Simple Summary
This study investigates the genetic diversity and genetic structure of the leaf beetle Platycorynus peregrinus in Thailand using mitochondrial COI gene sequences. Specimens were collected from multiple locations across the country. The results revealed significant genetic diversity both within and among populations, suggesting possible local adaptation and population structure. These findings provide valuable insights into the evolutionary potential of P. peregrinus and support the development of more effective pest management and control strategies in the future.
Abstract
Platycorynus peregrinus (Herbst, 1783) is a leaf beetle of agricultural importance, yet its genetic diversity and population structure remain poorly understood. We analyzed mitochondrial cytochrome c oxidase subunit I (COI) sequences from 147 individuals across 19 populations in Thailand. Forty-five haplotypes were identified, showing high haplotype diversity (0.942) and moderate nucleotide diversity (0.00562). Significant genetic differentiation (ΦST = 0.0000–0.7857) was strongly associated with geographical distance, and this population structure was further supported by AMOVA (FCT = 0.21925, p < 0.001). Neutrality tests and mismatch distribution analyses revealed signals of population expansion, alongside evidence of localized differentiation. The substantial genetic variability observed suggests high dispersal ability and possible cryptic lineages. These insights not only advance understanding of the evolutionary dynamics of P. peregrinus but also provide valuable guidance for developing sustainable pest management strategies.
1. Introduction
Platycorynus peregrinus (Herbst, 1783) is a species of leaf beetle belonging to the family Chrysomelidae, subfamily Eumolpinae. This beetle is primarily phytophagous and secondarily graminivorous, feeding on a variety of host plants [1]. The primary host plants for P. peregrinus are members of the genus Calotropis, commonly known as the crown flower tree, a toxic shrub characterized by its milky latex. Notably, P. peregrinus has been observed visiting more frequently on Calotropis gigantea, suggesting a strong preference for species within the Calotropis genus [2]. In addition to Calotropis, this beetle has also been reported feeding on various species of Digitaria (a genus of grasses), indicating a broader host range. Moreover, P. peregrinus is known to feed on several agriculturally important crops, including eggplant, okra, tomato, and potato [1]. This dietary versatility suggests that while the beetle has preferred host plants, it is capable of adapting to a variety of plant species, some of which are of economic significance. Both the adult and larval stages of this beetle feed on the leaves of these plants [1].
This leaf beetle is widely distributed in the tropical and subtropical regions, particularly in South and Southeast Asia [2]. Heavy infestations cause severe defoliation, reducing plant vigor and negatively impacting their ecological and economic value [1]. Although this species is agriculturally important, research on its biology and population genetics remains scarce. Knowledge of the genetic diversity of pest species is essential not only for understanding evolutionary processes and population structure, but also for informing pest management strategies [3]. Investigating its genetic variation can provide critical insights into its evolutionary history, population structure, and adaptability to diverse environments. Such information is important for predicting its potential spread and for developing evidence-based management strategies [4,5].
For example, a study on the invasive agricultural pest Thrips palmi Karny, 1925 in China demonstrated the importance of population genetic analysis for understanding pest distribution and informing control strategies [6]. Similarly, research on the African fig fly, Zaprionus indianus Gupta, 1970, showed that invasive populations can experience reductions in genetic diversity, which may affect their adaptability and fitness [7]. More recently, a study in Thailand reported the genetic variation in the seed bug Spilostethus pandurus (Scopoli, 1763), an agriculturally important insect. The results highlighted the species’ adaptability and potential for local differentiation, while the low divergence from populations in other continents suggested ongoing gene flow [8]. Collectively, these studies underscore the crucial role of genetic data in guiding pest management and control efforts.
Mitochondrial DNA (mtDNA) has become a valuable tool in genetic studies, particularly in identifying genetic variation within and between species [9]. The mitochondrial cytochrome c oxidase subunit I (COI) gene is widely used as a genetic marker in population genetics and phylogeography due to its relatively high mutation rate and maternal inheritance [10]. COI has been extensively employed in the DNA barcoding of species, enabling researchers to distinguish between species, and determining the possibility of cryptic species [11]. Studying the COI gene can provide insights into genetic differentiation between geographically isolated populations, reflecting patterns of gene flow, migration, and historical biogeography.
This study aims to examine the genetic diversity, genetic structure and population demographic history of P. peregrinus in Thailand by using the mitochondrial COI gene as a genetic marker. We seek to assess the genetic variability within populations and determine the degree of genetic differentiation between geographically distinct populations. The findings will contribute to the broader understanding of the population genetics of P. peregrinus and provide a baseline for future studies on its evolutionary history. This research will also contribute to the growing body of literature that utilizes mitochondrial markers to assess genetic variation in insect populations, thereby enhancing our understanding of genetic diversity across species and regions.
