Genetic Characterization of Caiman crocodilus (Crocodilia: Alligatoridae) on Gorgona Island, Colombia
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
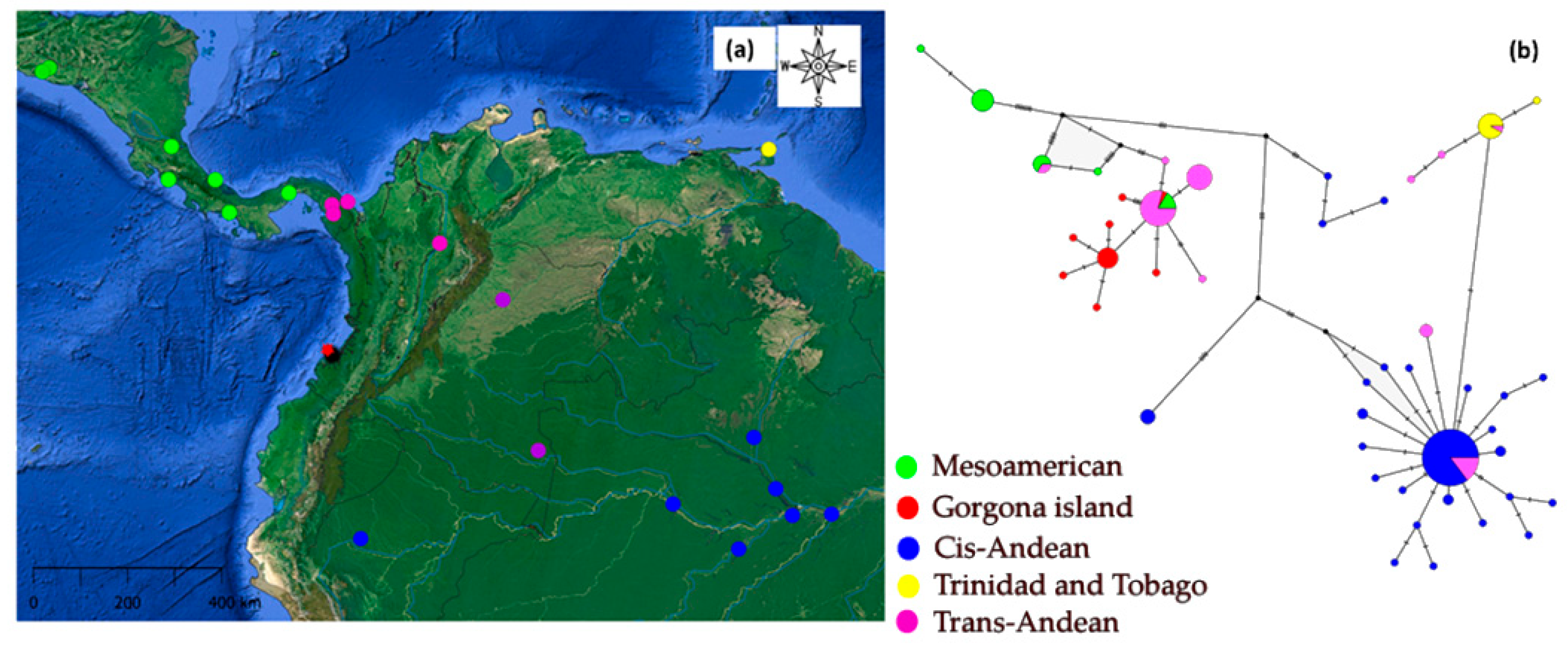
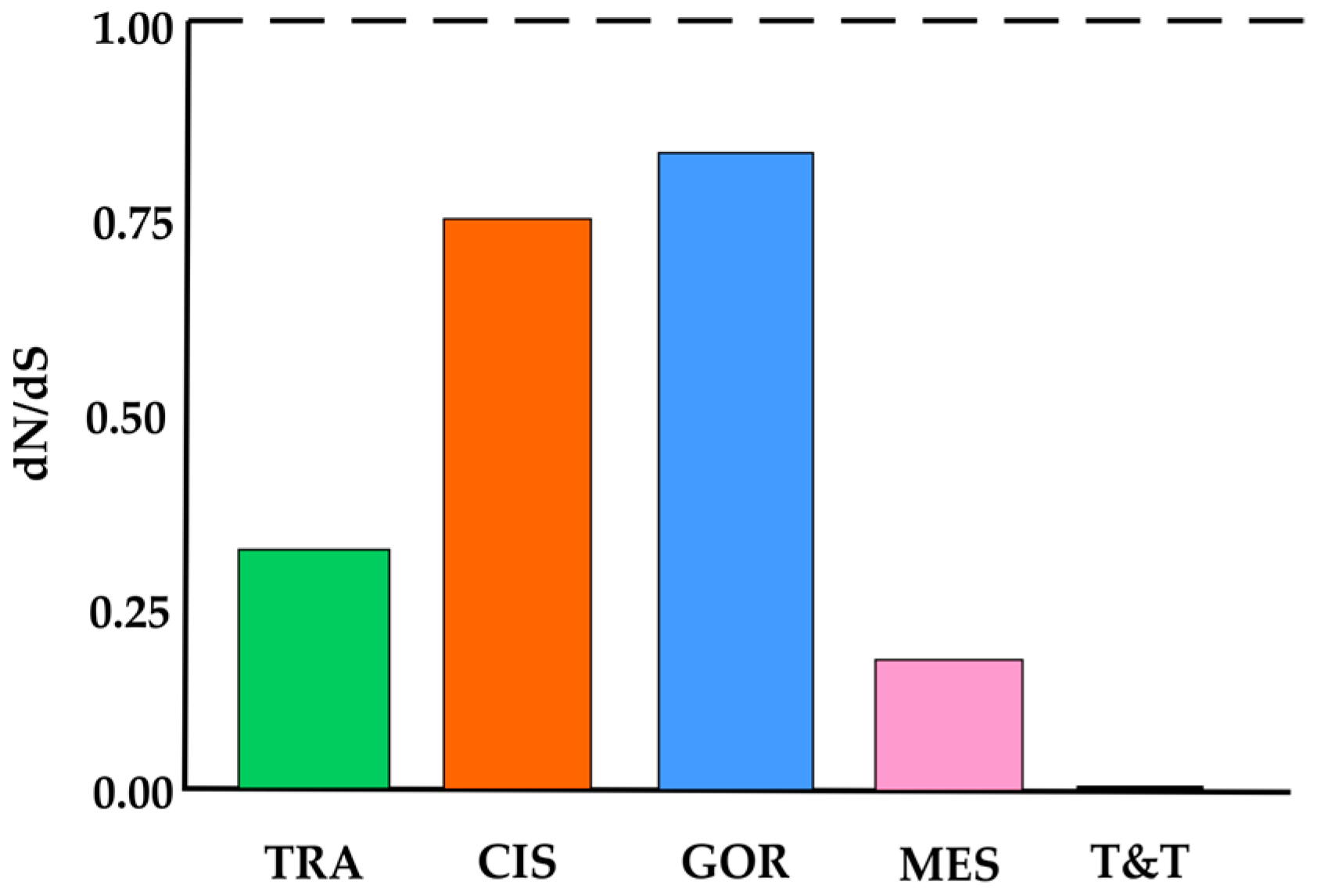
3.1. Genetic Variation
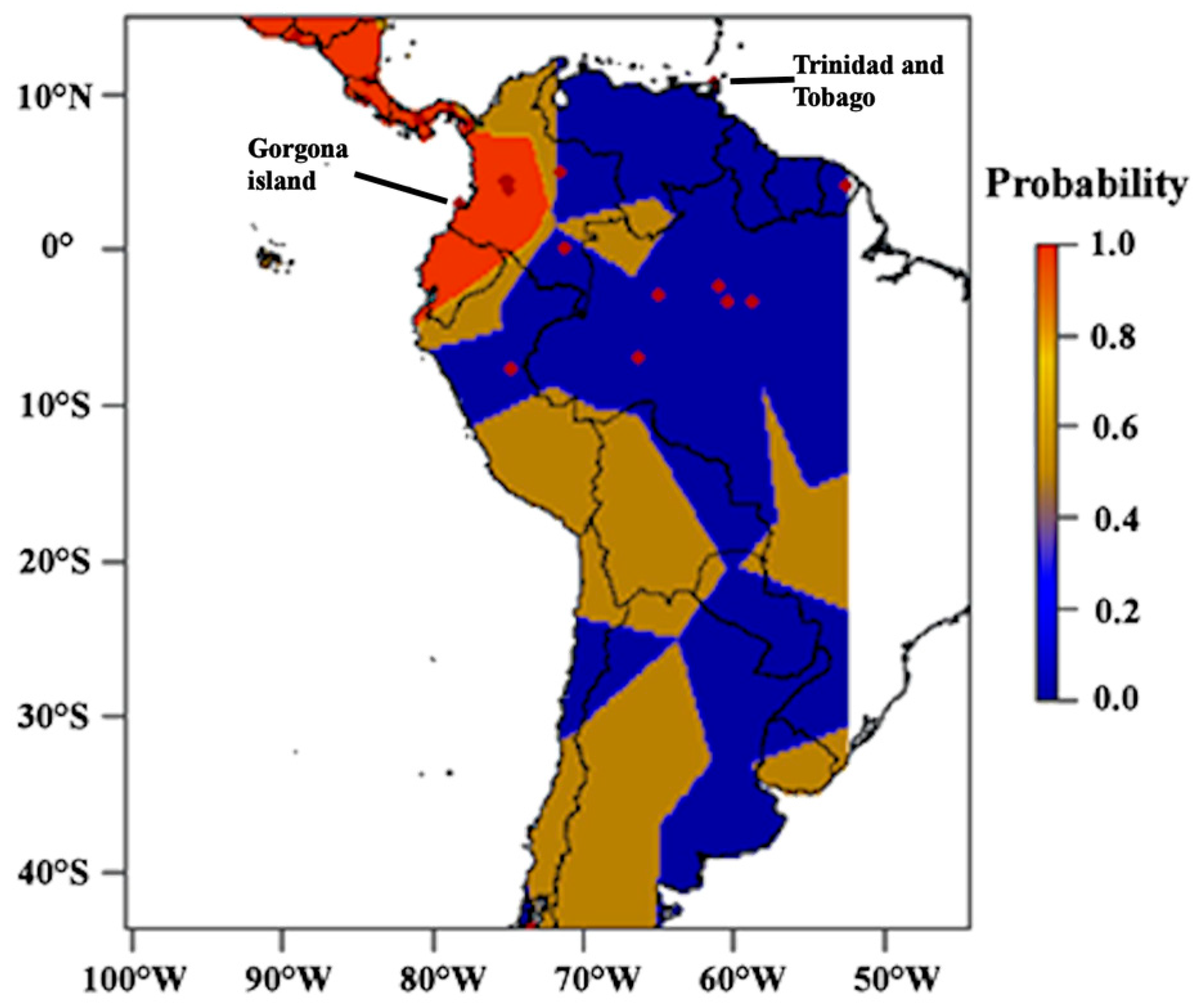
3.2. Genetic Structure
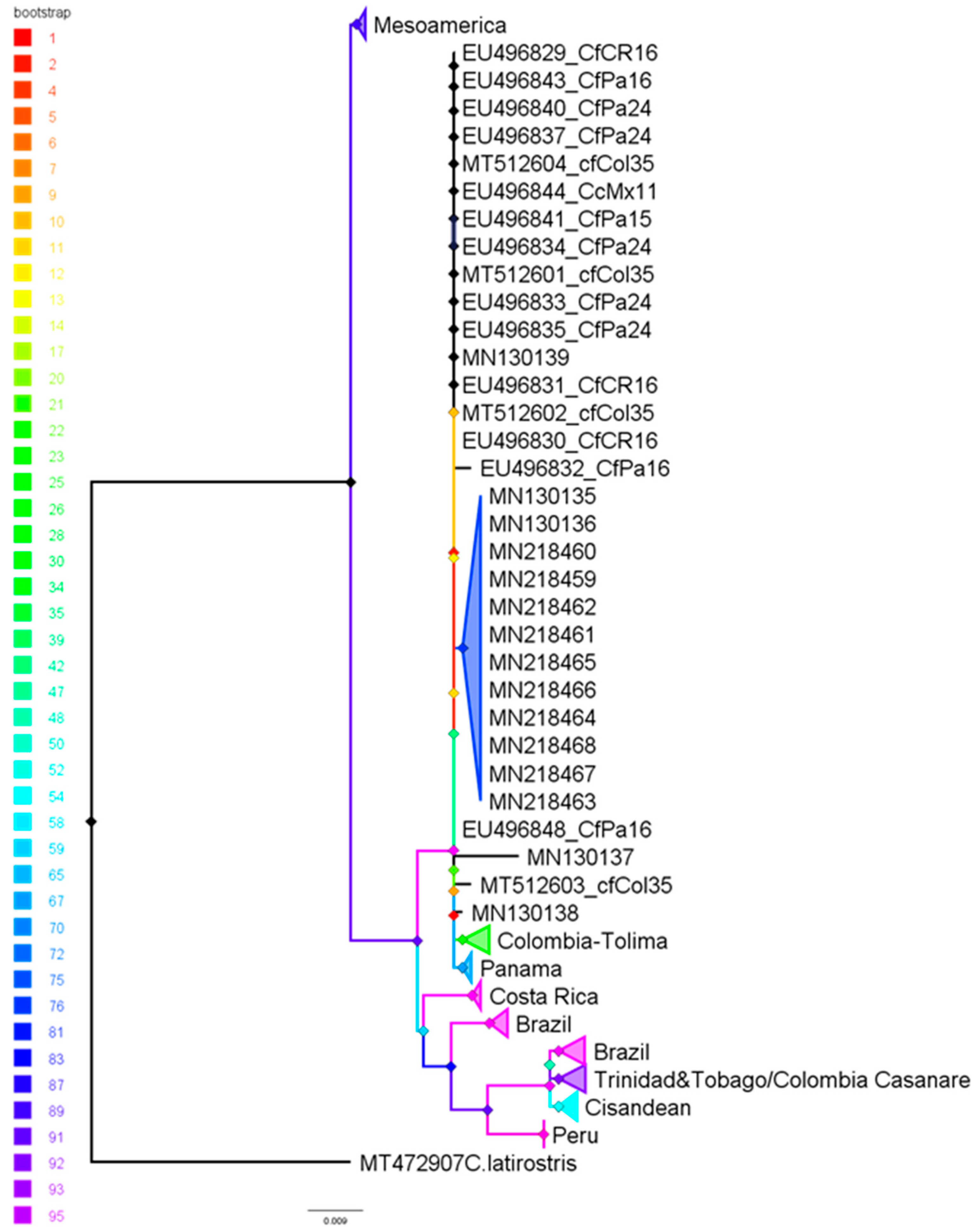
3.3. Phylogenetic Inference
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Myers, N.; Mittermeier, R.; Mittermeier, C.; da Fonseca, G.A.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Martin, S. Global diversity of crocodiles (Crocodilia, Reptilia) in freshwater. In Freshwater Animal Diversity Assessment. Developments in Hydrobiology; Balian, E.V., Lévêque, C., Segers, H., Martens, K., Eds.; Springer: Dordrecht, The Netherlands, 2007; Volume 198, pp. 587–591. [Google Scholar]
- Bartlett, R.; Bartlett, P. Reptiles and Amphibians of the Amazon; University Press of Florida: Gainesville, FL, USA, 2003; 291p. [Google Scholar]
- Díaz, J.M.; Pinzón, J.H.; Perdomo, A.M.; Barrios, L.M.; López-Victoria, M. Generalidades. In Gorgona Marina: Contribución al Conocimiento de una Isla Única; Barrios, L.M., López-Victoria, M., Eds.; Invemar: Gaira, Colombia, 2001; Volume 7, pp. 17–26. [Google Scholar]
- Giraldo, A.; Valencia, B. Isla Gorgona: Paraíso de Biodiversidad y Ciencia; Programa Editorial Universidad del Valle: Cali, Colombia, 2012; 223p. [Google Scholar]
