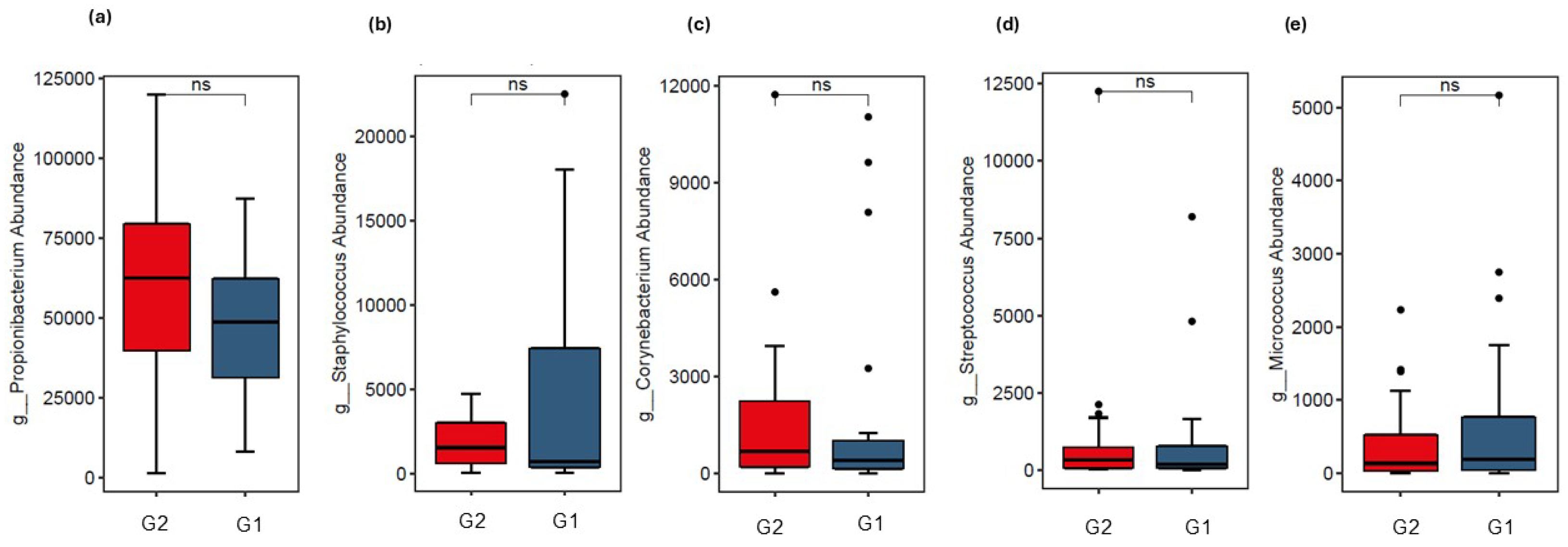
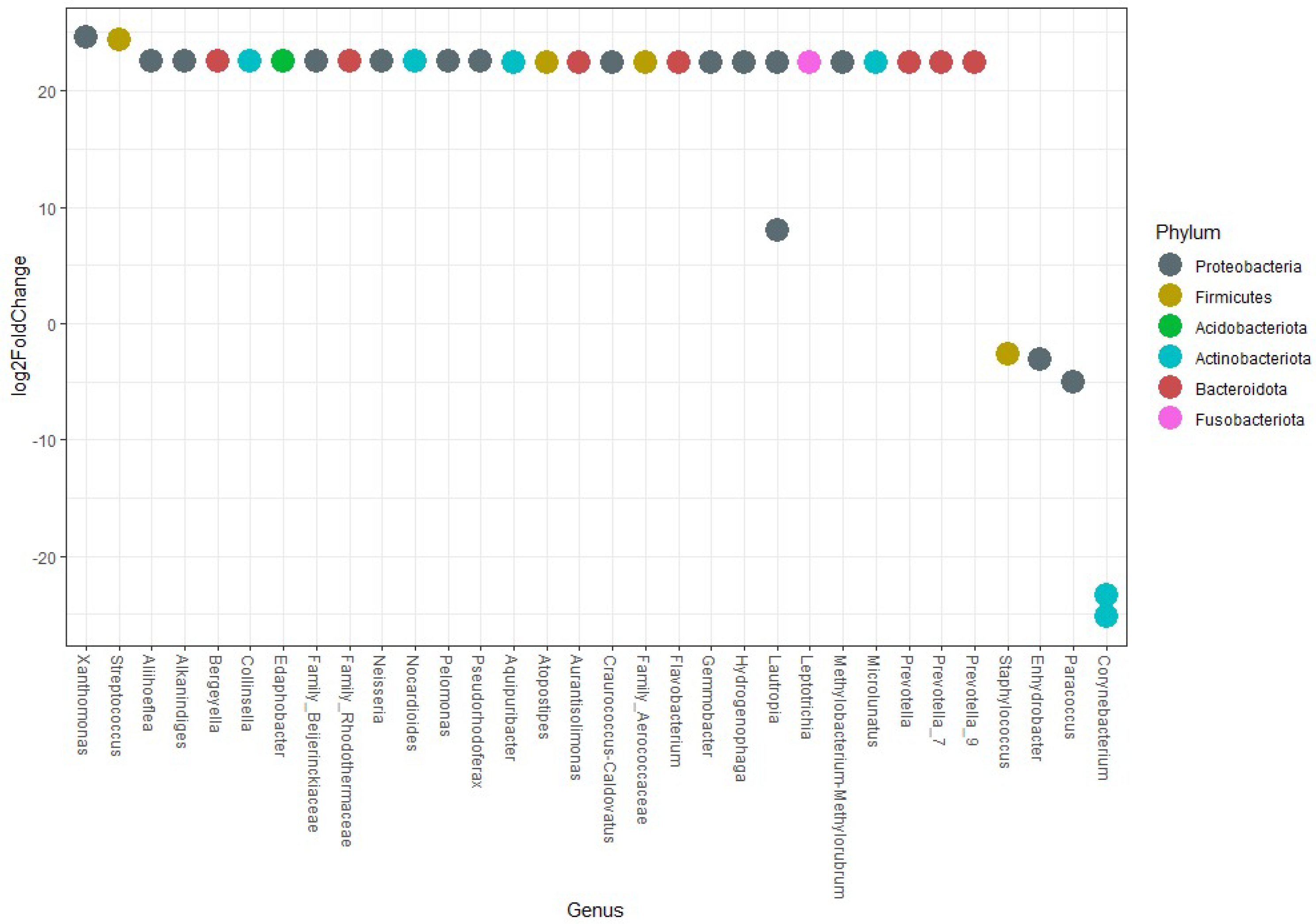
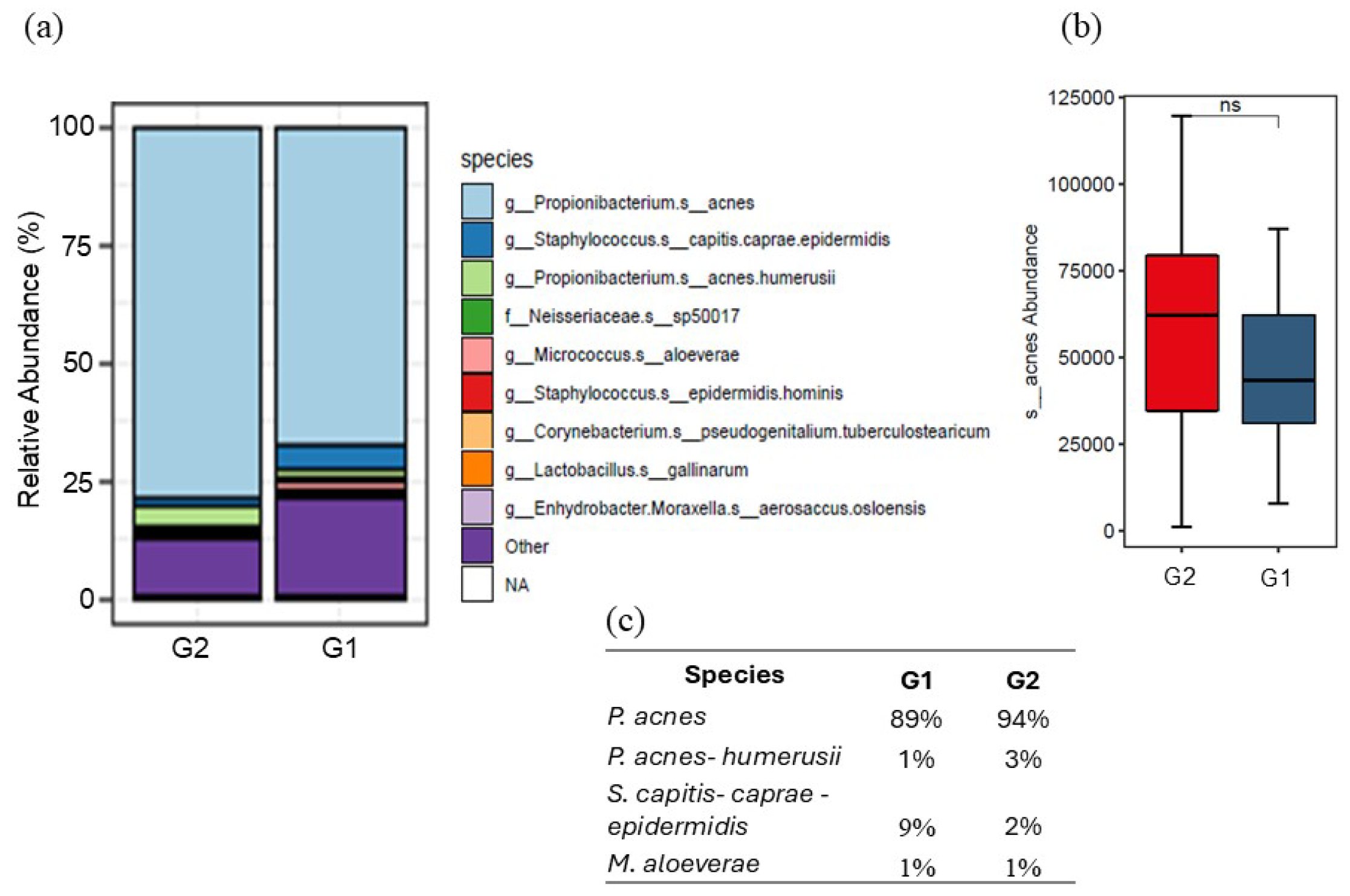
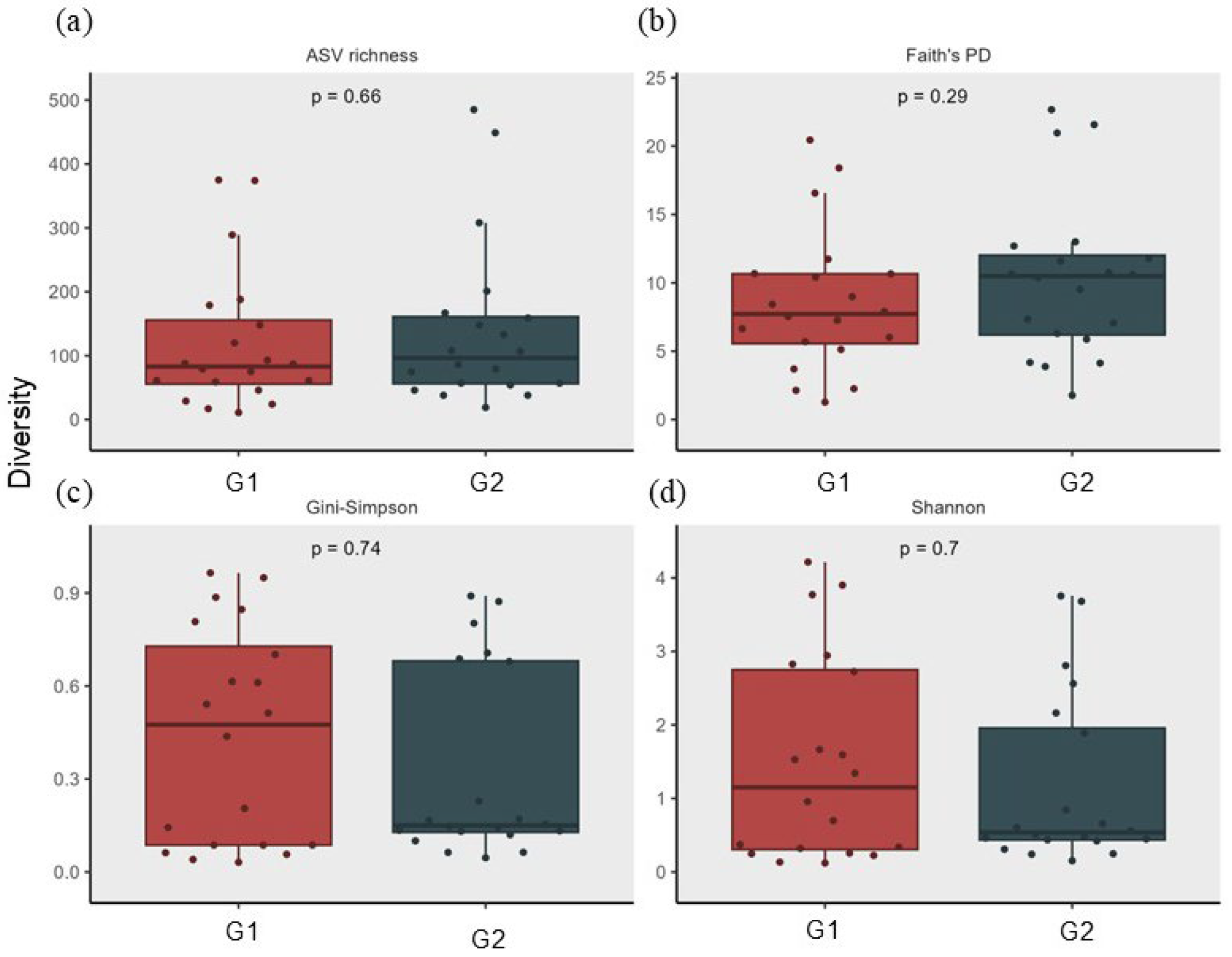
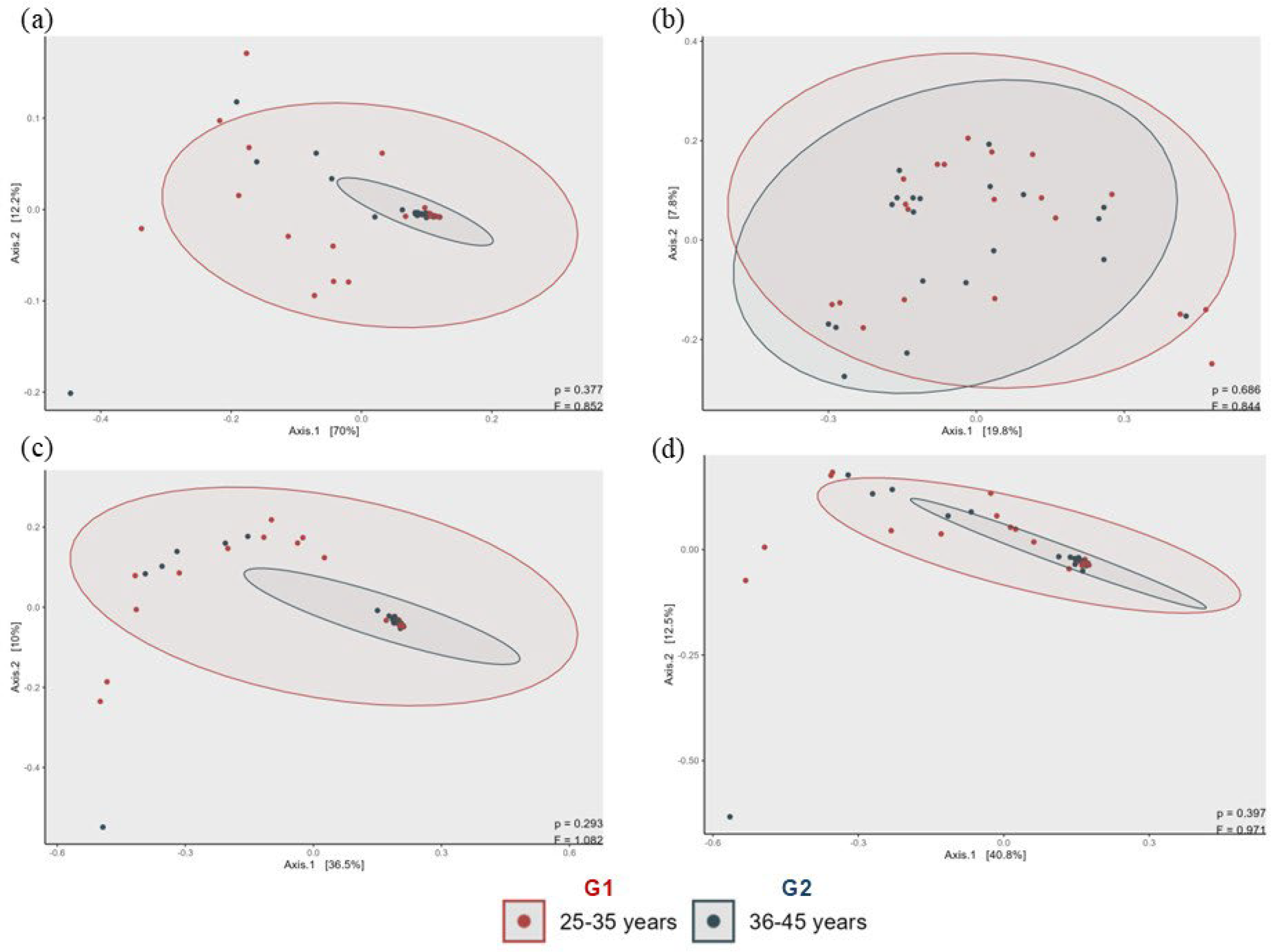
3.2. Analysis of Skin Microbiota by Metagenomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IBGE | Brazilian Institute of Geography and Statistics |