2. Materials and Methods
2.1. Sample Collection and Molecular Analysis
We collected 147 individuals of P. peregrinus (Figure 1) by hand picking from their host tree, Calotropis spp. (crown flower tree) from 19 localities in Thailand (Table 1 and Figure 2). The beetles were immobilized by chilling on ice for approximately 10 min, then immediately transferred to 80% ethanol for preservation. The leaf beetles were randomly selected for species identification by morphology using key to species of the family Chrysomelidae of Thailand reported by [12]. Total DNA was individually extracted from the left foreleg of each leaf beetle using E.Z.N.A.® Tissue DNA kit (Omega bio-tek, Norcross, GA, USA) following the manufacturer’s protocol. The remaining body was preserved as a voucher specimen for morphological reference if needed in the future. DNA samples were kept at −20 °C for further molecular analysis. Partial sequence of the COI fragment was amplified and sequenced using primers and PCR conditions as published by [13]. The PCR products were electrophoresed in 1% agarose gels and visualized with GelRedTM Nucleic Acid Gel Stain (Biotium, Inc., Hayward, CA, USA). The amplified band was cut and purified by using E.Z.N.A.® Gel Extraction kit (Omega bio-tek, Norcross, GA, USA). The purified PCR products were sent for DNA sequencing at ATGC Co., Ltd., Thailand.

Figure 1.
Adult of leaf beetle Platycorynus peregrinus in lateral (left) and dorsal (right) views.

Table 1.
Sampling localities and associated details for Platycorynus peregrinus populations collected in Thailand.
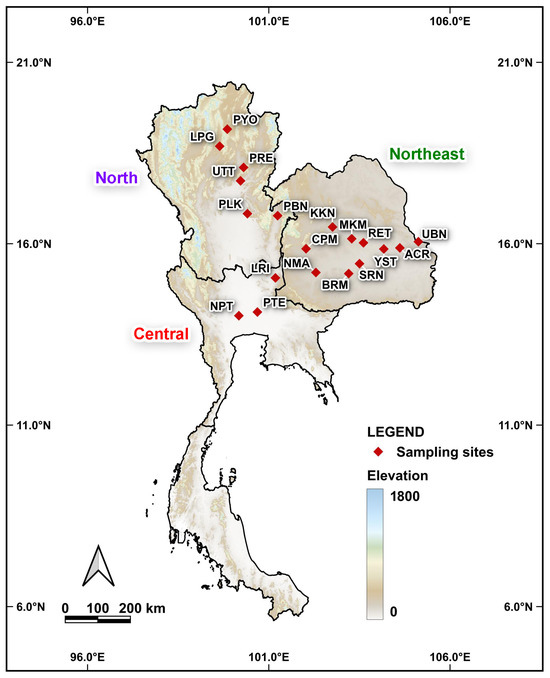
Figure 2.
Map showing the sampling localities of Platycorynus peregrinus populations across Thailand. Each locality corresponds to the localities listed in Table 1.
2.2. Data Analysis
All COI sequences generated in this study were aligned using the ClustalW program [14] and manually edited in the BioEdit program [15]. Molecular diversity indices and haplotype data were generated using the DnaSp v5 program [16]. A mitochondrial haplotype network was constructed in the Network program version 10.2 (https://www.fluxus-engineering.com/, accessed on 15 July 2025) based on median-joining method [17] using all sequences generated in this study. Neutrality tests (Tajima’s D and Fu’s Fs), genetic differentiation (ΦST) analysis, mismatch distribution analysis, and Analysis of Molecular Variance (AMOVA) were conducted using the Arlequin program version 3.5.2.2 [18]. The resulting ΦST p-values were corrected for multiple comparisons using a Bonferroni correction to avoid type I errors following the formula αnew = α/n, where α = 0.05 and n = 171 is the number of possible pairwise comparisons between the 19 populations. We considered a p-value to be statistically significant if it was less than the adjusted threshold of αnew = 0.05/171 ≈ 0.00029. Furthermore, the effective population size (Ne) for each population was estimated, following the formula Ne = θ/2μ, where the theta (θ) was estimated in DnaSp v5 (accessed on 15 July 2025) and the mutation rate per site per generation (μ) was assumed to be 1.8 × 10−8 [19,20]. To assess the geographical distance separating population groups, an Isolation by Distance (IBD) analysis was conducted to evaluate the correlation between pairwise Euclidean geographical distances and genetic differentiation (ΦST) at the population level. This analysis was performed using the ecodist package [21] in R [22].