- Giraldo, A.; Diazgranados, M.C.; Gutiérez-Landázuri, C.F. Isla Gorgona, enclave estratégico para los esfuerzos de conservación en el Pacífico Oriental Tropical. Rev. Biol. Trop. 2014, 62, 1–12. [Google Scholar] [CrossRef]
- Medem, F. Los anfibios y reptiles de las islas Gorgona y Gorgonilla. In Gorgona; von Prahl, H., Guhl, F., Grögl, M., Eds.; Universidad de los Andes: Bogotá, Colombia, 1979; Volume 7, pp. 189–218. [Google Scholar]
- Londoño, B.N. Atributos Poblacionales de Caiman crocodilus en un Ambiente Insular del Pacífico Colombiano. Master’s Thesis, Universidad del Valle, Cali, Colombia, 2023. [Google Scholar]
- Balaguera-Reina, S.; González-Maya, J. Estructura poblacional, abundancia y uso del hábitat de Caiman crocodilus fuscus (Cope, 1868) en la Vía Parque Isla Salamanca, Caribe Colombiano. Rev. Biol. Mar. Oceanogr. 2009, 44, 145–152. [Google Scholar] [CrossRef]
- Forero-Medina, G.; Castaño-Mora, O.V.; Rodriguez-Melo, M. Ecología de Caiman crocodilus fuscus en San Andrés Isla, Colombia: Un estudio preliminar. Caldasia 2006, 28, 115–124. [Google Scholar]
- Mohammed, R.S. Distribution of the Caiman, Caiman crocodilus on Tobago. Living World J. Trinidad Tobago Field Nat. Club 2015, 66, 67. [Google Scholar]
- Venegas-Anaya, M.; Crawford, A.; Escobedo-Galván, A.; Sanjur, O.; Densmore, L., III; Bermingham, E. Mitochondrial DNA phylogeography of Caiman crocodilus in Mesoamerica and South America. J. Exp. Zool. Part A Ecol. Genet. Physiol. 2008, 309, 614–627. [Google Scholar] [CrossRef]