| G1 | Group 1 between 25 and 35 years old |
| G2 | Group 2 between 36 and 45 years old |
| ASV | amplicon sequence variant |
| SCINEXA | Score of Intrinsic and Extrinsic Skin Aging |
References
- Costa, F. Características anatômicas da pele humana e suas variações étnicas. J. Dermatol. Bras. 2019, 43, 254–267. [Google Scholar]
- ABIHPEC. Panorama do Setor de Higiene Pessoal no Brasil; ABIHPEC: São Paulo, Brazil, 2024. [Google Scholar]
- IBGE. Censo Demográfico 2010: Características Gerais da População, Religião e Pessoas Com Deficiência; Instituto Brasileiro de Geografia e Estatística: Palácio da Fazenda, Brazil, 2010. [Google Scholar]
- Matsuo, T. Impacto da exposição solar na pele das brasileiras. Rev. Bras. Dermatol. 2017, 92, 659–667. [Google Scholar]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Prescott, S.L.; Larcombe, D.L.; Logan, A.C.; West, C.; Burks, W.; Caraballo, L.; Levin, M.; Etten, E.; Van Horwitz, P.; Kozyrskyj, A.; et al. The skin microbiome: Impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organ. J. 2017, 10, 29. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- UCHIME Improves Sensitivity and Speed of Chimera Detection|Bioinformatics|Oxford Academic [Internet]. Available online: https://academic.oup.com/bioinformatics/article/27/16/2194/255262 (accessed on 1 August 2025).