2.3. Phylogenetic Tree Reconstruction
Phylogenetic trees were constructed using Bayesian inference (BI), Maximum likelihood (ML), and Neighbor joining (NJ) methods. Using the corrected Akaike information criterion (AIC), MrModeltest 2.4 [23] provided the best-fit evolutionary models for the COI marker. The GTR + I + G was shown to be the best-fit model for ML and BI phylogenetic analysis. For the BI analysis, we used BEAST X v10.5 [24] with 30,000,000 MCMC iterations, sampling every 1000 iterations. The run was checked for the convergence of the MCMC and effective sample sizes (ESS > 200) in TRACER v.1.7.2 [25]. We used the program Figtree v.1.4.4 [26] to visualize the trees after discarding the first 10% of each MCMC chain as burn-in. The ML and NJ were carried out in MEGA12 [27] with 1000 bootstrap replicates using the GTR + I + G and Kimura 2-parameter (K2P) models, respectively.
3. Results
3.1. Genetic Variation and Effective Population Size
Mitochondrial COI sequences of P. peregrinus generated in this study were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/, accessed on 15 July 2025) under accession no. PV400438–PV400482. Across 147 individuals from 19 populations, a total of 45 variable sites were detected, namely 17 singleton variable sites and 28 parsimony informative sites (Table S1). Based on those variations, 45 haplotypes were identified (Pp1–Pp45). Haplotype diversity (Hd) in each population varied between 0 and 0.972 ± 0.064 (average 0.942 ± 0.008). Similarly, the nucleotide diversity (Nd) varies between 0.0000 ± 0.000 and 0.0067 ± 0.0014 (average 0.00562 ± 0.0004) (Table 2). Populations such as PRE, NPT, PBN, LPG, RET, PLK, MKM, and NMA exhibited high haplotype diversity (Hd) (>0.900). Among the 45 haplotypes identified, 32 haplotypes were unique to a single population. PRE and MKM harbored the highest number (4) of unique haplotypes (Uh) (Table 2). Conversely, populations such as KKN, PYO, and UTT had no unique haplotypes, indicating shared haplotypes with other populations.

Table 2.
Molecular diversity indices of Platycorynus peregrinus from different geographical localities in Thailand, based on COI sequences analysis.
Estimates of the population mutation parameter (θ) and effective population size (Ne) based on COI sequences varied among populations (Table 2). θ ranged from 0.0000 in KKN (no polymorphism) to 0.00811 in PBN, while Ne spanned from 1.49 × 105 (ACR) to 9.01 × 105 (PBN), with an overall mean Ne of 14.0 × 105. Populations such as PBN, PRE, UBN, and NRT showed higher Ne, indicating larger effective sizes and greater genetic variation, whereas ACR, PTE, BRM, and CPM exhibited lower Ne, reflecting reduced diversity and stronger genetic drift. KKN showed Ne = 0, consistent with the absence of variation. These results highlight heterogeneity in genetic diversity, with most populations maintaining large effective sizes, while some display reduced variation likely due to local demographic processes.
3.2. Haplotype Network Analysis
A mitochondrial DNA haplotype network revealed a complex genetic network among the populations from different geographic regions (Figure 3). A total of 45 haplotypes were identified, with several haplotypes shared across multiple populations and others restricted to specific localities. Three predominant haplotypes (Pp1, Pp7, and Pp11) were the most frequent but only Pp7 and Pp11 are widely distributed while Pp1 was restricted to the northeast of Thailand. Among 45 identified haplotypes, only 7 were shared among geographic regions. The network exhibited a star-like topology, suggesting historical population expansion. Overall, the haplotype network highlights a high level of genetic diversity across populations, with evidence of both historical gene flow and localized differentiation. The distribution of haplotypes suggests a mix of shared ancestral lineages and recent genetic divergence among certain populations.
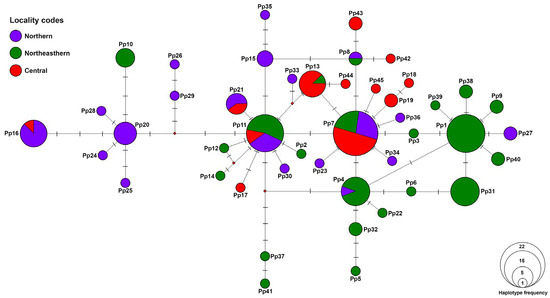
Figure 3.
A haplotype network constructed from COI haplotypes of Platycorynus peregrinus populations in Thailand. Each color represents a different region of sampling localities in Thailand (see Table 1 for details). The size of each circle corresponds to the number of individuals sharing that haplotype. Hash marks on the branches indicate the number of mutational steps between haplotypes.
3.3. Phylogenetic Analysis
A Bayesian phylogenetic tree based on COI haplotypes was constructed to further examine the genetic relationships among P. peregrinus haplotypes (Figure 4). The analysis recovered all 45 haplotypes (Pp1–Pp45) of P. peregrinus as a well-supported monophyletic clade, clearly separated from the outgroups (Platycorynus dejeani Bertoloni, 1849, Herpisticus bobadillae, and Polydrusus mollis Ström, 1768). Within P. peregrinus, the haplotypes were arranged in shallow branches with generally short internode distances, reflecting low sequence divergence among them. Several small clusters of haplotypes were observed with moderate to high support values (e.g., Pp5 and Pp32, Pp26 and Pp29, Pp41 and Pp37). These results are congruent with the haplotype network analysis (Figure 3), suggesting that P. peregrinus represents a single species with high haplotype diversity but shallow genealogical divergence.