- Amavet, P.S.; Pacheco-Sierra, G.; Uhart, M.M.; Prado, W.S.; Siroski, P.A. Phylogeographical analysis and phylogenetic inference based on the cytochrome b gene in the genus Caiman (Crocodylia: Alligatoridae) in Central and South America. Biol. J. Linn. Soc. 2023, 138, 289–303. [Google Scholar] [CrossRef]
- Palumbi, S.R. Nucleic acids II: The polymerase chain reaction. In Molecular Systematics, 2nd ed.; Hillis, D.M., Moritz, C., Mable, B.K., Eds.; Sinauer Associates: Sunderland, MA, USA, 1996; pp. 205–247. [Google Scholar]
- Arbor, A. Sequencher, Version 4.10; Gene Codes Corporation: Ann Arbor, MI, USA, 2010. Available online: https://www.genecodes.com (accessed on 14 March 2024).
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Vasconcelos, W.R.; Hrbek, T.; Da Silveira, R.; de Thoisy, B.; Marioni, B.; Farias, I.P. Population genetic analysis of Caiman crocodilus (Linnaeus, 1758) from South America. Genet. Mol. Biol. 2006, 29, 220–230. [Google Scholar] [CrossRef]
- Balaguera-Reina, S.; Vargas-Ramírez, M.; Ordóñez-Garza, N.; Hernández-González, F.; Densmore, L.D. Unveiling the mystery: Assessing the evolutionary trajectory of the Apaporis caiman population (Caiman crocodilus apaporiensis, Medem 1955) via mitochondrial molecular markers. Biol. J. Linn. Soc. 2020, 131, 163–171. [Google Scholar] [CrossRef]