- Kers, J.G.; Saccenti, E. The Power of Microbiome Studies: Some Considerations on Which Alpha and Beta Metrics to Use and How to Report Results. Front. Microbiol. 2022, 12, 796025. [Google Scholar] [CrossRef] [PubMed]
- Wilcoxon Signed Ranks Test-An Overview|ScienceDirect Topics [Internet]. Available online: https://www.sciencedirect.com/topics/medicine-and-dentistry/wilcoxon-signed-ranks-test (accessed on 25 July 2025).
- Elias, P.M. Stratum corneum defensive functions: An integrated view. J. Investig. Dermatol. 2005, 125, 183–200. [Google Scholar] [CrossRef] [PubMed]
- Schmid-Wendtner, M.H.; Korting, H.C. The pH of the skin surface and its impact on the barrier function. Ski. Pharmacol. Physiol. 2006, 19, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Jugé, R.; Rouaud-Tinguely, P.; Breugnot, J.; Servaes, K.; Grimaldi, C.; Roth, M.P.; Coppin, H.; Closs, B. Shift in skin microbiota of Western European women across aging. J. Appl. Microbiol. 2018, 125, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Shibagaki, N.; Suda, W.; Clavaud, C.; Bastien, P.; Takayasu, L.; Iioka, E.; Kurokawa, R.; Yamashita, N.; Hattori, Y.; Shindo, C.; et al. Aging-related changes in the diversity of women’s skin microbiomes associated with oral bacteria. Sci. Rep. 2017, 7, 10567. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bek-Thomsen, M.; Lomholt, H.B.; Scavenius, C.; Enghild, J.J.; Brüggemann, H. Proteome analysis of human sebaceous follicle infundibula extracted from healthy and acne-affected skin. PLoS ONE 2014, 9, e107908. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Howard, B.; Bascom, C.C.; Hu, P.; Binder, R.L.; Fadayel, G.; Huggins, T.G.; Jarrold, B.B.; Osborne, R.; Rocchetta, H.L.; Swift, D.; et al. Aging-Associated Changes in the Adult Human Skin Microbiome and the Host Factors that Affect Skin Microbiome Composition. J. Investig. Dermatol. 2022, 142, 1934–1946. [Google Scholar] [CrossRef] [PubMed]
- Elias, P.M.; Feingold, K.R. Lipids and the epidermal water barrier: Metabolism, regulation, and pathophysiology. Semin. Dermatol. 1992, 11, 176–182. [Google Scholar] [PubMed]
- Feingold, K.R. Thematic review series: Skin lipids. The role of epidermal lipids in cutaneous permeability barrier homeostasis. J. Lipid Res. 2007, 48, 2531–2546. [Google Scholar] [CrossRef] [PubMed]
- Knox, S.; O’Boyle, N.M. Skin lipids in health and disease: A review. Chem. Phys. Lipids 2021, 236, 105055. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, I.; Danby, F.W.; Ju, Q.; Wang, X.; Xiang, L.F.; Xia, L.; Chen, W.; Nagy, I.; Picardo, M.; Suh, D.H.; et al. New developments in our understanding of acne pathogenesis and treatment. Exp. Dermatol. 2009, 18, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Ogai, K.; Shibata, K.; Takahashi, N.; Ogura, K.; Okamoto, S.; Sugama, J. Amplicon-based skin microbiome profiles collected by tape stripping with different adhesive film dressings: A comparative study. BMC Microbiol. 2021, 21, 54. [Google Scholar] [CrossRef]