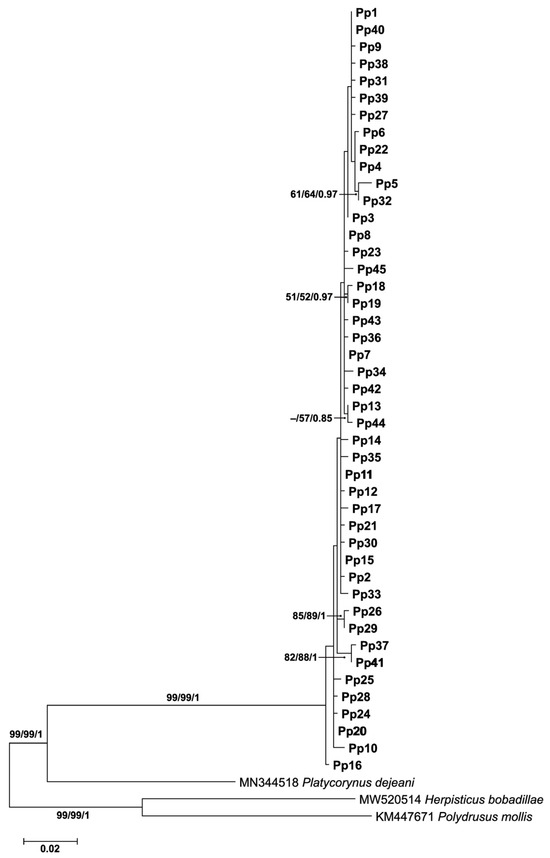
Figure 4.
Bayesian phylogenetic tree of 45 COI haplotypes (Pp1–Pp45) of Platycorynus peregrinus from Thailand. Support values at nodes represent Maximum Likelihood bootstrap, Neighbor-Joining bootstrap, and Bayesian posterior probability, respectively. The en dash (–) indicates supporting values less than 50. Outgroups include Platycorynus dejeani, Herpisticus bobadillae, and Polydrusus mollis.
3.4. Genetic Differentiation and Genetic Structure
Pairwise ΦST values among the 19 populations ranged from 0.0000 to 0.7857, indicating a wide range of genetic differentiation (Table 3). While some population pairs showed no significant genetic differentiation, others exhibited significant differentiation, as highlighted by the gray shading ΦST values in Table 3. The highest genetic difference was observed between CPM and KKN populations (ΦST = 0.7857, p < 0.00001), while consistently low and nonsignificant values were detected among MKM, NMA, SRN, and BRM populations. The table shows clusters of closely related populations with low ΦST values, while more divergent populations, especially KKN, display consistently higher ΦST values (Table 3), indicating substantial genetic differentiation. The statistical significance of the ΦST values is indicated by highlighting the genetic structure across the sampled range. A significant pattern of IBD was detected (Mantel test, r = 0.4596, p = 0.001), indicating a positive correlation between genetic differentiation (ΦST) and geographic Euclidean distance (Figure 5). This suggests that populations located farther apart are more genetically distinct, consistent with restricted gene flow over increasing distances.

Table 3.
Pairwise genetic differentiation (ΦST) among Platycorynus peregrinus populations from 19 different localities in Thailand based on COI sequence analysis.
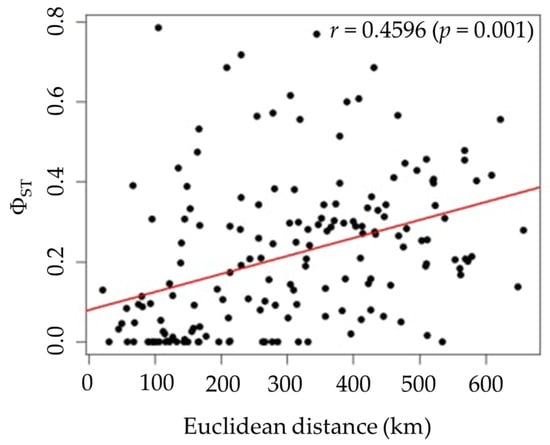
Figure 5.
Isolation-by-distance (IBD) by Mantel test based on COI sequences, showing the relationship between genetic differentiation (ΦST) and geographic Euclidean distance (km). r represents the correlation coefficient, and p represents the probability value (p-value) assessing statistical significance.