- Charif, D.; Lobry, J.R. SeqinR 1.0-2: A Contributed Package to the R Project for Statistical Computing Devoted to Biological Sequences Retrieval and Analysis. In Structural Approaches to Sequence Evolution; Bastolla, U., Porto, M., Roman, H.E., Vendruscolo, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 207–2323. [Google Scholar]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Guillot, G.; Mortier, F.; Estoup, A. Geneland: A computer package for landscape genetics. Mol. Ecol. 2005, 5, 712–715. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Dessauer, H.C.; Glenn, T.C.; Densmore, L.D. Studies on the molecular evolution of the Crocodylia: Footprints in the sands of time. J. Exp. Zool. 2002, 294, 302–311. [Google Scholar] [CrossRef]
- Roberto, I.J.; Bittencourt, P.S.; Muniz, F.L.; Hernández-Rangel, S.M.; Nóbrega, Y.C.; Ávila, R.W.; Souza, B.C.; Álvarez, G.; Miranda-Chumacero, G.; Campo, Z.; et al. Unexpected but unsurprising lineage diversity within the most widespread Neotropical crocodilian genus Caiman (Crocodylia, Alligatoridae). Syst. Biodivers. 2020, 18, 377–395. [Google Scholar] [CrossRef]
- Balaguera-Reina, S.A.; Konvalina, J.D.; Mohammed, R.S.; Gross, B.; Vazquez, R.; Moncada, J.F.; Ali, S.; Hoffman, E.A.; Densmore, L.D. From the river to the ocean: Mitochondrial DNA analyses provide evidence of spectacled caimans (Caiman crocodilus Linnaeus 1758) mainland–insular dispersal. Biol. J. Linn. Soc. 2021, 134, 486–497. [Google Scholar] [CrossRef]