- Perugini, P.; Grignani, C.; Condrò, G.; van der Hoeven, H.; Ratti, A.; Mondelli, A.; Colpani, A.; Bleve, M. Skin Microbiota: Setting up a Protocol to Evaluate a Correlation between the Microbial Flora and Skin Parameters. Biomedicines 2023, 11, 966. [Google Scholar] [CrossRef] [PubMed]
- Karoglan, A.; Paetzold, B.; Pereira de Lima, J.; Brüggemann, H.; Tüting, T.; Schanze, D.; Güell, M.; Gollnick, H. Safety and Efficacy of Topically Applied Selected Cutibacterium acnes Strains over Five Weeks in Patients with Acne Vulgaris: An Open-label, Pilot Study. Acta. Derm. Venereol. 2019, 99, 1253–1257. [Google Scholar] [CrossRef] [PubMed]
- Alan, M.; O’Neill, N.T.; Hayachi, A.; Williams, M.R.; Mills, R.H.; Gonzalez, D.J.; Gallo, R.L. Identification of a Human Skin Commensal Bacterium that Selectively Kills Cutibacterium acnes. J. Investig. Dermatol. 2020, 140, 1619–1628. [Google Scholar] [CrossRef]
- Robe, P.; Jarrin, C.; Zanchetta, C.; Dupont, J.; Chapuis, E.; Scandolera, A.; Auriol, D.; Reynaud, R. Rehabilitation of Skin Bacterial Counts to Assess the Short-Term Impact of Ingredients in Topical Applications—Presenting a Culture-Based Viability Score. Cosmetics 2023, 10, 50. [Google Scholar] [CrossRef]
- Shen, X.; Wang, C.; Zhou, X.; Zhou, W.; Hornburg, D.; Wu, S. Nonlinear dynamics of multi-omics profiles during human aging. Nat. Aging 2024, 4, 1619–1634. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siqueira, R.A.G.B.; Pequeno, A.L.V.; Santos, Y.R.; Morandi-Filho, R.; Lan, A.; Bagatin, E.; Leite-Silva, V.R.; Andreo-Filho, N.; Lopes, P.S. Metagenomic Analysis of the Skin Microbiota of Brazilian Women: How to Develop Anti-Aging Cosmetics Based on This Knowledge? Cosmetics 2025, 12, 165. https://doi.org/10.3390/cosmetics12040165
Siqueira RAGB, Pequeno ALV, Santos YR, Morandi-Filho R, Lan A, Bagatin E, Leite-Silva VR, Andreo-Filho N, Lopes PS. Metagenomic Analysis of the Skin Microbiota of Brazilian Women: How to Develop Anti-Aging Cosmetics Based on This Knowledge? Cosmetics. 2025; 12(4):165. https://doi.org/10.3390/cosmetics12040165
Chicago/Turabian StyleSiqueira, Raquel Allen Garcia Barbeto, Ana Luiza Viana Pequeno, Yasmin Rosa Santos, Romualdo Morandi-Filho, Alexandra Lan, Edileia Bagatin, Vânia Rodrigues Leite-Silva, Newton Andreo-Filho, and Patricia Santos Lopes. 2025. "Metagenomic Analysis of the Skin Microbiota of Brazilian Women: How to Develop Anti-Aging Cosmetics Based on This Knowledge?" Cosmetics 12, no. 4: 165. https://doi.org/10.3390/cosmetics12040165
APA StyleSiqueira, R. A. G. B., Pequeno, A. L. V., Santos, Y. R., Morandi-Filho, R., Lan, A., Bagatin, E., Leite-Silva, V. R., Andreo-Filho, N., & Lopes, P. S. (2025). Metagenomic Analysis of the Skin Microbiota of Brazilian Women: How to Develop Anti-Aging Cosmetics Based on This Knowledge? Cosmetics, 12(4), 165. https://doi.org/10.3390/cosmetics12040165