Analysis of Molecular Variance (AMOVA) demonstrated significant genetic differentiation among the three defined groups based on different regions in Thailand, namely northern, central, and northeastern, with FCT = 0.21925 (p < 0.001). Additionally, significant genetic variation was detected among populations within groups (FSC = 0.06520, p < 0.05) and among individuals within populations (FST = 0.27016, p < 0.001) (Table 4).

Table 4.
Analysis of Molecular Variance (AMOVA) based on COI sequences of Platycorynus peregrinus populations defined by three population groups corresponded to different regions in Thailand, namely northern, central, and northeastern regions.
3.5. Demographic History
Neutrality tests of all populations yielded significant negative values for both Tajima’s D (–1.65317, p = 0.01300) and Fu’s Fs (–25.93013, p < 0.00001), suggesting possible population expansion or purifying selection. The mismatch distribution analysis revealed a unimodal and smooth pattern of pairwise differences among sequences, indicating an excess of low-frequency polymorphisms and recent population expansion (Figure 6). The observed distribution did not differ significantly from the expected under sudden expansion model, suggesting that the population has experienced recent demographic expansion. Both sum of square deviation and Harpending’s Raggedness index tests were not statistically significant (Figure 6).
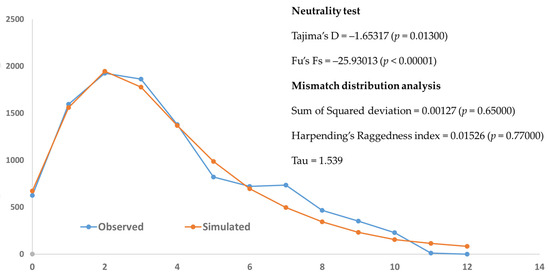
Figure 6.
Mismatch distribution graph of Platycorynus peregrinus based on COI sequence data. The solid blue line represents the observed frequency of pairwise nucleotide differences, while the solid orange line indicates the expected distribution under a model of sudden demographic expansion.
4. Discussion
High genetic diversity of P. peregrinus was observed in this study, suggesting considerable evolutionary potential that may facilitate adaptation to environmental changes, host plant defenses, and pest control measures. P. peregrinus is considered as a pest of various agriculturally important crops, including eggplant, okra, tomato, and potato [1]. A high diversity of plant species can lead to increased genetic variation in herbivorous beetle populations by providing a wider range of food sources and potential habitats, which can drive specialization and genetic differentiation among beetle populations [28]. Therefore, observation of the genetic variation in P. peregrinus in this study may indicate the presence of multiple host-adapted lineages. This idea is supported by a previous study on the leaf beetle Neochlamisus bebbianae (Brown, 1943), which identified two sympatric host-associated forms, one adapted to maple and the other to willow, and demonstrated that genetic differences related to host use resulted in reduced hybrid fitness, thereby promoting reproductive isolation between host-adapted lineages [29]. In this study, P. peregrinus were collected only from crown flower trees; thus, we cannot determine whether the observed genetic differentiation is related to host plant usage or not. Therefore, continued genetic monitoring and host-use studies are essential to better understand host plant usage and genetic differentiation which can be used to support the development of more targeted and effective pest management strategies.
The significant spatial genetic structure observed in the isolation-by-distance (IBD) analysis of P. peregrinus populations across Thailand suggests restricted gene flow among geographically separated populations. This pattern may be influenced by geographical distance, dispersal limitations, or natural barriers. Further analysis integrating landscape features and species distribution with genetic differentiation could elucidate their impacts on gene flow. Significant genetic differentiation, as indicated by ΦST values, reflects barriers to gene flow shaped by ecological or geographic factors. These patterns may be influenced by ecological factors such as host plant availability, climate variability, and habitat fragmentation. Similar patterns have been reported in the willow leaf beetle Chrysomela aeneicollis (Schaeffer, 1928), which exhibited significant genetic differentiation among populations correlated with variation in snowpack and temperature [30]. For instance, research on the oak wilt vector beetle Platypus quercivorus Murayama, 1925 revealed that populations exhibited genetic structuring influenced by geographical distance and landscape features. This suggests that natural barriers, such as mountain ranges or fragmented habitats, can limit dispersal and gene flow among beetle populations [31]. In Thailand, similar spatial genetic structuring has been reported in the jewel beetle Sternocera aequisignata Saunders, 1866 [32], the large brown cricket Tarbinskiellus portentosus (Lichtenstein 1796) [13], and the seed bug S. pandurus [8].
High genetic diversity might indicate the presence of cryptic species or incipient speciation within the pest population. Recognizing and understanding this cryptic diversity is crucial for effective pest management, as they may differ in behavior, host preference, or susceptibility to control measures. For instance, the whitefly Bemisia tabaci Gennadius, 1889 comprises multiple cryptic species with varying reproductive compatibilities and insecticide resistance levels, necessitating tailored management strategies [33]. Similarly, the onion thrips (Thrips tabaci Lindeman) consists of distinct lineages exhibiting reproductive isolation and differences in behavior, emphasizing the importance of accurate species identification for effective control [34]. In the context of biological control, the inadvertent introduction of cryptic species can lead to unintended ecological consequences, highlighting the need for thorough genetic assessments prior to agent release [35]. Therefore, integrating molecular tools into pest management programs is essential to detect cryptic diversity, inform targeted control measures, and prevent the misapplication of management strategies.