- Jiménez-Alonso, G.; Balaguera-Reina, S.A.; Hoyos, M.; Bloor, P. Phylogenetic and phylogeographic insights on Trans-Andean spectacled caiman populations in Colombia. Mar. Freshw. Res. 2023, 74, 1071–1080. [Google Scholar] [CrossRef]
- Gil-Vargas, D.L.; Sedano-Cruz, R.E. Genetic variation of avian malaria in the tropical Andes: A relationship with the spatial distribution of hosts. Malar. J. 2019, 18, 129. [Google Scholar] [CrossRef] [PubMed]
- Morrone, J. Cladistic biogeography of the Neotropical region: Identifying the main events in the diversification of the terrestrial biota. Cladistics 2014, 30, 202–214. [Google Scholar] [PubMed]
- Noguera-Urbano, E.A.; Escalante, T. Áreas de endemismo de los mamíferos (Mammalia) neotropicales. Acta Biol. Colomb. 2015, 20, 47–65. [Google Scholar] [CrossRef]
- Meyer, N.F.; Balkenhol, N.; Dutta, T.; Hofman, M.; Meyer, J.Y.; Ritchie, E.G.; Alley, C.; Beranek, C.; Bugir, C.K.; Callen, A.; et al. Beyond species counts for assessing, valuing, and conserving biodiversity: Response to Wallach et al. 2019. Conserv. Biol. 2021, 35, 369–372. [Google Scholar] [CrossRef] [PubMed]
- Holmes, I.A.; Mautz, W.J.; Davis Rabosky, A.R. Historical environment is reflected in modern population genetics and biogeography of an island endemic lizard (Xantusia riversiana reticulata). PLoS ONE 2016, 11, e0163738. [Google Scholar]
- Trumbo, D.R.; Funk, W.C.; Pauly, G.B.; Robertson, J.M. Conservation genetics of an island-endemic lizard: Low Ne and the critical role of intermediate temperatures for genetic connectivity. Conserv. Genet. 2021, 22, 783–797. [Google Scholar] [CrossRef]