A primary limitation of this study is the relatively low number of samples per site, which may reduce the resolution of population genetic patterns. In addition, reliance on the COI marker alone may limit the detection of finer-scale genetic differentiation, as shown in the paper wasp Polistes fuscatus (Fabricius, 1793) [36]. Therefore, future studies incorporating additional nuclear markers could provide more robust insights into population differentiation. We also observed a high frequency of unique haplotypes, suggesting local adaptation to environmental factors such as climate, host plant availability, and predation. Similar habitat-driven genetic differentiation has been reported in other herbivorous insects, such as the seed bug S. pandurus [8]. In addition, historical demographic events like bottlenecks, founder effects, and expansions may have influenced the current genetic structure by reducing diversity and later promoting new mutations [37].
We also detected a signal of population expansion in P. peregrinus populations in Thailand. However, information on generation time is unavailable, and thus the timing of the population expansion cannot be estimated. Studies in Thailand and Southeast Asia have reported that many insect species have undergone recent population expansion in response to Pleistocene climatic and environmental fluctuations, including the black fly Simulium chumpornense Takaoka & Kuvangkadilok, 2000 [38], the fruit fly Bactrocera latifrons (Hendel, 1915) [39], Anopheles Meigen, 1818 mosquitoes [40,41], and the buffalo fly Haematobia exigua Meijere, 1906 [42]. Therefore, we suspect that the population expansion signal observed in P. peregrinus may also represent a response of this species to Pleistocene climatic changes.
Lastly, our current finding on genetic variation and population structure of P. peregrinus populations in Thailand provides valuable insights not only into their evolutionary dynamics but also into practical pest management. The observed population structure and genetic differentiation suggest that local populations may respond differently to control measures, highlighting the need for region-specific management strategies. Moreover, understanding gene flow among populations can inform the deployment of targeted interventions, such as monitoring high-risk areas for rapid infestations and optimizing the timing of biological or chemical control to limit the spread of genetically distinct populations.
5. Conclusions
In conclusion, the level of genetic variation observed within and among populations of P. peregrinus in the present study suggests the possible presence of cryptic diversity, which may complicate pest management efforts. The beetle’s dispersal ability and adaptability emphasize the importance of using molecular tools in monitoring and management programs to detect hidden diversity and enhance future control strategies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology14091266/s1, Table S1: Variable sites in the COI sequences among 45 haplotypes of Platycorynus peregrinus examined in this study.
Author Contributions
Conceptualization, W.S. (Weerachai Saijuntha) and C.T.; methodology, S.K., W.P., S.N., T.C. and J.S.; software, C.J., W.S. (Weerachai Saijuntha) and W.T.; validation, W.S. (Weerachai Saijuntha), N.P. and P.P.; formal analysis, W.S. (Weerachai Saijuntha), C.J., W.T., W.S. (Warong Suksavate) and P.P.; investigation, S.K., W.P., N.P. and W.S. (Weerachai Saijuntha); resources, S.K., W.P., S.N., T.C. and J.S.; writing—original draft preparation, S.K. and W.S. (Weerachai Saijuntha); writing—review and editing, W.S. (Weerachai Saijuntha), C.T., N.P. and P.P.; visualization, C.J. and W.S. (Warong Suksavate); supervision, W.S. (Weerachai Saijuntha) and C.T.; project administration, W.S. (Weerachai Saijuntha); funding acquisition, S.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by Mahasarakham University (Grant number 6724004/2567).
Institutional Review Board Statement
The ethical standards of this research were approved by Institutional Animal Care and Use Committee, Mahasarakham University (IACUC-MSU-24/2022, approved on 27 October 2022).
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are available upon request.
Acknowledgments
We would like to acknowledge all individuals who helped collect samples for this project.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Prasad, P.; Singh, B.K. Studies on the food and food preference in Platycorynus peregrinus (Coleoptera: Chrysomelidae). In Advances in Zoology, Environmental Degradation and Biodiversity, Proceedings of the 10th All India Congress of Zoology and National Symposium on Environmental Degradation and Animal Biodiversity the Problems of India and Its Remedial Measures, Bihar, India, 14–18 October 1998; Magadh University: Bihar, India, 2000; pp. 122–125. [Google Scholar]
- Wijeweera, W.P.S.N.; Senaratne, K.A.D.W.; Dhileepan, K.; de Silva, M.P.K.S.K. Insect diversity on Calotropis gigantea (L.) in Sri Lanka. Ceylon J. Sci. 2022, 51, 121–128. [Google Scholar] [CrossRef]
- Sethuraman, A.; Janzen, F.J.; Weisrock, D.W.; Obrycki, J.J. Insights from Population Genomics to Enhance and Sustain Biological Control of Insect Pests. Insects 2020, 11, 462. [Google Scholar] [CrossRef]
- Kaewkrajang, N.; Grootaert, P.; Boonrotpong, S. Genetic Variation of the Long-Legged Flies Phacaspis mitis Complex (Diptera: Dolichopodidae) in Peninsular Thailand Inferred from Three Mitochondrial Genes. J. Insect Sci. 2018, 18, 6. [Google Scholar] [CrossRef] [PubMed]
- Mintara, R.; Pramual, P. Common but not connected: High genetic structure and cryptic genetic diversity in the ubiquitous biting midge Culicoides peregrinus Kieffer. Trop. Biomed. 2023, 40, 363–369. [Google Scholar] [CrossRef]
- Cao, L.J.; Gao, Y.F.; Gong, Y.J.; Chen, J.C.; Chen, M.; Hoffmann, A.; Wei, S.J. Population analysis reveals genetic structure of an invasive agricultural thrips pest related to invasion of greenhouses and suitable climatic space. Evol. Appl. 2019, 12, 1868–1880. [Google Scholar] [CrossRef] [PubMed]
- Comeault, A.A.; Wang, J.; Tittes, S.; Isbell, K.; Ingley, S.; Hurlbert, A.H.; Matute, D.R. Genetic Diversity and Thermal Performance in Invasive and Native Populations of African Fig Flies. Mol. Biol. Evol. 2020, 37, 1893–1906. [Google Scholar] [CrossRef] [PubMed]
- Pilap, W.; Pradit, N.; Jaroenchaiwattanachote, C.; Saijuntha, J.; Kongbuntad, W.; Tawong, W.; Tantrawatpan, C.; Saijuntha, W. Unveiling Genetic Variation in the Seed Bug Spilostethus pandurus (Scopoli, 1763) (Hemiptera: Lygaeidae) in Thailand Using Mitochondrial CO1 Sequence. Biology 2025, 14, 1022. [Google Scholar] [CrossRef]
- Avise, J.C. Molecular Markers, Natural History, and Evolution; Springer: New York, NY, USA, 1994. [Google Scholar]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. Available online: http://www.barcodinglife.org (accessed on 15 July 2025). [CrossRef]
- Kimoto, S.; Gressitt, J.L. Chrysomelidae (Coleoptera) of Thailand, Cambodia, Laos, and Vietnam. III. Eumolpinae. Esakia 1982, 18, 1–141. [Google Scholar] [CrossRef]
- Pradit, N.; Saijuntha, W.; Pilap, W.; Suksavate, W.; Agatsuma, T.; Jongsomchai, K.; Kongbuntad, W.; Tantrawatpan, C. Genetic variation of Tarbinskiellus portentosus (Lichtenstein 1796) (Orthoptera: Gryllidae) in mainland Southeast Asia examined by mitochondrial DNA sequences. Int. J. Trop. Insect Sci. 2022, 42, 955–964. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, F.; Wu, J.; Ye, S.; Xu, Y.; Liu, Y. Fine-Scale Genetic Structure of Curculio chinensis (Coleoptera: Curculionidae) Based on Mitochondrial COI: The Role of Host Specificity and Spatial Distance. Insects 2024, 15, 116. [Google Scholar] [CrossRef]
- Toju, H. Natural selection drives the fine-scale divergence of a coevolutionary arms race involving a long-mouthed weevil and its obligate host plant. BMC Evol. Biol. 2009, 9, 273. [Google Scholar] [CrossRef]
- Goslee, S.C.; Urban, D.L. The ecodist Package for Dissimilarity-based Analysis of Ecological Data. J. Stat. Softw. 2007, 22, 1–19. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Nylander, J.A.A. MrModeltest v2. Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Baele, G.; Ji, X.; Hassler, G.W.; McCrone, J.T.; Shao, Y.; Zhang, Z.; Holbrook, A.J.; Lemey, P.; Drummond, A.J.; Rambaut, A.; et al. BEAST X for Bayesian phylogenetic, phylogeographic and phylodynamic inference. Nat. Methods 2025, 22, 1653–1656. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Rambaut, A. Figtree ver 1.4.4; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, Scotland, 2018. [Google Scholar]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Laukkanen, L.; Mutikainen, P.; Muola, A.; Leimu, R. Plant-Species Diversity Correlates with Genetic Variation of an Oligophagous Seed Predator. PLoS ONE 2014, 9, e94105. [Google Scholar] [CrossRef]