| Region | N | H | Hd | π | Tajima’s D | p |
|---|---|---|---|---|---|---|
| Trans-Andean | 34 | 17 | 0.7219 | 0.012189 ± 0.006434 | 0.32835 | 0.70700 |
| Gorgona | 15 | 8 | 0.7195 | 0.003751 ± 0.002525 | −2.021300 | 0.01200 |
| Trinity and Tobago | 12 | 2 | 0.1667 | 0.000730 ± 0.000750 | −1.62929 | 0.04600 |
| Cis-Andean | 118 | 32 | 0.7333 | 0.011278 ± 0.005861 | −0.90917 | 0.19000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Londoño, N.; Sedano-Cruz, R.E.; Giraldo, A. Genetic Characterization of Caiman crocodilus (Crocodilia: Alligatoridae) on Gorgona Island, Colombia. Biology 2025, 14, 1227. https://doi.org/10.3390/biology14091227
Londoño N, Sedano-Cruz RE, Giraldo A. Genetic Characterization of Caiman crocodilus (Crocodilia: Alligatoridae) on Gorgona Island, Colombia. Biology. 2025; 14(9):1227. https://doi.org/10.3390/biology14091227
Chicago/Turabian StyleLondoño, Natalia, Raúl Ernesto Sedano-Cruz, and Alan Giraldo. 2025. "Genetic Characterization of Caiman crocodilus (Crocodilia: Alligatoridae) on Gorgona Island, Colombia" Biology 14, no. 9: 1227. https://doi.org/10.3390/biology14091227
APA StyleLondoño, N., Sedano-Cruz, R. E., & Giraldo, A. (2025). Genetic Characterization of Caiman crocodilus (Crocodilia: Alligatoridae) on Gorgona Island, Colombia. Biology, 14(9), 1227. https://doi.org/10.3390/biology14091227