- Egan, S.P.; Funk, D.J. Ecologically dependent postmating isolation between sympatric host forms of Neochlamisus bebbianae leaf beetles. Proc. Natl. Acad. Sci. USA 2009, 106, 19426–19431. [Google Scholar] [CrossRef]
- Keller, A.G.; Dahlhoff, E.P.; Bracewell, R.; Chatla, K.; Bachtrog, D.; Rank, N.E.; Williams, C.M. Multi-locus genomic signatures of local adaptation to snow across the landscape in California populations of a willow leaf beetle. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2023, 290, 20230630. [Google Scholar] [CrossRef]
- Shoda-Kagaya, E.; Saito, S.; Okada, M.; Nozaki, A.; Nunokawa, K.; Tsuda, Y. Genetic structure of the oak wilt vector beetle Platypus quercivorus: Inferences toward the process of damaged area expansion. BMC Ecol. 2010, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Thaenasa, A.; Pradit, N.; Pilap, W.; Jaroenchaiwattanachote, C.; Wongpakam, K.; Inkhavilay, K.; Saijuntha, J.; Tawong, W.; Suksavate, W.; Tantrawatpan, C.; et al. Exploring the Genetic Diversity of the Jewel Beetles Sternocera aequisignata Saunders, 1866, and S. ruficornis Saunders, 1866 (Coleoptera: Buprestidae) in Thailand and Lao PDR. Insects 2025, 16, 322. [Google Scholar] [CrossRef] [PubMed]
- Boykin, L.M.; Armstrong, K.F.; Kubatko, L.; De Barro, P. Species delimitation and global biosecurity. Evol. Bioinform. 2012, 8, 1–37. [Google Scholar] [CrossRef]
- Király, K.D.; Ladányi, M.; Fail, J. Reproductive Isolation in the Cryptic Species Complex of a Key Pest: Analysis of Mating and Rejection Behaviour of Onion Thrips (Thrips tabaci Lindeman). Biology 2022, 11, 396. [Google Scholar] [CrossRef] [PubMed]
- Paterson, I.D.; Mangan, R.; Downie, D.A.; Coetzee, J.A.; Hill, M.P.; Burke, A.M.; Downey, P.O.; Henry, T.J.; Compton, S.G. Two in one: Cryptic species discovered in biological control agent populations using molecular data and crossbreeding experiments. Ecol. Evol. 2016, 6, 6139–6150. [Google Scholar] [CrossRef]
- Bluher, S.E.; Miller, S.E.; Sheehan, M.J. Fine-Scale Population Structure but Limited Genetic Differentiation in a Cooperatively Breeding Paper Wasp. Genome Biol. Evol. 2020, 12, 701–714. [Google Scholar] [CrossRef]
- Excoffier, L.; Ray, N. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol. 2008, 23, 347–351. [Google Scholar] [CrossRef]
- Pramual, P.; Jomkumsing, P.; Wathasith, P.; Wongpakam, K. Population structure and population history of the black fly Simulium chumpornense (Diptera: Simuliidae) from Thailand. Acta Trop. 2022, 227, 106301. [Google Scholar] [CrossRef] [PubMed]
- Meeyen, K.; Nanork Sopaladawan, P.; Pramual, P. Population structure, population history and DNA barcoding of fruit fly Bactrocera latifrons (Hendel) (Diptera: Tephritidae). Entomol. Sci. 2014, 17, 219–230. [Google Scholar] [CrossRef]
- O’Loughlin, S.M.; Okabayashi, T.; Honda, M.; Kitazoe, Y.; Kishino, H.; Somboon, P.; Sochantha, T.; Nambanya, S.; Saikia, P.K.; Dev, V.; et al. Complex population history of two Anopheles dirus mosquito species in Southeast Asia suggests the influence of Pleistocene climate change rather than human-mediated effects. J. Evol. Biol. 2008, 21, 1555–1569. [Google Scholar] [CrossRef] [PubMed]
- Morgan, K.; O’Loughlin, S.M.; Chen, B.I.N.; Linton, Y.M.; Thongwat, D.; Somboon, P.; Fong, M.Y.; Butlin, R.; Verity, R.; Prakash, A.; et al. Comparative phylogeography reveals a shared impact of pleistocene environmental change in shaping genetic diversity within nine Anopheles mosquito species across the Indo-Burma biodiversity hotspot. Mol. Ecol. 2011, 20, 4533–4549. [Google Scholar] [CrossRef] [PubMed]
- Low, V.L.; Tan, T.K.; Prakash, B.K.; Vinnie-Siow, W.Y.; Tay, S.T.; Masmeatathip, R.; Hadi, U.K.; Lim, Y.A.L.; Chen, C.D.; Norma-Rashid, Y.; et al. Contrasting evolutionary patterns between two haplogroups of Haematobia exigua (Diptera: Muscidae) from the mainland and islands of Southeast Asia. Sci. Rep. 2017, 7, 5871. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).